Abstract
Background
Clear-cell renal-cell carcinoma (ccRCC) is one of the leading causes of tumour-related death worldwide. Methyltransferase-like 14 (METTL14) is reported to regulate m6A modification in cancers. The aim of this study is to investigate the biological function and molecular mechanism of METTL14 in the pathogenesis of ccRCC.
Methods
Quantitative real-time PCR (qRT-PCR), western blot and immunohistochemical (IHC) assays were used to detect the expression of METTL14 and Pten. METTL14 overexpression or knockdown was used in the in vitro and in vivo studies to investigate the biological functions of METTL14. m6A-RNA immunoprecipitation and RNA immunoprecipitation were used to investigate the m6A modification mediated by METTL14.
Results
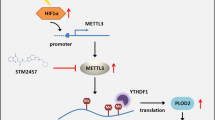
METTL14 expression was significantly down-regulated in ccRCC tissues. Functionally, upregulation of METTL14 inhibited ccRCC cells proliferation and migration in vitro. METTL14 overexpression significantly inhibited the activation of the phosphoinositide 3 kinase (PI3K)/AKT signalling pathway. Furthermore, phosphate and tension homology deleted on chromosome ten (Pten) is a target of METTL14. Overexpression of METTL14 increased the m6A enrichment of Pten, and promoted Pten expression. METTL14-enhanced Pten mRNA stability was dependent on YTHDF1.
Conclusions
METTL14-mediated m6A modification of Pten mRNA inhibited tumour progression, suggesting that METTL14 might be a potential prognostic biomarker and effective therapeutic target for ccRCC.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 24 print issues and online access
$259.00 per year
only $10.79 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout









Similar content being viewed by others
Data availability
The data used to support the findings of this study are available from the corresponding author upon request.
References
Akhtar M, Al-Bozom IA, Al Hussain, T. Molecular and metabolic basis of clear cell carcinoma of the kidney. Adv Anat Pathol. 2018;25:189–96.
Cheville JC, Lohse CM, Zincke H, Weaver AL, Blute ML. Comparisons of outcome and prognostic features among histologic subtypes of renal cell carcinoma. Am J surgical Pathol. 2003;27:612–24.
Lopez JI, Guarch R, Larrinaga G, Corominas-Cishek A, Orozco R. Cell heterogeneity in clear cell renal cell carcinoma. APMIS: Acta Pathologica, Microbiologica, et Immunologica Scandinavica. 2013;121:1187–91.
Luo Y, Shen D, Chen L, Wang G, Liu X, Qian K, et al. Identification of 9 key genes and small molecule drugs in clear cell renal cell carcinoma. Aging. 2019;11:6029–52.
Desrosiers R, Friderici K, Rottman F. Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proc Natl Acad Sci USA. 1974;71:3971–5.
Wei CM, Gershowitz A, Moss B. Methylated nucleotides block 5’ terminus of HeLa cell messenger RNA. Cell. 1975;4:379–86.
Sun T, Wu R, Ming L. The role of m6A RNA methylation in cancer. Biomedicine Pharmacother = Biomedecine Pharmacotherapie. 2019;112:108613.
Zhao W, Qi X, Liu L, Ma S, Liu J, Wu J, et al. Epigenetic regulation of m(6)A modifications in human cancer. Mol Ther Nucleic Acids. 2020;19:405–12.
Weng H, Huang H, Wu H, Qin X, Zhao BS, Dong L, et al. METTL14 inhibits hematopoietic stem/progenitor differentiation and promotes leukemogenesis via mRNA m(6)A modification. Cell Stem Cell. 2018;22:191–205.e199.
Śledź P, Jinek M. Structural insights into the molecular mechanism of the m(6)A writer complex. eLife. 2016;5:e18434.
Xu K, Yang Y, Feng GH, Sun BF, Chen JQ, Li YF, et al. Mettl3-mediated m(6)A regulates spermatogonial differentiation and meiosis initiation. Cell Res. 2017;27:1100–14.
Zhuang M, Li X, Zhu J, Zhang J, Niu F, Liang F, et al. The m6A reader YTHDF1 regulates axon guidance through translational control of Robo3.1 expression. Nucleic Acids Res. 2019;47:4765–77.
Alarcón CR, Lee H, Goodarzi H, Halberg N, Tavazoie SF. N6-methyladenosine marks primary microRNAs for processing. Nature. 2015;519:482–5.
Chen J, Fang X, Zhong P, Song Z, Hu X. N6-methyladenosine modifications: interactions with novel RNA-binding proteins and roles in signal transduction. RNA Biol. 2019;16:991–1000.
Han J, Wang JZ, Yang X, Yu H, Zhou R, Lu HC, et al. METTL3 promote tumor proliferation of bladder cancer by accelerating pri-miR221/222 maturation in m6A-dependent manner. Mol Cancer. 2019;18:110.
Xu D, Shao W, Jiang Y, Wang X, Liu Y, Liu X, et al. FTO expression is associated with the occurrence of gastric cancer and prognosis. Oncol Rep. 2017;38:2285–92.
Nishizawa Y, Konno M, Asai A, Koseki J, Kawamoto K, Miyoshi N, et al. Oncogene c-Myc promotes epitranscriptome m(6)A reader YTHDF1 expression in colorectal cancer. Oncotarget. 2018;9:7476–86.
Chen J, Yu K, Zhong G, Shen W. Identification of a m(6)A RNA methylation regulators-based signature for predicting the prognosis of clear cell renal carcinoma. Cancer Cell Int. 2020;20:157.
Zhang QJ, Luan JC, Song LB, Cong R, Ji CJ, Zhou X, et al. m6A RNA methylation regulators correlate with malignant progression and have potential predictive values in clear cell renal cell carcinoma. Exp Cell Res. 2020;392:112015.
Polivka J Jr., Janku F. Molecular targets for cancer therapy in the PI3K/AKT/mTOR pathway. Pharmacol Therapeutics. 2014;142:164–75.
Guo H, German P, Bai S, Barnes S, Guo W, Qi X, et al. The PI3K/AKT pathway and renal cell carcinoma. J Genet Genomics = Yi chuan xue bao. 2015;42:343–53.
Lin X, Zhou X, Liu D, Yun L, Zhang L, Chen X, et al. MicroRNA-29 regulates high-glucose-induced apoptosis in human retinal pigment epithelial cells through PTEN. In vitro Cell Dev Biol Animal. 2016;52:419–26.
Shinde SR, Maddika S. PTEN modulates EGFR late endocytic trafficking and degradation by dephosphorylating Rab7. Nat Commun. 2016;7:10689.
Zhao W, Cui Y, Liu L, Ma X, Qi X, Wang Y, et al. METTL3 facilitates oral squamous cell carcinoma tumorigenesis by enhancing c-Myc stability via YTHDF1-mediated m(6)A modification. Mol Ther Nucleic Acids. 2020;20:1–12.
Wang X, Zhao BS, Roundtree IA, Lu Z, Han D, Ma H, et al. N(6)-methyladenosine modulates messenger RNA translation efficiency. Cell. 2015;161:1388–99.
Wang X, Wang T, Chen C, Wu Z, Bai P, Li S, et al. Serum exosomal miR-210 as a potential biomarker for clear cell renal cell carcinoma. J Cell Biochem. 2018; https://doi.org/10.1002/jcb.27347.
Ghatalia P, Zibelman MR, Geynisman DM, Plimack ER. Evolving landscape of the treatment of metastatic clear cell renal cell carcinoma. Clin Adv Hematol Oncol. 2018;16:677–86.
Wang Q, Chen C, Ding Q, Zhao Y, Wang Z, Chen J, et al. METTL3-mediated m(6)A modification of HDGF mRNA promotes gastric cancer progression and has prognostic significance. Gut. 2020;69:1193–205.
Yang X, Zhang S, He C, Xue P, Zhang L, He Z, et al. METTL14 suppresses proliferation and metastasis of colorectal cancer by down-regulating oncogenic long non-coding RNA XIST. Mol Cancer. 2020;19:46.
Brown JA, Kinzig CG, DeGregorio SJ, Steitz JA. Methyltransferase-like protein 16 binds the 3’-terminal triple helix of MALAT1 long noncoding RNA. Proc Natl Acad Sci USA. 2016;113:14013–8.
Chen S, Li Y, Zhi S, Ding Z, Wang W, Peng Y, et al. WTAP promotes osteosarcoma tumorigenesis by repressing HMBOX1 expression in an m(6)A-dependent manner. Cell Death Dis. 2020;11:659.
Chen X, Xu M, Xu X, Zeng K, Liu X, Pan B, et al. METTL14-mediated N6-methyladenosine modification of SOX4 mRNA inhibits tumor metastasis in colorectal cancer. Mol Cancer. 2020;19:106.
Deng X, Su R, Weng H, Huang H, Li Z, Chen J. RNA N6-methyladenosine modification in cancers: current status and perspectives. Cell Res. 2018;28:507–17.
Yang G, Sun Z, Zhang N. Reshaping the role of m6A modification in cancer transcriptome: a review. Cancer Cell Int. 2020;20:353.
Tao Z, Zhao Y, Chen X. Role of methyltransferase-like enzyme 3 and methyltransferase-like enzyme 14 in urological cancers. PeerJ. 2020;8:e9589.
Wang Q, Zhang H, Chen Q, Wan Z, Gao X. Identification of METTL14 in kidney renal clear cell carcinoma using bioinformatics analysis. Disease Markers. 2019;2019:5648783.
Sun T, Wu Z, Wang X, Wang Y, Hu X, Qin W, et al. LNC942 promoting METTL14-mediated m(6)A methylation in breast cancer cell proliferation and progression. Oncogene. 2020;39:5358–72.
Chen X, Xu M, Xu X, Zeng K, Liu X, Pan B, et al. METTL14-mediated N6-methyladenosine modification of SOX4 mRNA inhibits tumor metastasis in colorectal cancer. Mol Cancer. 2020;19:106 https://doi.org/10.1186/s12943-020-01220-7.
Xu T, Gao S, Ruan H, Liu J, Liu Y, Liu D, et al. METTL14 acts as a potential regulator of tumor immune and progression in clear cell renal cell carcinoma. Front Genet. 2021;12:609174.
Pasha M, Sivaraman SK, Frantz R, Agouni A, Munusamy S. Metformin induces different responses in clear cell renal cell carcinoma caki cell lines. Biomolecules. 2019;9:113.
O’Reilly KE, Rojo F, She QB, Solit D, Mills GB, Smith D, et al. mTOR inhibition induces upstream receptor tyrosine kinase signaling and activates Akt. Cancer Res. 2006;66:1500–8.
Shaw RJ, Cantley LC. Ras, PI(3)K and mTOR signalling controls tumour cell growth. Nature. 2006;441:424–30.
Zhou J, Wang J, Hong B, Ma K, Xie H, Li L, et al. Gene signatures and prognostic values of m6A regulators in clear cell renal cell carcinoma - a retrospective study using TCGA database. Aging. 2019;11:1633–47.
Aoki M, Fujishita T. Oncogenic roles of the PI3K/AKT/mTOR axis. Curr Top Microbiol Immunol. 2017;407:153–89.
Jhanwar-Uniyal M, Wainwright JV, Mohan AL, Tobias ME, Murali R, Gandhi CD, et al. Diverse signaling mechanisms of mTOR complexes: mTORC1 and mTORC2 in forming a formidable relationship. Adv Biol Regul. 2019;72:51–62.
Ma L, Zhang R, Li D, Qiao T, Guo X. Fluoride regulates chondrocyte proliferation and autophagy via PI3K/AKT/mTOR signaling pathway. Chem-Biol Interact. 2021;349:109659.
Huang H, Weng H, Sun W, Qin X, Shi H, Wu H, et al. Recognition of RNA N(6)-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat Cell Biol. 2018;20:285–95.
Xiao Y, Thakkar KN. The m(6)A RNA demethylase FTO is a HIF-independent synthetic lethal partner with the VHL tumor suppressor. Proc Natl Acad Sci USA. 2020;117:21441–9.
Funding
The study was supported by the National Natural Science Foundation of China (31560327 and 31560325). We would like to thank all the researchers and study participants for their contributions.
Author information
Authors and Affiliations
Contributions
All authors contributed to this review with conception and design, literature review, drafting and critical revision, editing and approval of the final version.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The consent agreement was signed and approved by the Ethics Committee of The Affiliated Hospital of Zunyi Medical University. All patients involved in the present study didn’t receive chemotherapy or radiotherapy before the surgery.
Consent to publish
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Zhang, L., Luo, X. & Qiao, S. METTL14-mediated N6-methyladenosine modification of Pten mRNA inhibits tumour progression in clear-cell renal cell carcinoma. Br J Cancer 127, 30–42 (2022). https://doi.org/10.1038/s41416-022-01757-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41416-022-01757-y
This article is cited by
-
Nucleic acid and protein methylation modification in renal diseases
Acta Pharmacologica Sinica (2024)
-
The role of RNA-modifying proteins in renal cell carcinoma
Cell Death & Disease (2024)
-
RNA N6-methyladenosine modifications in urological cancers: from mechanism to application
Nature Reviews Urology (2024)
-
N6-methyladenosine methylation in kidney injury
Clinical Epigenetics (2023)
-
Role of m6A modification in regulating the PI3K/AKT signaling pathway in cancer
Journal of Translational Medicine (2023)


