Abstract
Background
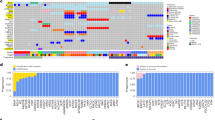
Deleted in colorectal cancer (DCC) encodes a transmembrane dependence receptor and is frequently mutated in melanoma. The associations of DCC mutation with chromosomal instability and immunotherapeutic efficacy in melanoma are largely uncharacterised.
Methods
We performed an integrated study based on biological experiments and multi-dimensional data types, including genomic, transcriptomic and clinical immune checkpoint blockade (ICB)-treated melanoma cohorts from public databases.
Results
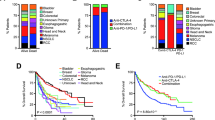
DCC mutation was significantly correlated with the tumour mutational burden (TMB) in The Cancer Genome Atlas (TCGA), International Cancer Genome Consortium (ICGC) and ICB-treated melanoma cohorts. DCC expression levels were correlated with DNA damage response and repair (DDR) pathways responsive to irradiation (IR) in the Malme-3M and SK-MEL-2 cell lines. In the TCGA cohort, DCC-mutated samples presented more neoantigens, higher proportions of infiltrating antitumour immunocytes and lower proportions of infiltrating pro-tumour immunocytes than DCC wild-type samples. DCC-mutated samples were significantly enriched in activated immune response and DDR pathways. Furthermore, patients harbouring mutated DCC treated with ICB showed remarkable clinical benefits in terms of the response rate and overall survival.
Conclusions
Somatic mutations in DCC are associated with improved clinical outcomes in ICB-treated melanoma patients. Once further validated, the DCC mutational status can improve patient selection for clinical practice and future study enrolment.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 24 print issues and online access
$259.00 per year
only $10.79 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The datasets used in this study are available from the TCGA, ICGC and CCLE repositories. The ICB-treated melanoma datasets generated during the current study are available in six previous studies and their supplementary data files as described in the ‘Genomic and clinical data’ subsection. All data supporting the conclusions of this study have been included within the article and the Supplemental Data.
Code availability
The computer codes used to generate results that support the paper’s conclusions are available from the corresponding author upon reasonable request.
Materials availability
All materials supporting the conclusions of this study have been included in the article and the Supplemental Data.
References
Larkin J, Chiarion-Sileni V, Gonzalez R, Grob JJ, Cowey CL, Lao CD, et al. Combined nivolumab and ipilimumab or monotherapy in untreated melanoma. N Engl J Med. 2015;373:23–34.
Robert C, Schachter J, Long GV, Arance A, Grob JJ, Mortier L, et al. Pembrolizumab versus ipilimumab in advanced melanoma. N Engl J Med. 2015;372:2521–32.
Schadendorf D, Hodi FS, Robert C, Weber JS, Margolin K, Hamid O, et al. Pooled analysis of long-term survival data from phase II and phase III trials of ipilimumab in unresectable or metastatic melanoma. J Clin Oncol. 2015;33:1889–94.
Wolchok JD, Chiarion-Sileni V, Gonzalez R, Rutkowski P, Grob J-J, Cowey CL, et al. Overall survival with combined nivolumab and ipilimumab in advanced melanoma. N Engl J Med. 2017;377:1345–56.
Liu D, Schilling B, Liu D, Sucker A, Livingstone E, Jerby-Amon L, et al. Integrative molecular and clinical modeling of clinical outcomes to PD1 blockade in patients with metastatic melanoma. Nat Med. 2019;25:1916–27.
Chan TA, Yarchoan M, Jaffee E, Swanton C, Quezada SA, Stenzinger A, et al. Development of tumor mutation burden as an immunotherapy biomarker: utility for the oncology clinic. Ann Oncol. 2019;30:44–56.
Snyder A, Makarov V, Merghoub T, Yuan J, Zaretsky JM, Desrichard A, et al. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N Engl J Med. 2014;371:2189–99.
Van Allen EM, Miao D, Schilling B, Shukla SA, Blank C, Zimmer L, et al. Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. Science. 2015;350:207–11.
Mouw KW, Goldberg MS, Konstantinopoulos PA, D’Andrea AD. DNA damage and repair biomarkers of immunotherapy response. Cancer Discov. 2017;7:675–93.
Tang H, Wang Y, Chlewicki LK, Zhang Y, Guo J, Liang W, et al. Facilitating T cell infiltration in tumor microenvironment overcomes resistance to PD-L1 blockade. Cancer Cell. 2016;29:285–96.
Tumeh PC, Harview CL, Yearley JH, Shintaku IP, Taylor EJ, Robert L, et al. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature. 2014;515:568–71.
Topalian SL, Taube JM, Anders RA, Pardoll DM. Mechanism-driven biomarkers to guide immune checkpoint blockade in cancer therapy. Nat Rev Cancer. 2016;16:275–87.
Fearon ER, Cho KR, Nigro JM, Kern SE, Simons JW, Ruppert JM, et al. Identification of a chromosome 18q gene that is altered in colorectal cancers. Science. 1990;247:49–56.
Shibata D, Reale MA, Lavin P, Silverman M, Fearon ER, Steele G Jr, et al. The DCC protein and prognosis in colorectal cancer. N Engl J Med. 1996;335:1727–32.
Mehlen P, Fearon ER. Role of the dependence receptor DCC in colorectal cancer pathogenesis. J Clin Oncol. 2004;22:3420–8.
Goldschneider D, Mehlen P. Dependence receptors: a new paradigm in cell signaling and cancer therapy. Oncogene. 2010;29:1865–82.
Mehlen P, Tauszig-Delamasure S. Dependence receptors and colorectal cancer. Gut. 2014;63:1821–9.
Gibert B, Mehlen P. Dependence receptors and cancer: addiction to trophic ligands. Cancer Res. 2015;75:5171–5.
Krauthammer M, Kong Y, Ha BH, Evans P, Bacchiocchi A, McCusker JP, et al. Exome sequencing identifies recurrent somatic RAC1 mutations in melanoma. Nat Genet. 2012;44:1006–14.
Boussouar A, Tortereau A, Manceau A, Paradisi A, Gadot N, Vial J, et al. Netrin-1 and its receptor DCC are causally implicated in melanoma progression. Cancer Res. 2020;80:747–56.
Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SAJR, Behjati S, Biankin AV, et al. Signatures of mutational processes in human cancer. Nature. 2013;500:415–21.
Alexandrov LB, Kim J, Haradhvala NJ, Huang MN, Tian Ng AW, Wu Y, et al. The repertoire of mutational signatures in human cancer. Nature. 2020;578:94–101.
Boland CR, Goel A. Microsatellite instability in colorectal cancer. Gastroenterology. 2010;138:2073–87.e3.
Le DT, Durham JN, Smith KN, Wang H, Bartlett BR, Aulakh LK, et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science. 2017;357:409–13.
Ghandi M, Huang FW, Jané-Valbuena J, Kryukov GV, Lo CC, McDonald ER, et al. Next-generation characterization of the Cancer Cell Line Encyclopedia. Nature. 2019;569:503–8.
Hugo W, Zaretsky JM, Sun L, Song C, Moreno BH, Hu-Lieskovan S, et al. Genomic and transcriptomic features of response to anti-PD-1 therapy in metastatic melanoma. Cell. 2016;165:35–44.
Zaretsky JM, Garcia-Diaz A, Shin DS, Escuin-Ordinas H, Hugo W, Hu-Lieskovan S, et al. Mutations associated with acquired resistance to PD-1 blockade in melanoma. N Engl J Med. 2016;375:819–29.
Riaz N, Havel JJ, Makarov V, Desrichard A, Urba WJ, Sims JS, et al. Tumor and microenvironment evolution during immunotherapy with nivolumab. Cell. 2017;171:934–49.e16.
Roh W, Chen P-L, Reuben A, Spencer CN, Prieto PA, Miller JP, et al. Integrated molecular analysis of tumor biopsies on sequential CTLA-4 and PD-1 blockade reveals markers of response and resistance. Sci Transl Med. 2017;9:eaah3560.
Ramos AH, Lichtenstein L, Gupta M, Lawrence MS, Pugh TJ, Saksena G, et al. Oncotator: cancer variant annotation tool. Hum Mutat. 2015;36:E2423–9.
Knijnenburg TA, Wang L, Zimmermann MT, Chambwe N, Gao GF, Cherniack AD, et al. Genomic and molecular landscape of DNA damage repair deficiency across The Cancer Genome Atlas. Cell Rep. 2018;23:239–54.e6.
Wang H, Song M. Ckmeans.1d.dp: optimal k-means clustering in one dimension by dynamic programming. R J. 2011;3:29–33.
Kim J, Mouw KW, Polak P, Braunstein LZ, Kamburov A, Kwiatkowski DJ, et al. Somatic ERCC2 mutations are associated with a distinct genomic signature in urothelial tumors. Nat Genet. 2016;48:600–6.
Yarchoan M, Johnson BA 3rd, Lutz ER, Laheru DA, Jaffee EM. Targeting neoantigens to augment antitumour immunity. Nat Rev Cancer. 2017;17:209–22.
Rooney MS, Shukla SA, Wu CJ, Getz G, Hacohen N. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell. 2015;160:48–61.
Shukla SA, Rooney MS, Rajasagi M, Tiao G, Dixon PM, Lawrence MS, et al. Comprehensive analysis of cancer-associated somatic mutations in class I HLA genes. Nat Biotechnol. 2015;33:1152–8.
Charoentong P, Finotello F, Angelova M, Mayer C, Efremova M, Rieder D, et al. Pan-cancer immunogenomic analyses reveal genotype-immunophenotype relationships and predictors of response to checkpoint blockade. Cell Rep. 2017;18:248–62.
Newman AM, Liu CL, Green MR, Gentles AJ, Feng W, Xu Y, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12:453–7.
Pardoll DM. The blockade of immune checkpoints in cancer immunotherapy. Nat Rev Cancer. 2012;12:252–64.
Burugu S, Dancsok AR, Nielsen TO. Emerging targets in cancer immunotherapy. Semin Cancer Biol. 2018;52:39–52.
Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–50.
Liberzon A, Subramanian A, Pinchback R, Thorvaldsdottir H, Tamayo P, Mesirov JP. Molecular signatures database (MSigDB) 3.0. Bioinformatics. 2011;27:1739–40.
Skidmore ZL, Wagner AH, Lesurf R, Campbell KM, Kunisaki J, Griffith OL, et al. GenVisR: genomic visualizations in R. Bioinformatics. 2016;32:3012–4.
Lee JH, Paull TT. Direct activation of the ATM protein kinase by the Mre11/Rad50/Nbs1 complex. Science. 2004;304:93–6.
Carson CT, Schwartz RA, Stracker TH, Lilley CE, Lee DV, Weitzman MD. The Mre11 complex is required for ATM activation and the G2/M checkpoint. EMBO J. 2003;22:6610–20.
Gatei M, Scott SP, Filippovitch I, Soronika N, Lavin MF, Weber B, et al. Role for ATM in DNA damage-induced phosphorylation of BRCA1. Cancer Res. 2000;60:3299–304.
Prakash R, Zhang Y, Feng W, Jasin M. Homologous recombination and human health: the roles of BRCA1, BRCA2, and associated proteins. Cold Spring Harb Perspect Biol. 2015;7:a016600.
Chen YQ, Hsieh JT, Yao F, Fang B, Pong RC, Cipriano SC, et al. Induction of apoptosis and G2/M cell cycle arrest by DCC. Oncogene. 1999;18:2747–54.
Castets M, Broutier L, Molin Y, Brevet M, Chazot G, Gadot N, et al. DCC constrains tumour progression via its dependence receptor activity. Nature. 2011;482:534–7.
Hedrick L, Cho KR, Fearon ER, Wu TC, Kinzler KW, Vogelstein B. The DCC gene product in cellular differentiation and colorectal tumorigenesis. Genes Dev. 1994;8:1174–83.
Bernet A, Fitamant J. Netrin-1 and its receptors in tumour growth promotion. Expert Opin Ther Targets. 2008;12:995–1007.
Yang L, Garbe DS, Bashaw GJ. A frazzled/DCC-dependent transcriptional switch regulates midline axon guidance. Science. 2009;324:944–7.
Neuhaus-Follini A, Bashaw GJ. The intracellular domain of the Frazzled/DCC receptor is a transcription factor required for commissural axon guidance. Neuron. 2015;87:751–63.
Fitamant J, Guenebeaud C, Coissieux M-M, Guix C, Treilleux I, Scoazec J-Y, et al. Netrin-1 expression confers a selective advantage for tumor cell survival in metastatic breast cancer. Proc Natl Acad Sci USA. 2008;105:4850–5.
Grandin M, Mathot P, Devailly G, Bidet Y, Ghantous A, Favrot C, et al. Inhibition of DNA methylation promotes breast tumor sensitivity to netrin-1 interference. EMBO Mol Med. 2016;8:863–77.
Paradisi A, Creveaux M, Gibert B, Devailly G, Redoulez E, Neves D, et al. Combining chemotherapeutic agents and netrin-1 interference potentiates cancer cell death. EMBO Mol Med. 2013;5:1821–34.
Acknowledgements
The authors would like to thank all patients with melanoma who kindly donated samples for the TCGA and ICGC projects. The authors sincerely appreciated all of the data-sharing platforms, such as the TCGA, ICGC and CCLE. The authors would like to thank the colleagues from our department for their assistance.
Funding
This work was supported by the National Natural Science Foundation of China (81372429 to Lujun Zhao).
Author information
Authors and Affiliations
Contributions
YL had full access to perform experiments and statistical analysis in this study. LZ conceived and designed this study. All authors performed acquisition, analyses, or interpretation of data. YL drafted of the manuscript under close supervision and critical revision of LZ. All authors contributed paper writing and proofreading.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
Not applicable.
Consent to publish
Not applicable.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Li, Y., Wang, Q., Chen, Y. et al. Somatic mutations in DCC are associated with genomic instability and favourable outcomes in melanoma patients treated with immune checkpoint inhibitors. Br J Cancer 127, 1411–1423 (2022). https://doi.org/10.1038/s41416-022-01921-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41416-022-01921-4