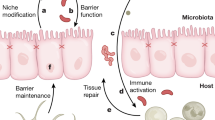
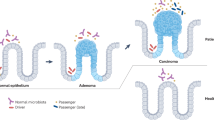
Abstract
Background
Distinct sets of microbes contribute to colorectal cancer (CRC) initiation and progression. Some occur due to the evolving intestinal environment but may not contribute to disease. In contrast, others may play an important role at particular times during the tumorigenic process. Here, we describe changes in the microbiota and host over the course of azoxymethane (AOM)-induced tumorigenesis.
Methods
Mice were administered AOM or PBS and were euthanised 8, 12, 24 and 48 weeks later. Samples were analysed using 16S rRNA gene sequencing, UPLC-MS and qRT-PCR.
Results
The microbiota and bile acid profile showed distinct changes at each timepoint. The inflammatory response became apparent at weeks 12 and 24. Moreover, significant correlations between individual taxa, cytokines and bile acids were detected. One co-abundance group (CAG) differed significantly between PBS- and AOM-treated mice at week 24. Correlation analysis also revealed significant associations between CAGs, bile acids and the bile acid transporter, ASBT. Aberrant crypt foci and adenomas were first detectable at weeks 24 and 48, respectively.
Conclusion
The observed changes precede host hyperplastic transformation and may represent early therapeutic targets for the prevention or management of CRC at specific timepoints in the tumorigenic process.

This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 24 print issues and online access
$259.00 per year
only $10.79 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
The datasets generated and/or analysed during the current study are available from the corresponding author on reasonable request.
References
Keum N, Giovannucci E. Global burden of colorectal cancer: emerging trends, risk factors and prevention strategies. Nat Rev Gastroenterol Hepatol. 2019;16:713–32.
Yamagishi H, Kuroda H, Imai Y, Hiraishi H. Molecular pathogenesis of sporadic colorectal cancers. Chin J Cancer. 2016;35:4.
Flemer B, Lynch DB, Brown JM, Jeffery IB, Ryan FJ, Claesson MJ, et al. Tumour-associated and non-tumour-associated microbiota in colorectal cancer. Gut. 2017;66:633–43.
Scott AJ, Alexander JL, Merrifield CA, Cunningham D, Jobin C, Brown R, et al. International Cancer Microbiome Consortium consensus statement on the role of the human microbiome in carcinogenesis. Gut. 2019;68:1624–32.
Dulal S, Keku TO. Gut microbiome and colorectal adenomas. Cancer J. 2014;20:225–31.
Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, et al. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–8.
Ahn J, Sinha R, Pei Z, Dominianni C, Wu J, Shi J, et al. Human gut microbiome and risk for colorectal cancer. J Natl Cancer Inst. 2013;105:1907–11.
Mangifesta M, Mancabelli L, Milani C, Gaiani F, de’Angelis N, de’Angelis GL, et al. Mucosal microbiota of intestinal polyps reveals putative biomarkers of colorectal cancer. Sci Rep. 2018;8:13974.
Ohigashi S, Sudo K, Kobayashi D, Takahashi O, Takahashi T, Asahara T, et al. Changes of the intestinal microbiota, short chain fatty acids, and fecal pH in patients with colorectal cancer. Dig Dis Sci. 2013;58:1717–26.
Alrawi SJ, Schiff M, Carroll RE, Dayton M, Gibbs JF, Kulavlat M, et al. Aberrant crypt foci. Anticancer Res. 2006;26:107–19.
Hong BY, Ideta T, Lemos BS, Igarashi Y, Tan Y, DiSiena M, et al. Characterization of mucosal dysbiosis of early colonic neoplasia. NPJ Precis Oncol. 2019;3:29.
Keane JM, Joyce SA, Gahan CGM, Hyland NP, Houston A. Microbial metabolites as molecular mediators of host-microbe symbiosis in colorectal cancer. Results Probl Cell Differ. 2020;69:581–603.
Ocvirk S, O’Keefe SJ. Influence of bile acids on colorectal cancer risk: potential mechanisms mediated by diet-gut microbiota interactions. Curr Nutr Rep. 2017;6:315–22.
Earnest DL, Holubec H, Wali RK, Jolley CS, Bissonette M, Bhattacharyya AK, et al. Chemoprevention of azoxymethane-induced colonic carcinogenesis by supplemental dietary ursodeoxycholic acid. Cancer Res. 1994;54:5071–4.
Adam JK, Odhav B, Bhoola KD. Immune responses in cancer. Pharm Ther. 2003;99:113–32.
Zuo T, Ng SC. The gut microbiota in the pathogenesis and therapeutics of inflammatory bowel disease. Front Microbiol. 2018;9:2247.
Arthur JC, Perez-Chanona E, Mühlbauer M, Tomkovich S, Uronis JM, Fan T-J, et al. Intestinal inflammation targets cancer-inducing activity of the microbiota. Science 2012;338:120–3.
Lucas C, Barnich N, Nguyen HTT. Microbiota, inflammation and colorectal cancer. Int J Mol Sci. 2017;18:1310.
Mendes MCS, Paulino DSM, Brambilla SR, Camargo JA, Persinoti GF, Carvalheira JBC. Microbiota modification by probiotic supplementation reduces colitis associated colon cancer in mice. World J Gastroenterol. 2018;24:1995–2008.
Bent R, Moll L, Grabbe S, Bros M. Interleukin-1 beta-a friend or foe in malignancies? Int J Mol Sci. 2018;19:2155.
Scheller J, Chalaris A, Schmidt-Arras D, Rose-John S. The pro- and anti-inflammatory properties of the cytokine interleukin-6. Biochim Biophys Acta. 2011;1813:878–88.
Wang X, Lin Y. Tumor necrosis factor and cancer, buddies or foes? Acta Pharm Sin. 2008;29:1275–88.
Claesson MJ, Jeffery IB, Conde S, Power SE, O’Connor EM, Cusack S, et al. Gut microbiota composition correlates with diet and health in the elderly. Nature. 2012;488:178–84.
Joyce SA, MacSharry J, Casey PG, Kinsella M, Murphy EF, Shanahan F, et al. Regulation of host weight gain and lipid metabolism by bacterial bile acid modification in the gut. Proc Natl Acad Sci USA. 2014;111:7421–6.
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods (San Diego, Calif). 2001;25:402–8.
Rosenberg DW, Giardina C, Tanaka T. Mouse models for the study of colon carcinogenesis. Carcinogenesis. 2009;30:183–96.
Chen W, Liu F, Ling Z, Tong X, Xiang C. Human intestinal lumen and mucosa-associated microbiota in patients with colorectal cancer. PLoS ONE. 2012;7:e39743.
Fang CY, Chen JS, Hsu BM, Hussain B, Rathod J, Lee KH. Colorectal cancer stage-specific fecal bacterial community fingerprinting of the Taiwanese population and underpinning of potential taxonomic biomarkers. Microorganisms. 2021;9:1548.
Liu L, Yang M, Dong W, Liu T, Song X, Gu Y, et al. Gut dysbiosis and abnormal bile acid metabolism in colitis-associated cancer. Gastroenterol Res Pr. 2021;2021:6645970.
Le Gall G, Guttula K, Kellingray L, Tett AJ, Ten Hoopen R, Kemsley EK, et al. Metabolite quantification of faecal extracts from colorectal cancer patients and healthy controls. Oncotarget. 2018;9:33278–89.
Ai D, Pan H, Li X, Gao Y, Liu G, Xia LC. Identifying gut microbiota associated with colorectal cancer using a zero-inflated lognormal model. Front Microbiol. 2019;10:826.
Feng Q, Liang S, Jia H, Stadlmayr A, Tang L, Lan Z, et al. Gut microbiome development along the colorectal adenoma–carcinoma sequence. Nat Commun. 2015;6:6528.
Scanlan PD, Shanahan F, Clune Y, Collins JK, O’Sullivan GC, O’Riordan M, et al. Culture-independent analysis of the gut microbiota in colorectal cancer and polyposis. Environ Microbiol. 2008;10:789–98.
Zou J, Shen Y, Chen M, Zhang Z, Xiao S, Liu C, et al. Lizhong decoction ameliorates ulcerative colitis in mice via modulating gut microbiota and its metabolites. Appl Microbiol Biotechnol. 2020;104:5999–6012.
Jones-Hall YL, Kozik A, Nakatsu C. Ablation of tumor necrosis factor is associated with decreased inflammation and alterations of the microbiota in a mouse model of inflammatory bowel disease. PLoS ONE. 2015;10:e0119441.
Rausch P, Steck N, Suwandi A, Seidel JA, Künzel S, Bhullar K, et al. Expression of the blood-group-related gene B4galnt2 alters susceptibility to salmonella infection. PLoS Pathog. 2015;11:e1005008–e.
Jia W, Rajani C, Xu H, Zheng X. Gut microbiota alterations are distinct for primary colorectal cancer and hepatocellular carcinoma. Protein Cell. 2021;12:374–93.
Hu Y, Le Leu RK, Christophersen CT, Somashekar R, Conlon MA, Meng XQ, et al. Manipulation of the gut microbiota using resistant starch is associated with protection against colitis-associated colorectal cancer in rats. Carcinogenesis. 2016;37:366–75.
Kemis JH, Linke V, Barrett KL, Boehm FJ, Traeger LL, Keller MP, et al. Genetic determinants of gut microbiota composition and bile acid profiles in mice. PLoS Genet. 2019;15:e1008073.
Zarrinpar A, Chaix A, Xu ZZ, Chang MW, Marotz CA, Saghatelian A, et al. Antibiotic-induced microbiome depletion alters metabolic homeostasis by affecting gut signaling and colonic metabolism. Nat Commun. 2018;9:2872.
Grosser G, Müller SF, Kirstgen M, Döring B, Geyer J. Substrate specificities and inhibition pattern of the solute carrier family 10 members NTCP, ASBT and SOAT. Front Mol Biosci. 2021;8:689757.
Tian Y, Gui W, Koo I, Smith PB, Allman EL, Nichols RG, et al. The microbiome modulating activity of bile acids. Gut Microbes. 2020;11:979–96.
Ju T, Kong JY, Stothard P, Willing BP. Defining the role of Parasutterella, a previously uncharacterized member of the core gut microbiota. ISME J. 2019;13:1520–34.
Devlin AS, Fischbach MA. A biosynthetic pathway for a prominent class of microbiota-derived bile acids. Nat Chem Biol. 2015;11:685–90.
Liu HX, Rocha CS, Dandekar S, Wan YJ. Functional analysis of the relationship between intestinal microbiota and the expression of hepatic genes and pathways during the course of liver regeneration. J Hepatol. 2016;64:641–50.
He T, Cheng X, Xing C. The gut microbial diversity of colon cancer patients and the clinical significance. Bioengineered. 2021;12:7046–60.
Bailey AM, Zhan L, Maru D, Shureiqi I, Pickering CR, Kiriakova G, et al. FXR silencing in human colon cancer by DNA methylation and KRAS signaling. Am J Physiol Gastrointest Liver Physiol. 2014;306:G48–58.
Fu T, Coulter S, Yoshihara E, Oh TG, Fang S, Cayabyab F, et al. FXR regulates intestinal cancer stem cell proliferation. Cell 2019;176:1098–112.e18.
Hagi T, Geerlings SY, Nijsse B, Belzer C. The effect of bile acids on the growth and global gene expression profiles in Akkermansia muciniphila. Appl Microbiol Biotechnol. 2020;104:10641–53.
Theriot CM, Bowman AA, Young VB. Antibiotic-induced alterations of the gut microbiota alter secondary bile acid production and allow for Clostridium difficile spore germination and outgrowth in the large intestine. mSphere. 2016;1:e00045-15.
Ovadia C, Perdones-Montero A, Spagou K, Smith A, Sarafian MH, Gomez-Romero M, et al. Enhanced microbial bile acid deconjugation and impaired ileal uptake in pregnancy repress intestinal regulation of bile acid synthesis. Hepatology 2019;70:276–93.
Wei W, Wang H-F, Zhang Y, Zhang Y-L, Niu B-Y, Yao S-K. Altered metabolism of bile acids correlates with clinical parameters and the gut microbiota in patients with diarrhea-predominant irritable bowel syndrome. World J Gastroenterol. 2020;26:7153–72.
Palm NW, de Zoete MR, Cullen TW, Barry NA, Stefanowski J, Hao L, et al. Immunoglobulin A coating identifies colitogenic bacteria in inflammatory bowel disease. Cell. 2014;158:1000–10.
Miyauchi E, Kim SW, Suda W, Kawasumi M, Onawa S, Taguchi-Atarashi N, et al. Gut microorganisms act together to exacerbate inflammation in spinal cords. Nature. 2020;585:102–6.
Umar S. Citrobacter infection and Wnt signaling. Curr Colorectal Cancer Rep. 2012;8 https://doi.org/10.1007/s11888-012-0143-4.
da Silva Duarte V, Dos Santos Cruz BC, Tarrah A, Sousa Dias R, de Paula Dias Moreira L, Lemos Junior WJF, et al. Chemoprevention of DMH-induced early colon carcinogenesis in male BALB/c mice by administration of Lactobacillus Paracasei DTA81. Microorganisms. 2020;8:1994.
Zeng H, Ishaq SL, Liu Z, Bukowski MR. Colonic aberrant crypt formation accompanies an increase of opportunistic pathogenic bacteria in C57BL/6 mice fed a high-fat diet. J Nutr Biochem. 2018;54:18–27.
Zhang W, An Y, Qin X, Wu X, Wang X, Hou H, et al. Gut microbiota-derived metabolites in colorectal cancer: the bad and the challenges. Front Oncol. 2021;11:739648.
Wang X, Huycke MM. Extracellular superoxide production by Enterococcus faecalis promotes chromosomal instability in mammalian cells. Gastroenterology 2007;132:551–61.
Liu Y, Zhang S, Zhou W, Hu D, Xu H, Ji G. Secondary bile acids and tumorigenesis in colorectal cancer. Front Oncol. 2022;12:813745.
Peters BA, Dominianni C, Shapiro JA, Church TR, Wu J, Miller G, et al. The gut microbiota in conventional and serrated precursors of colorectal cancer. Microbiome. 2016;4:69.
Rosshart SP, Vassallo BG, Angeletti D, Hutchinson DS, Morgan AP, Takeda K, et al. Wild mouse gut microbiota promotes host fitness and improves disease resistance. Cell. 2017;171:1015–28.e13.
Leystra AA, Clapper ML. Gut microbiota influences experimental outcomes in mouse models of colorectal cancer. Genes (Basel). 2019;10:900.
Wu Y, Jiao N, Zhu R, Zhang Y, Wu D, Wang AJ, et al. Identification of microbial markers across populations in early detection of colorectal cancer. Nat Commun. 2021;12:3063.
Zhang YK, Zhang Q, Wang YL, Zhang WY, Hu HQ, Wu HY, et al. A Comparison study of age and colorectal cancer-related gut bacteria. Front Cell Infect Microbiol. 2021;11:606490.
Lavelle A, Nancey S, Reimund JM, Laharie D, Marteau P, Treton X, et al. Fecal microbiota and bile acids in IBD patients undergoing screening for colorectal cancer. Gut Microbes. 2022;14:2078620.
Geller LT, Barzily-Rokni M, Danino T, Jonas OH, Shental N, Nejman D, et al. Potential role of intratumor bacteria in mediating tumor resistance to the chemotherapeutic drug gemcitabine. Science. 2017;357:1156–60.
Acknowledgements
We acknowledge Pat Casey for his assistance with the animal studies. Graphical abstract was created with BioRender.com.
Funding
This work was supported by the APC Innovation Platform. APC Microbiome Ireland is a research institute funded by Science Foundation Ireland (SFI) through the Irish Governments National Development Plan (Grant SFI/12/RC/2273).
Author information
Authors and Affiliations
Contributions
JMK acquired data and played an important role in interpreting the results and drafted the manuscript. CJW, PC and KB acquired data. SM helped to design the work that led to the submission. PDC helped draft the manuscript, acquired data, and/or played an important role in interpreting the results. SAJ, CGMG, AH and NPH conceived and designed the work that led to the submission, played an important role in interpreting the results and drafted the manuscript. All authors approved the final version and agreed to be accountable for all aspects of the work.
Corresponding author
Ethics declarations
Competing interests
The authors are not aware of any competing interests that might be perceived as affecting the findings of this study.
Ethics approval and consent to participate
Animal experiments were conducted in accordance with the regulations and guidelines of the Irish Department of Health following approval by the University College Cork Animal Experimentation Ethics Committee (2011/023).
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Keane, J.M., Walsh, C.J., Cronin, P. et al. Investigation of the gut microbiome, bile acid composition and host immunoinflammatory response in a model of azoxymethane-induced colon cancer at discrete timepoints. Br J Cancer 128, 528–536 (2023). https://doi.org/10.1038/s41416-022-02062-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41416-022-02062-4
This article is cited by
-
Metformin facilitates anti-PD-L1 efficacy through the regulation of intestinal microbiota
Genes & Immunity (2023)
-
Anise (Pimpinella anisum L.) attenuates azoxymethane-induced colorectal cancer by antioxidant, anti-inflammatory, and anti-apoptotic pathways in rats
Environmental Science and Pollution Research (2023)