Abstract
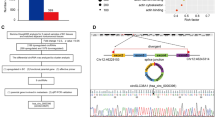
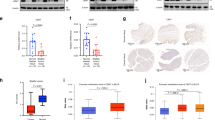
COMMD10, a member of COMMD protein, has been proved to target p65 NF-kappaB (nuclear factor-kappaB) subunit and reduce its nuclear translocation, thereby leading to the inactivation of NF-kappaB pathway and suppression of colorectal cancer invasion and metastasis. The aim of this study is to explore its expression pattern and tissue distribution in human normal tissues and other tumor tissues and to investigate the relevant mechanism. We firstly provided the expression profile and histological distribution of COMMD10 in various BALB/c mice tissues and identified the biological distribution of COMMD10 in different kinds of human normal and tumor tissues. We verified the expression profile of COMMD10 using TCGA database. The interacting genes of COMMD10 were predicted by using STRING using. Finally, we performed database, and the microRNAs targeting COMMD10 were predicted using miRDB, miRWalk, TargetScan and microRNA. GO and KEGG pathway analyses were performed to predict the biological function of COMMD10 and its interacting genes. mRNA expression of COMMD10 showed the highest level in the lung and spleen, and the lowest level in the heart and brain. Immunohistochemistry detection revealed that COMMD10 was expressed in different tissues with different degrees and was was located mainly in the cytoplasm. Subsequently, we showed that COMMD10 displayed various degrees of expression in different human normal tissues that mainly located in cytoplasm, while COMMD10 of liver cells resided in both nucleus and cytoplasm. All the tumor tissues except breast small cell carcinoma, breast phyllodes tumor, lung adenocarcinoma, thymoma, cervical cancer and bladder urothelial carcinoma showed that COMMD10 was positive staining in cytoplasm. Kaplan–Meier plotter indicated that renal clear cell carcinoma patients with increased expression level of COMMD10 exhibited longer survival. STRING database revealed that COMMD10 had 41 interacting genes, and data from 4 different databases indicated that hsa-miR-590-3p may be the potential regulator of COMMD10. GO analysis demonstrated that COMMD10 and its interacting genes were mainly enriched in Cullin-RING ubiquitin ligase complexes, binding and transport of copper ions, the transport and steady-state maintenance of copper ions, transcription, translation and transport of proteins, and negatively regulate the activity of NF-kappaB transcription factors. KEGG pathway showed that COMMD10 and its interacting genes were mainly involved in renal cell carcinoma, HIF-1 signaling pathways, ubiquitination-mediated proteolysis, endocytosis and mineral absorption. COMMD10 may play a tumor suppressive role in renal clear cell carcinoma through the miR-590-3p-COMMD10-Cul2-RBX1-NF-κB/HIF/NRF2 pathway and regulate the chemotherapy resistance of various tumor cells to cisplatin.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108.
McGuire S. World Cancer Report 2014. Geneva, Switzerland: World Health Organization, International Agency for Research on Cancer, WHO Press, 2015. Adv Nutr. 2016;7:418–9.
Bartuzi P, Hofker MH, van de Sluis B. Tuning NF-kappaB activity: a touch of COMMD proteins. Biochim Biophys Acta. 2013;1832:2315–21.
Burstein E, Hoberg JE, Wilkinson AS, Rumble JM, Csomos RA, Komarck CM, et al. COMMD proteins, a novel family of structural and functional homologs of MURR1. J Biol Chem. 2005;280:22222–32.
Maine GN, Mao X, Komarck CM, Burstein E. COMMD1 promotes the ubiquitination of NF-kappaB subunits through a cullin-containing ubiquitin ligase. EMBO J. 2007;26:436–47.
van De Sluis B, Rothuizen J, Pearson PL, van Oost BA, Wijmenga C. Identification of a new copper metabolism gene by positional cloning in a purebred dog population. Hum Mol Genet. 2002;11:165–73.
Li H, Burstein E. COMMD1 regulates inflammation and colitis-associated cancer progression. Oncoimmunology. 2014;3:e947891.
Riera-Romo M. COMMD1: a multifunctional regulatory protein. J Cell Biochem. 2018;119:34–51.
Matsuda H, Hamet P, Tremblay J. Hypertension-related, calcium-regulated gene (HCaRG/COMMD5) and kidney diseases: HCaRG accelerates tubular repair. J Nephrol. 2014;27:351–60.
Gkiafi Z, Panayotou G. Comparative proteomic analysis implicates COMMD proteins as Epstein-Barr virus targets in the BL41 Burkitt’s lymphoma cell line. J Proteome Res. 2011;10:2959–68.
Yang SS, Li XM, Yang M, Ren XL, Hu JL, Zhu XH, et al. FMNL2 destabilises COMMD10 to activate NF-kappaB pathway in invasion and metastasis of colorectal cancer. Br J Cancer. 2017;117:1164–75.
Maine GN, Burstein E. COMMD proteins: COMMing to the scene. Cell Mol Life Sci. 2007;64:1997–2005.
Maine GN, Burstein E. COMMD proteins and the control of the NF kappa B pathway. Cell Cycle. 2007;6:672–6.
Jia Z, Xu C, Shen J, Xia T, Yang J, He Y. The natural compound celastrol inhibits necroptosis and alleviates ulcerative colitis in mice. Int Immunopharmacol. 2015;29:552–9.
Taura M, Kudo E, Kariya R, Goto H, Matsuda K, Hattori S, et al. COMMD1/Murr1 reinforces HIV-1 latent infection through IkappaB-alpha stabilization. J Virol. 2015;89:2643–58.
van de Sluis B, Muller P, Duran K, Chen A, Groot AJ, Klomp LW, et al. Increased activity of hypoxia-inducible factor 1 is associated with early embryonic lethality in Commd1 null mice. Mol Cell Biol. 2007;27:4142–56.
Li H, Koo Y, Mao X, Sifuentes-Dominguez L, Morris LL, Jia D, et al. Endosomal sorting of Notch receptors through COMMD9-dependent pathways modulates Notch signaling. J Cell Biol. 2015;211:605–17.
Grubb RR, Choyke PL, Pinto PA, Linehan WM, Walther MM. Management of von Hippel-Lindau-associated kidney cancer. Nat Clin Pract Urol. 2005;2:248–55.
Linehan WM, Vasselli J, Srinivasan R, Walther MM, Merino M, Choyke P, et al. Genetic basis of cancer of the kidney: disease-specific approaches to therapy. Clin Cancer Res. 2004;10(18 Pt 2):6282S–6289S.
Wang X, Qu H, Dong Y, Wang G, Zhen Y, Zhang L. Targeting signal-transducer-and-activator-of-transcription 3 sensitizes human cutaneous melanoma cells to BRAF inhibitor. Cancer Biomark. 2018;23:67–77.
Kaelin WJ. The von Hippel-Lindau tumor suppressor gene and kidney cancer. Clin Cancer Res. 2004;10(18 Pt 2):6290S–6295S.
Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–8.
Pang H, Zheng Y, Zhao Y, Xiu X, Wang J. miR-590-3p suppresses cancer cell migration, invasion and epithelial-mesenchymal transition in glioblastoma multiforme by targeting ZEB1 and ZEB2. Biochem Biophys Res Commun. 2015;468:739–45.
Zhang J, Jin M, Chen X, Zhang R, Huang Y, Liu H, et al. Loss of PPM1F expression predicts tumour recurrence and is negatively regulated by miR-590-3p in gastric cancer. Cell Prolif. 2018;51:e12444.
Sun ZQ, Shi K, Zhou QB, Zeng XY, Liu J, Yang SX, et al. MiR-590-3p promotes proliferation and metastasis of colorectal cancer via Hippo pathway. Oncotarget. 2017;8:58061–71.
Kuo MT, Chen HH, Song IS, Savaraj N, Ishikawa T. The roles of copper transporters in cisplatin resistance. Cancer Metastas- Rev. 2007;26:71–83.
Chang C, Hu Y, Hogan SL, Mercke N, Gomez M, O’Bryant C, et al. Pharmacogenomic variants may influence the urinary excretion of novel kidney injury biomarkers in patients receiving cisplatin. Int J Mol Sci. 2017;18:1333.
Leonhardt K, Gebhardt R, Mossner J, Lutsenko S, Huster D. Functional interactions of Cu-ATPase ATP7B with cisplatin and the role of ATP7B in the resistance of cells to the drug. J Biol Chem. 2009;284:7793–802.
Acknowledgements
This project was supported by the National Natural Science Foundation of China (Grant Nos. 81602685, 81672992); the Science and Technology Projects in Guangdong Province (Grant Nos. 2018-1201-SF-0019, 2012B031800262); and the Natural Science Foundation of Guangdong Province (Grant Nos. 2017A030313486, S2013040013505). We thank State Key Laboratory of Oncology in Southern Medical University for providing experimental platform.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethics statement
The study was approved by the Institutional Ethics Committee of Southern Medical University. All the participants provided their written informed consent to be included in the study. The animal experiments were approved by the Institutional Animal Care and Use Committee at the Southern Medical University and performed in accordance with the “Guidelines for Experimental Animals” of the Ministry of Science and Technology (Beijing, China). The 4-week-old male BALB/c mice were purchased from the Experimental Animal Center of Southern Medical University, license No.: SCXK (Guangdong) 2011-0015. The BALB/c mice need not to be grouped, and hence we did not have randomization or blind methods.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Fan, Y., Zhang, L., Sun, Y. et al. Expression profile and bioinformatics analysis of COMMD10 in BALB/C mice and human. Cancer Gene Ther 27, 216–225 (2020). https://doi.org/10.1038/s41417-019-0087-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41417-019-0087-9
This article is cited by
-
Targeting the COMMD4–H2B protein complex in lung cancer
British Journal of Cancer (2023)
-
Exploring prognostic value and regulation network of PPP1R1A in hepatocellular carcinoma
Human Cell (2022)
-
SFTPA1 is a potential prognostic biomarker correlated with immune cell infiltration and response to immunotherapy in lung adenocarcinoma
Cancer Immunology, Immunotherapy (2022)
-
Expression profile of SYNE3 and bioinformatic analysis of its prognostic value and functions in tumors
Journal of Translational Medicine (2020)
-
Defining COMMD4 as an anti-cancer therapeutic target and prognostic factor in non-small cell lung cancer
British Journal of Cancer (2020)