Abstract
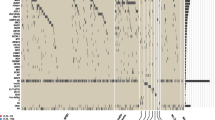
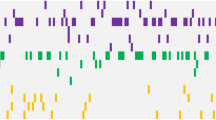
Mutations in FMS-like tyrosine kinase 3 (FLT3) gene occur frequently in acute myeloid leukemia (AML) and are rare in acute lymphoblastic leukemia (ALL). We aimed to analyze the incidence and characteristics of FLT3 mutations in ALL. Amplicon-targeted next-generation sequencing of 58 genes was performed on 1571 patients (AML, n = 829; ALL, n = 742). FLT3 mutations were identified in 5.12% (38/742) of ALL patients. Four types of FLT3 mutations were disclosed, including internal tandem duplication (ITD), tyrosine kinase domain (TKD), juxtamembrane insertion and deletion (JM-INDEL), and juxtamembrane point mutation (JM-PM), which were respectively identified in 1.21, 1.89, 0.67, and 1.89% of patients. The incidence of FLT3-JM-PM (1.89 vs 0.48%, P = 0.009) and the proportion of TKD non-D835 mutations that accounted for the total TKD mutations (57.14 vs 18.18%, P = 0.013) were significantly higher in ALL when compared with AML. FLT3-JM-INDEL were mainly found in B-ALL. In addition, FLT3-JM-INDEL and FLT3-JM-PM were first reported in patients with B-ALL. Patients with FLT3 mutations besides of ITD and/or TKD had a potential response to tyrosine kinase inhibitors. We showed that the mutation spectrum of FLT3 gene in ALL is distinct from AML that will facilitated an in-depth understand of the pathogenesis and provide a guidance for treatment.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
DiNardo CD, Cortes JE. Mutations in AML: prognostic and therapeutic implications. Hematol Am Soc Hematol Educ Program. 2016;2016:348–55.
Vardiman JW, Thiele J, Arber DA, Brunning RD, Borowitz MJ, Porwit A, et al. The 2008 revision of the World Health Organization (WHO) classification of myeloid neoplasms and acute leukemia: rationale and important changes. Blood. 2009;114:937–51.
Zhang Y, Wang F, Chen X, Zhang Y, Wang M, Liu H, et al. CSF3R Mutations are frequently associated with abnormalities of RUNX1, CBFB, CEBPA, and NPM1 genes in acute myeloid leukemia. Cancer. 2018;124:3329–38.
Chen X, Wang F, Zhang Y, Wang M, Tian W, Teng W, et al. Retrospective analysis of 36 fusion genes in 2479 Chinese patients of de novo acute lymphoblastic leukemia. Leuk Res. 2018;72:99–104.
Li M, Datto M, Duncavage E, Kulkarni S, Lindeman N, Roy S, et al. Standards and Guidelines for the Interpretation and Reporting of Sequence Variants in Cancer: A Joint Consensus Recommendation of the Association for molecular pathology, American Society of Clinical Oncology, and College of American Pathologists. J Mol Diagn. 2017;19:4–23.
Elyamany G, Awad M, Alsuhaibani O, Fadalla K, Al Sharif O, Al Shahrani M, et al. FLT3 internal tandem duplication and D835 mutations in patients with acute lymphoblastic leukemia and its clinical significance. Mediterr J Hematol Infect Dis. 2014;6:e2014038.
Yamamoto Y, Kiyoi H, Nakano Y, Suzuki R, Kodera Y, Miyawaki S, et al. Activating mutation of D835 within the activation loop of FLT3 in human hematologic malignancies. Blood. 2001;97:2434–9.
Whitman SP, Ruppert AS, Radmacher MD, Mrózek K, Paschka P, Langer C, et al. FLT3 D835/I836 mutations are associated with poor disease-free survival and a distinct gene-expression signature among younger adults with de novo cytogenetically normal acute myeloid leukemia lacking FLT3 internal tandem duplications. Blood. 2008;111:1552–9.
Alkhayat N, Elborai Y, Al Sharif O, Al Shahrani M, Alsuhaibani O, Awad M, et al. Cytogenetic profile and gene mutations of childhood acute lymphoblastic leukemia. Clin Med Insights Oncol. 2017;11:1179554917721710.
Matsuno N, Nanri T, Kawakita T, Mitsuya H, Asou N. A novel FLT3 activation loop mutation N841K in acute myeloblastic leukemia. Leukemia. 2005;19:480–1.
Li C, Liu L, Liang L, Xia Z, Li Z, Wang X, et al. AMG 925 is a dual FLT3/CDK4 inhibitor with the potential to overcome FLT3 inhibitor resistance in acute myeloid leukemia. Mol Cancer Ther. 2015;14:375–83.
Kindler T, Breitenbuecher F, Kasper S, Estey E, Giles F, Feldman E, et al. Identification of a novel activating mutation (Y842C) within the activation loop of FLT3 in patients with acute myeloid leukemia (AML). Blood. 2005;105:335–40.
Jiang J, Paez J, Lee J, Bo R, Stone R, DeAngelo D, et al. Identifying and characterizing a novel activating mutation of the FLT3 tyrosine kinase in AML. Blood. 2004;104:1855–8.
Smith C, Zhang C, Lin K, Lasater E, Zhang Y, Massi E, et al. Characterizing and overriding the structural mechanism of the quizartinib-resistant FLT3 “Gatekeeper” F691L mutation with PLX3397. Cancer Discov. 2015;5:668–79.
Smith C, Lasater E, Zhu X, Lin K, Stewart W, Damon L, et al. Activity of ponatinib against clinically-relevant AC220-resistant kinase domain mutants of FLT3-ITD. Blood. 2013;121:3165–71.
Janke H, Pastore F, Schumacher D, Herold T, Hopfner KP, Schneider S, et al. Activating FLT3 mutants show distinct gain-of-function phenotypes in vitro and a characteristic signaling pathway profile associated with prognosis in acute myeloid leukemia. PLoS ONE. 2014;9:e89560.
Grundler R, Thiede C, Miething C, Steudel C, Peschel C, Duyster J. Sensitivity toward tyrosine kinase inhibitors varies between different activating mutations of the FLT3 receptor. Blood. 2003;102:646–51.
Kesarwani M, Huber E, Azam M. Overcoming AC220 resistance of FLT3-ITD by SAR302503. Blood Cancer J. 2013;3:e138.
Cools J, Mentens N, Furet P, Fabbro D, Clark J, Griffin J, et al. Prediction of resistance to small molecule FLT3 inhibitors: implications for molecularly targeted therapy of acute leukemia. Cancer Res. 2004;64:6385–9.
Man C, Fung T, Ho C, Han H, Chow H, Ma A, et al. Sorafenib treatment of FLT3-ITD(+) acute myeloid leukemia: favorable initial outcome and mechanisms of subsequent nonresponsiveness associated with the emergence of a D835 mutation. Blood. 2012;119:5133–43.
Chatain N, Perera R, Rossetti G, Rossa J, Carloni P, Schemionek M, et al. Rare FLT3 deletion mutants may provide additional treatment options to patients with AML: an approach to individualized medicine. Leukemia. 2015;29:2434–8.
Sandhöfer N, Bauer J, Reiter K, Dufour A, Rothenberg M, Konstandin NP, et al. The new and recurrent FLT3 juxtamembrane deletion mutation shows a dominant negative effect on the wild-type FLT3 receptor. Sci Rep. 2016;6:28032.
Meshinchi S, Stirewalt D, Alonzo T, Boggon T, Gerbing R, Rocnik J, et al. Structural and numerical variation of FLT3/ITD in pediatric AML. Blood. 2008;111:4930–3.
Reindl C, Bagrintseva K, Vempati S, Schnittger S, Ellwart J, Wenig K, et al. Point mutations in the juxtamembrane domain of FLT3 define a new class of activating mutations in AML. Blood. 2006;107:3700–7.
Fröhling S, Scholl C, Levine R, Loriaux M, Boggon T, Bernard O, et al. Identification of driver and passenger mutations of FLT3 by high-throughput DNA sequence analysis and functional assessment of candidate alleles. Cancer Cell. 2007;12:501–13.
Syampurnawati M, Tatsumi E, Furuta K, Hayashi Y. Four novel point mutations in exons 12, 13, and 14 of the FLT3 gene. Leuk Res. 2007;31:877.
Stirewalt D, Meshinchi S, Kussick S, Sheets K, Pogosova-Agadjanyan E, Willman C, et al. Novel FLT3 point mutations within exon 14 found in patients with acute myeloid leukaemia. Br J Haematol. 2004;124:481–4.
Heiss E, Masson K, Sundberg C, Pedersen M, Sun J, Bengtsson S, et al. Identification of Y589 and Y599 in the juxtamembrane domain of Flt3 as ligand-induced autophosphorylation sites involved in binding of Src family kinases and the protein tyrosine phosphatase SHP2. Blood. 2006;108:1542–50.
Rocnik J, Okabe R, Yu J, Lee B, Giese N, Schenkein D, et al. Roles of tyrosine 589 and 591 in STAT5 activation and transformation mediated by FLT3-ITD. Blood. 2006;108:1339–45.
Tarlock K, Hansen ME, Hylkema T, Ries R, Farrar JE, Auvil JG, et al. Discovery and functional validation of novel pediatric specific FLT3 activating mutations in acute myeloid leukemia: results from the COG/NCI target initiative. Blood. 2015;126:87–87.
Döhner H, Estey E, Grimwade D, Amadori S, Appelbaum FR, Büchner T, et al. Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood. 2017;129:424–47.
O’Donnell MR, Tallman MS, Abboud CN, Altman JK, Appelbaum FR, Arber DA, et al. Acute myeloid leukemia, version 3.2017, NCCN clinical practice guidelines in oncology. J Natl Compr Cancer Netw. 2017;15:926–57.
Levis M. Midostaurin approved for FLT3-mutated AML. Blood. 2017;129:3403–6.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Zhang, Y., Zhang, Y., Wang, F. et al. The mutational spectrum of FLT3 gene in acute lymphoblastic leukemia is different from acute myeloid leukemia. Cancer Gene Ther 27, 81–88 (2020). https://doi.org/10.1038/s41417-019-0120-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41417-019-0120-z
This article is cited by
-
CD19/CD22 chimeric antigen receptor T-cell therapy for refractory acute B-cell lymphoblastic leukemia with FLT3-ITD mutations
Bone Marrow Transplantation (2020)