Abstract
Background
Large cell lung carcinoma (LCLC) is an exceptionally aggressive disease with a poor prognosis. At present, little is known about the molecular pathology of LCLC.
Methods
Ultra-deep sequencing of cancer-related genes and exome sequencing were used to detect the LCLC mutational in 118 tumor-normal pairs. The cell function test was employed to confirm the potential carcinogenic mutation of PI3K pathway.
Results
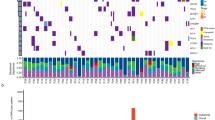
The mutation pattern is determined by the predominance of A > C mutations. Genes with a significant non-silent mutation frequency (FDR) < 0.05) include TP53 (47.5%), EGFR (13.6%) and PTEN (12.1%). Moreover, PI3K signaling (including EGFR, FGRG4, ITGA1, ITGA5, and ITGA2B) is the most mutated pathway, influencing 61.9% (73/118) of the LCLC samples. The cell function test confirmed that the potential carcinogenic mutation of PI3K pathway had a more malignant cell function phenotype. Multivariate analysis further revealed that patients with the PI3K signaling pathway mutations have a poor prognosis (P = 0.007).
Conclusions
These results initially identified frequent mutation of PI3K signaling pathways in LCLC and indicate potential targets for the treatment of this fatal type of LCLC.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 24 print issues and online access
$259.00 per year
only $10.79 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request. Whole-exome sequencing data and target sequencing data from this study are available for download through the NCBI Sequence Read Archive under accession number PRJNA639383, PRJNA639657, and PRJNA643364. These submission will be released upon publication. Release of BioProject or BioSamples is also triggered by the release of submitted data.
References
Schwendenwein A, Megyesfalvi Z, Barany N, Valko Z, Bugyik E, Lang C, et al. Molecular profiles of small cell lung cancer subtypes: therapeutic implications. Mol Ther Oncolytics. 2021;20:470–83.
Liu SH, Hsu KW, Lai YL, Lin YF, Chen FH, Peng PH, et al. Systematic identification of clinically relevant miRNAs for potential miRNA-based therapy in lung adenocarcinoma. Mol Ther Nucleic Acids. 2021;25:1–10.
Nicholson AG, Tsao MS, Beasley MB, Borczuk AC, Brambilla E, Cooper WA, et al. The 2021 WHO classification of lung tumors: impact of advances since 2015. J Thorac Oncol. 2022;17:362–87.
Lin G, Qi K, Liu B, Liu H, Li J. A nomogram prognostic model for large cell lung cancer: analysis from the Surveillance, Epidemiology and End Results Database. Transl Lung Cancer Res. 2021;10:622–35.
Sugár S, Bugyi F, Tóth G, Pápay J, Kovalszky I, Tornóczky T, et al. Proteomic analysis of lung cancer types-a pilot study. Cancers. 2022;14:2629.
Ramos-Paradas J, Gómez-Sánchez D, Rosado A, Ucero AC, Ferrer I, García-Luján R, et al. Comprehensive characterization of human lung large cell carcinoma identifies transcriptomic signatures with potential implications in response to immunotherapy. J Clin Med. 2022;11:1500.
Karlsson A, Brunnström H, Micke P, Veerla S, Mattsson J, La Fleur L, et al. Gene expression profiling of large cell lung cancer links transcriptional phenotypes to the new histological WHO 2015 classification. J Thorac Oncol. 2017;12:1257–67.
Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–20.
do Valle ÍF, Giampieri E, Simonetti G, Padella A, Manfrini M, Ferrari A, et al. Optimized pipeline of MuTect and GATK tools to improve the detection of somatic single nucleotide polymorphisms in whole-exome sequencing data. BMC Bioinforma. 2016;17:341.
Ye K, Schulz MH, Long Q, Apweiler R, Ning Z. Pindel: a pattern growth approach to detect break points of large deletions and medium sized insertions from paired-end short reads. Bioinformatics. 2009;25:2865–71.
Cingolani P. Variant annotation and functional prediction: SnpEff. Methods Mol Biol. 2022;2493:289–314.
Flanagan SE, Patch AM, Ellard S. Using SIFT and PolyPhen to predict loss-of-function and gain-of-function mutations. Genet Test Mol Biomark. 2010;14:533–7.
Choi Y, Chan AP. PROVEAN web server: a tool to predict the functional effect of amino acid substitutions and indels. Bioinformatics. 2015;31:2745–7.
Vaser R, Adusumalli S, Leng SN, Sikic M, Ng PC. SIFT missense predictions for genomes. Nat Protoc. 2016;11:1–9.
Lawrence MS, Stojanov P, Polak P, Kryukov GV, Cibulskis K, Sivachenko A, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499:214–8.
Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30.
Bailey MH, Tokheim C, Porta-Pardo E, Sengupta S, Bertrand D, Weerasinghe A, et al. Comprehensive characterization of cancer driver genes and mutations. Cell. 2018;173:371–85.
Tian X, Gu T, Lee MH, Dong Z. Challenge and countermeasures for EGFR targeted therapy in non-small cell lung cancer. Biochim Biophys Acta Rev Cancer. 2022;1877:188645.
Yu N, Hwang M, Lee Y, Song BR, Kang EH, Sim H, et al. Patient-derived cell-based pharmacogenomic assessment to unveil underlying resistance mechanisms and novel therapeutics for advanced lung cancer. J Exp Clin Cancer Res. 2023;42:37.
Wang L, Ren Z, Yu B, Tang J. Development of nomogram based on immune-related gene FGFR4 for advanced non-small cell lung cancer patients with sensitivity to immune checkpoint inhibitors. J Transl Med. 2021;19:22.
Gao G, Cui L, Zhou F, Jiang T, Wang W, Mao S, et al. Special issue “The advance of solid tumor research in China”: FGFR4 alterations predict efficacy of immune checkpoint inhibitors in nonsmall cell lung cancer. Int J Cancer. 2023;152:79–89.
Zhou C, Li S, Bin K, Qin G, Pan P, Ren D, et al. ITGA2 overexpression inhibits DNA repair and confers sensitivity to radiotherapies in pancreatic cancer. Cancer Lett. 2022;547:215855.
Wang J, Wren JD, Ding Y, Chen J, Mittal N, Xu C, et al. EWI2 promotes endolysosome-mediated turnover of growth factor receptors and integrins to suppress lung cancer. Cancer Lett. 2022;536:215641.
Mei S, Xu Q, Hu Y, Tang R, Feng J, Zhou Y, et al. Integrin β3-PKM2 pathway-mediated aerobic glycolysis contributes to mechanical ventilation-induced pulmonary fibrosis. Theranostics. 2022;12:6057–68.
Acknowledgements
We would like to thank Prof. Wen Li for data analysis and critical discussion of the manuscript.
Funding
his study was supported partly by the National Natural Science Foundation of China (82272766, 81702243 and 81472202), Construction of Clinical Medical Center for Tumor Biological Samples in Nantong (HS2016004), Natural Science Foundation of Shanghai (21140903500), Basic Medical Research Program of Navy Military Medical University Affiliated Changhai Hospital (2021JCMS11), Program of Navy Military Medical University (2022MS019), Program of Key Research and Development Program of Hunan Province (2021NK2026), and Key Program of Hunan Provincial Department of Science and Technology (2021JJ30060 and 2020WK2020).
Author information
Authors and Affiliations
Contributions
LKH, JBL, CYW and DF conceived and directed the study. JHG, YSM, JWL, JH, GXJ, LKH, HML, CYW and DF contributed to the project design. JHG, YSM, LKH, JH, GXJ, HML, WW, XD, QYF, LKH and DF performed experiments. YSM, JWL, JH, HML, JBL, CYW and DF performed bioinformatics data analysis. LKH, YSM, GXJ, JBL, CYW, and DF contributed samples, data and comments on the manuscript. JHG, YSM, JWL, LKH, JH, HML, CYW, and DF analyzed and interpreted data. YSM, LKH, JH, JWL, GXJ, JBL, CYW, and DF contributed reagents, materials and/or analysis tools. LKH, YSM, and DF wrote the manuscript. JHG, YSM, JWL, GXJ, JH and HML contributed equally to this work. All authors contributed to the final version of the manuscript and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
The study was approved by the Ethics Committee of Tongji University School of Medicine and Shanghai Pulmonary Hospital (K15-199). Each participant provided their written informed consent to participate in this study.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Guo, JH., Ma, YS., Lin, JW. et al. Whole-exome and targeted gene sequencing of large-cell lung carcinoma reveals recurrent mutations in the PI3K pathway. Br J Cancer 129, 366–373 (2023). https://doi.org/10.1038/s41416-023-02301-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41416-023-02301-2