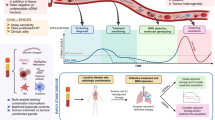
Abstract
Introduction
Small cell lung cancer (SCLC) is an aggressive malignancy with no established biomarkers. Schlafen 11(SLFN11), a DNA/RNA helicase that sensitises cancer cells to DNA-damaging agents, has emerged as a promising predictive biomarker for several drug classes including platinum and PARP inhibitors. Detection of SLFN11 in circulating tumour cells (CTCs) may provide a valuable alternative to tissue sampling.
Methods
SLFN11 expression was evaluated in tumour samples and characterised in circulating tumour cells (CTC) longitudinally to determine its potential role as a biomarker of response.
Results
Among 196 SCLC tumours, 51% expressed SLFN11 by IHC. In addition, 20/29 extra-thoracic high-grade neuroendocrine tumours expressed SLFN11 expression. In 64 blood samples from 42 SCLC patients, 83% (53/64) of samples had detectable CTCs, and SLFN11-positive CTCs were detected in 55% (29/53). Patients actively receiving platinum treatment had the lowest number of CTCs and a lower percentage of SLFN11-positive CTCs (p = 0.014). Analysis from patients with longitudinal samples suggest a decrease in CTC number and in SLFN11 expression that correlates with clinical response.
Conclusions
SLFN11 levels can be monitored in CTCs from SCLC patients using non-invasive liquid biopsies. The ability to detect SLFN11 in CTCs from SCLC patients adds a valuable tool for the detection and longitudinal monitoring of this promising biomarker.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 24 print issues and online access
$259.00 per year
only $10.79 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Data availability
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
References
Allison Stewart C, Tong P, Cardnell RJ, Sen T, Li L, Gay CM, et al. Dynamic variations in epithelial-to-mesenchymal transition (EMT), ATM, and SLFN11 govern response to PARP inhibitors and cisplatin in small cell lung cancer. Oncotarget. 2017;8:28575–87.
Sousa FG, Matuo R, Tang SW, Rajapakse VN, Luna A, Sander C, et al. Alterations of DNA repair genes in the NCI-60 cell lines and their predictive value for anticancer drug activity. DNA Repair (Amst). 2015;28:107–15.
Zoppoli G, Regairaz M, Leo E, Reinhold WC, Varma S, Ballestrero A, et al. Putative DNA/RNA helicase Schlafen-11 (SLFN11) sensitizes cancer cells to DNA-damaging agents. Proc Natl Acad Sci USA. 2012;109:15030–5.
Pietanza MC, Waqar SN, Krug LM, Dowlati A, Hann CL, Chiappori A, et al. Randomized, double-blind, phase II study of temozolomide in combination with either veliparib or placebo in patients with relapsed-sensitive or refractory small-cell lung cancer. J Clin Oncol. 2018;36:2386–94.
Byers, LA, Bentsion, D, Gans, S, Penkov, K, Son, C, Sibille, A et al. Veliparib in combination with carboplatin and etoposide in patients with treatment-naive extensive-stage small cell lung cancer: a phase 2 randomized study. Clin Cancer Res. 2021; https://doi.org/10.1158/1078-0432.CCR-20-4259.
Zhang, B, Ramkumar, K, Cardnell, RJ, Gay, CM, Stewart, CA, Wang, WL et al. A wake-up call for cancer DNA damage: the role of Schlafen 11 (SLFN11) across multiple cancers. Br J Cancer. 2021; https://doi.org/10.1038/s41416-021-01476-w.
Winkler, C, King, M, Berthe, J, Ferraioli, D, Garuti, A, Grillo, F et al. SLFN11 captures cancer-immunity interactions associated with platinum sensitivity in high-grade serous ovarian cancer. JCI Insight 2021;6:e146098.
Conteduca V, Ku SY, Puca L, Slade M, Fernandez L, Hess J, et al. SLFN11 Expression in advanced prostate cancer and response to platinum-based chemotherapy. Mol Cancer Ther. 2020;19:1157–64.
Gay CM, Stewart CA, Park EM, Diao L, Groves SM, Heeke S, et al. Patterns of transcription factor programs and immune pathway activation define four major subtypes of SCLC with distinct therapeutic vulnerabilities. Cancer Cell. 2021;39:346–360.e347.
Ireland AS, Micinski AM, Kastner DW, Guo B, Wait SJ, Spainhower KB, et al. MYC Drives temporal evolution of small cell lung cancer subtypes by reprogramming neuroendocrine fate. Cancer Cell. 2020;38:60–78.e12.
Stewart CA, Gay CM, Xi Y, Sivajothi S, Sivakamasundari V, Fujimoto J, et al. Single-cell analyses reveal increased intratumoral heterogeneity after the onset of therapy resistance in small-cell lung cancer. Nat Cancer. 2020;1:423–36.
Simpson KL, Stoney R, Frese KK, Simms N, Rowe W, Pearce SP, et al. A biobank of small cell lung cancer CDX models elucidates inter- and intratumoral phenotypic heterogeneity. Nat Cancer. 2020;1:437–51.
Gardner EE, Lok BH, Schneeberger VE, Desmeules P, Miles LA, Arnold PK, et al. Chemosensitive relapse in small cell lung cancer proceeds through an EZH2-SLFN11 Axis. Cancer Cell. 2017;31:286–99.
Hou JM, Krebs MG, Lancashire L, Sloane R, Backen A, Swain RK, et al. Clinical significance and molecular characteristics of circulating tumor cells and circulating tumor microemboli in patients with small-cell lung cancer. J Clin Oncol. 2012;30:525–32.
Hodgkinson CL, Morrow CJ, Li Y, Metcalf RL, Rothwell DG, Trapani F, et al. Tumorigenicity and genetic profiling of circulating tumor cells in small-cell lung cancer. Nat Med. 2014;20:897–903.
Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, et al. The cancer cell line encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–7.
Armstrong AJ, Halabi S, Luo J, Nanus DM, Giannakakou P, Szmulewitz RZ, et al. Prospective multicenter validation of androgen receptor splice variant 7 and hormone therapy resistance in high-risk castration-resistant prostate cancer: The PROPHECY Study. J Clin Oncol. 2019;37:1120–9.
Puca, L, Gavyert, K, Sailer, V, Conteduca, V, Dardenne, E, Sigouros, M et al. Delta-like protein 3 expression and therapeutic targeting in neuroendocrine prostate cancer. Sci Transl Med. 2019;11:eaav0891.
Scher HI, Armstrong AJ, Schonhoft JD, Gill A, Zhao JL, Barnett E, et al. Development and validation of circulating tumour cell enumeration (Epic Sciences) as a prognostic biomarker in men with metastatic castration-resistant prostate cancer. Eur J Cancer. 2021;150:83–94.
Scher HI, Lu D, Schreiber NA, Louw J, Graf RP, Vargas HA, et al. Association of AR-V7 on circulating tumor cells as a treatment-specific biomarker with outcomes and survival in castration-resistant prostate cancer. JAMA Oncol. 2016;2:1441–9.
Schonhoft JD, Zhao JL, Jendrisak A, Carbone EA, Barnett ES, Hullings MA, et al. Morphology-predicted large-scale transition number in circulating tumor cells identifies a chromosomal instability biomarker associated with poor outcome in castration-resistant prostate cancer. Cancer Res. 2020;80:4892–903.
Brown, LC, Halabi, S, Schonhoft, JD, Yang, Q, Luo, J, Nanus, DM et al. Circulating tumor cell chromosomal instability and neuroendocrine phenotype by immunomorphology and poor outcomes in men with mCRPC treated with abiraterone or enzalutamide. Clin Cancer Res. 2021; https://doi.org/10.1158/1078-0432.CCR-20-3471.
Werner SL, Graf RP, Landers M, Valenta DT, Schroeder M, Greene SB, et al. Analytical validation and capabilities of the epic CTC platform: enrichment-free circulating tumour cell detection and characterization. J Circ Biomark. 2015;4:3.
Beltran H, Jendrisak A, Landers M, Mosquera JM, Kossai M, Louw J, et al. The initial detection and partial characterization of circulating tumor cells in neuroendocrine prostate cancer. Clin Cancer Res. 2016;22:1510–9.
Dorantes-Heredia R, Ruiz-Morales JM, Cano-Garcia F. Histopathological transformation to small-cell lung carcinoma in non-small cell lung carcinoma tumors. Transl Lung Cancer Res. 2016;5:401–12.
Travis WD, Brambilla E, Burke AP, Marx A, Nicholson AG. Introduction to The 2015 World Health Organization Classification of Tumors of the Lung, Pleura, Thymus, and Heart. J Thorac Oncol. 2015;10:1240–2.
Travis WD, Brambilla E, Nicholson AG, Yatabe Y, Austin JHM, Beasley MB, et al. The 2015 World Health Organization Classification of Lung Tumors: Impact of Genetic, Clinical and Radiologic Advances Since the 2004 Classification. J Thorac Oncol. 2015;10:1243–60.
R Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing, V., Austria. 2020; https://www.R-project.org/.
Scher HI, Graf RP, Schreiber NA, Jayaram A, Winquist E, McLaughlin B, et al. Assessment of the validity of nuclear-localized androgen receptor splice variant 7 in circulating tumor cells as a predictive biomarker for castration-resistant prostate cancer. JAMA Oncol. 2018;4:1179–86.
Murai J, Feng Y, Yu GK, Ru Y, Tang SW, Shen Y, et al. Resistance to PARP inhibitors by SLFN11 inactivation can be overcome by ATR inhibition. Oncotarget. 2016;7:76534–50.
Travis WD. Update on small cell carcinoma and its differentiation from squamous cell carcinoma and other non-small cell carcinomas. Mod Pathol. 2012;25:S18–30.
Thunnissen E, Borczuk AC, Flieder DB, Witte B, Beasley MB, Chung JH, et al. The use of immunohistochemistry improves the diagnosis of small cell lung cancer and its differential diagnosis. An International Reproducibility Study in a Demanding Set of Cases. J Thorac Oncol. 2017;12:334–46.
Tay RY, Fernandez-Gutierrez F, Foy V, Burns K, Pierce J, Morris K, et al. Prognostic value of circulating tumour cells in limited-stage small-cell lung cancer: analysis of the concurrent once-daily versus twice-daily radiotherapy (CONVERT) randomised controlled trial. Ann Oncol. 2019;30:1114–20.
Shishido SN, Carlsson A, Nieva J, Bethel K, Hicks JB, Bazhenova L, et al. Circulating tumor cells as a response monitor in stage IV non-small cell lung cancer. J Transl Med. 2019;17:294.
Kiniwa Y, Nakamura K, Mikoshiba A, Ashida A, Akiyama Y, Morimoto A, et al. Usefulness of monitoring circulating tumor cells as a therapeutic biomarker in melanoma with BRAF mutation. BMC Cancer. 2021;21:287.
Frese KK, Simpson KL, Dive C. Small cell lung cancer enters the era of precision medicine. Cancer Cell. 2021;39:297–9.
Qu, S, Fetsch, P, Thomas, A, Pommier, Y, Schrump, DS, Miettinen, MM et al. Molecular Subtypes of Primary SCLC Tumors and Their Associations With Neuroendocrine and Therapeutic Markers. J Thorac Oncol. 2021; https://doi.org/10.1016/j.jtho.2021.08.763.
Winkler C, Armenia J, Jones GN, Tobalina L, Sale MJ, Petreus T, et al. SLFN11 informs on standard of care and novel treatments in a wide range of cancer models. Br J Cancer. 2021;124:951–62.
Mezzadra R, de Bruijn M, Jae LT, Gomez-Eerland R, Duursma A, Scheeren FA, et al. SLFN11 can sensitize tumor cells towards IFN-gamma-mediated T cell killing. PLoS ONE. 2019;14:e0212053.
Mavrommatis E, Fish EN, Platanias LC. The schlafen family of proteins and their regulation by interferons. J Interferon Cytokine Res. 2013;33:206–10.
Acknowledgements
We would like to acknowledge the patients who participated in this study and volunteered for blood draws (under protocol LAB10-0442) to help further our understanding of SCLC. We would also like to thank Patrice Hartsfield and Heather Napoleon from the protocol blood collection team. This work was supported by: The NIH/NCI CCSG P30-CA016672 (Bioinformatics Shared Resource); NIH/NCI R01-CA207295; NIH/NCI U01-CA213273; NIH/NCI R50-CA243698; The University of Texas-Southwestern and MD Anderson Lung SPORE (5 P50 CA070907); NIH T32 training grant (T32-CA009666); and CPRIT Early Clinical Investigator Award. and through generous philanthropic contributions to The University of Texas MD Anderson Lung Cancer Moon Shot Program and to the Byers Lab; The Andrew Sabin Family Fellowship; The Abell Hangar Foundation; the LUNGevity Foundation Career Development Award, and the Rexanna Foundation for Fighting Lung Cancer.
Funding
This work was supported by NIH T32 CA009666 (BZ); NIH/NCI R50-CA243698 (CS); University of Texas-Southwestern and MD Anderson Lung SPORE (5 P50 CA070907), The University of Texas MD Anderson Lung Cancer Moon Shot Program, The Andrew Sabin Family Fellowship, The Abell Hangar Foundation; the LUNGevity Foundation Career Development Award, the Rexanna Foundation for Fighting Lung Cancer, and CPRIT Early Clinical Investigator Award (CG); NIH/NCI U01-CA213273 and NIH R01-CA207295 (LB).
Author information
Authors and Affiliations
Contributions
BZ, CAS, RJC, LAB and CMG: conceptualised this study. BZ, CAS, QW, PR, JF, LS, RW, LF, AJ, CG, JDS, JB and JJ contributed to methodology development, data curation and analyses of the study. VN, PA, LH, HT, AA, MN, JZ, WW and IW provided administration and resources support. JW, RW, LAB and CMG supervised and provided funding for the study. All authors contributed to the writing, editing and review of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
IW received honorarium from Genentech/Roche, Bayer, Bristol-Myers Squibb, AstraZeneca, Pfizer, HTG Molecular, Asuragen, Merck, GlaxoSmithKline, Guardant Health, Flame, Novartis, Sanofi, Daiichi Sankyo, Amgen, Oncocyte, and MSD; and received research support from Genentech, HTG Molecular, DepArray, Merck, Bristol-Myers Squibb, Medimmune, Adaptive, Adaptimmune, EMD Serono, Pfizer, Takeda, Amgen, Karus, Johnson & Johnson, Bayer, Iovance, 4D, Novartis, and Akoya. JZ served on advisory board for AstraZeneca, Novartis, Johnson and Johnson, Geneplus and received speaker’s fees from BMS, OrigMed, Innovent, grants from Merck, Novartis, Johnson and Johnson from outside the submitted work JS received stock options from Epic Sciences at the time of this work. MN reports research funding to institution: Mirati, Novartis, Checkmate, Ziopharm, AstraZeneca, Pfizer, Genentech; consulted for Mirati, Merk and served on advisory board for MSD; meal expenses from Ziopharm. LB received research funding from AstraZeneca, GenMab, Sierra Oncology, ToleroPharmaceuticals; served as an advisor/consultant for AstraZeneca, GenMab, Sierra Oncology, PharmaMar, AbbVie, Bristol-Myers Squibb, Alethia, Merck, Pfizer, Jazz Pharmaceuticals, Genentech, Debiopharm Group. CG received research funding from AstraZeneca; served as an adviser for Bristol-Myers Squibb, Jazz Pharmaceuticals, AstraZeneca, Kisoji; and served on the Speaker’s Bureau for AstraZeneca, Beigene.
Ethics approval and consent to participate
Participants underwent informed written consent to Institutional Review Board (IRB)-approved protocol LAB10-0442 (“Evaluation of blood-based test for the detection of circulating tumour cells and circulating proteins and microRNAs and molecular analysis for polymorphisms and mutations”). The study was performed in accordance with the Declaration of Helsinki.
Consent to publish
Not applicable.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Zhang, B., Stewart, C.A., Wang, Q. et al. Dynamic expression of Schlafen 11 (SLFN11) in circulating tumour cells as a liquid biomarker in small cell lung cancer. Br J Cancer 127, 569–576 (2022). https://doi.org/10.1038/s41416-022-01811-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41416-022-01811-9