Abstract
Background
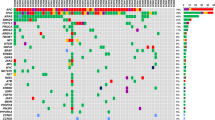
Targeted sequencing of circulating tumour DNA (ctDNA) is a promising tool to monitor dynamic changes in the variant allele frequencies (VAF) of genomic alterations and predict clinical outcomes in patients with advanced urothelial carcinoma (UC).
Methods
We performed targeted sequencing of 182 serial ctDNA samples from 53 patients with advanced UC.
Results
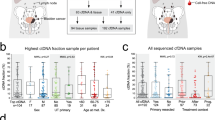
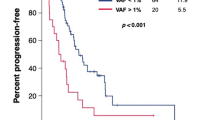
Serial ctDNA-derived metrics predicted the clinical outcomes in patients with advanced UC. Combining serial ctDNA aggregate VAF (aVAF) values with clinical factors, including age, sex, and liver metastasis, improved the performance of prognostic models. An increase of the ctDNA aVAF by ≥1 in serial ctDNA samples predicted disease progression within 6 months in 90% of patients. The majority of patients with aVAFs ≤0.7 in three consecutive ctDNA samples achieved durable clinical responses (≥6 months).
Conclusions
Serial ctDNA analysis predicts disease progression and enables dynamic monitoring to guide precision medicine in patients with advanced UC.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 24 print issues and online access
$259.00 per year
only $10.79 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
Data access and responsibility: NJV and BMF had full access to all the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis. Data sharing policy: Data are available for bona fide researchers upon request from the authors.
References
Robertson AG, Kim J, Al-Ahmadie H, Bellmunt J, Guo G, Cherniack AD, et al. Comprehensive molecular characterization of muscle-invasive bladder. Cancer Cell. 2017;171:540–556.e25.
Sailer V, Eng KW, Zhang T, Bareja R, Pisapia DJ, Sigaras A, et al. Integrative molecular analysis of patients with advanced and metastatic cancer. JCO Precis Oncol. 2019;3:1–12.
Faltas BM, Prandi D, Tagawa ST, Molina AM, Nanus DM, Sternberg C, et al. Clonal evolution of chemotherapy-resistant urothelial carcinoma. Nat Genet. 2016;48:1490–9.
Diehl F, Schmidt K, Choti MA, Romans K, Goodman S, Li M, et al. Circulating mutant DNA to assess tumor dynamics. Nat Med. 2008;14:985–90.
Razavi P, Li BT, Brown DN, Jung B, Hubbell E, Shen R, et al. High-intensity sequencing reveals the sources of plasma circulating cell-free DNA variants. Nat Med. 2019;25:1928–37.
Bettegowda C, Sausen M, Leary RJ, Kinde I, Wang Y, Agrawal N, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med. 2014;6:224ra24–224ra24.
Lanman RB, Mortimer SA, Zill OA, Sebisanovic D, Lopez R, Blau S, et al. Analytical and clinical validation of a digital sequencing panel for quantitative, highly accurate evaluation of cell-free circulating tumor DNA. PLoS ONE. 2015;10:1–27.
Agarwal N, Pal SK, Hahn AW, Nussenzveig RH, Pond GR, Gupta SV, et al. Characterization of metastatic urothelial carcinoma via comprehensive genomic profiling of circulating tumor DNA. Cancer. 2018;124:2115–24.
Birkenkamp-Demtröder K, Nordentoft I, Christensen E, Høyer S, Reinert T, Vang S, et al. Genomic alterations in liquid biopsies from patients with bladder cancer. Eur Urol. 2016;70:75–82.
Christensen E, Birkenkamp-Demtröder K, Sethi H, Shchegrova S, Salari R, Nordentoft I, et al. Early detection of metastatic relapse and monitoring of therapeutic efficacy by ultra-deep sequencing of plasma cell-free DNA in patients with urothelial bladder carcinoma. J Clin Oncol. 2019;37:1547–57.
Powles TB, Assaf ZJ, Davarpanah N, Hussain M, Oudard S, Gschwend JE, et al. Clinical outcomes in post-operative ctDNA-positive muscle-invasive urothelial carcinoma (MIUC) patients after atezolizumab adjuvant therapy. Ann Oncol. 2020;31:S1417.
Frenel JS, Carreira S, Goodall J, Roda D, Perez-Lopez R, Tunariu N, et al. Serial next-generation sequencing of circulating cell-free DNA evaluating tumor clone response to molecularly targeted drug administration. Clin Cancer Res. 2015;21:4586–96.
Patel KM, Van Der Vos KE, Smith CG, Mouliere F, Tsui D, Morris J, et al. Association of plasma and urinary mutant DNA with clinical outcomes in muscle invasive bladder cancer. Sci Rep. 2017;7:1–12.
Grivas P, Lalani A-KA, Pond GR, Nagy RJ, Faltas B, Agarwal N, et al. Circulating tumor DNA alterations in advanced urothelial carcinoma and association with clinical outcomes: a pilot study. Eur Urol Oncol. 2020;3:695–9.
Hilke FJ, Muyas F, Admard J, Kootz B, Nann D, Welz S, et al. Dynamics of cell-free tumour DNA correlate with treatment response of head and neck cancer patients receiving radiochemotherapy. Radiother Oncol. 2020;151:182–9.
Vandekerkhove G, Lavoie J, Annala M, Murtha AJ, Sundahl N, Walz S, et al. Plasma ctDNA is a tumor tissue surrogate and enables clinical-genomic stratification of metastatic bladder cancer. Nat Commun. 2021;12:184.
Powles T, Carroll D, Chowdhury S, Gravis G, Joly F, Carles J, et al. An adaptive, biomarker-directed platform study of durvalumab in combination with targeted therapies in advanced urothelial cancer. Nat Med. 2021;27:793–801.
Raja R, Kuziora M, Brohawn PZ, Higgs BW, Gupta A, Dennis PA, et al. Early reduction in ctDNA predicts survival in patients with lung and bladder cancer treated with durvalumab. Clin Cancer Res. 2018;24:6212–22.
Sundahl N, Vandekerkhove G, Decaestecker K, Meireson A, De Visschere P, Fonteyne V, et al. Randomized phase 1 trial of pembrolizumab with sequential versus concomitant stereotactic body radiotherapy in metastatic urothelial carcinoma. Eur Urol. 2019;75:707–11.
Powles T, Assaf ZJ, Davarpanah N, Banchereau R, Szabados BE, Yuen KC, et al. ctDNA guiding adjuvant immunotherapy in urothelial carcinoma. Nature. 2021;595:432–7.
Odegaard JI, Vincent JJ, Mortimer S, Vowles JV, Ulrich BC, Banks KC, et al. Validation of a plasma-based comprehensive cancer genotyping assay utilizing orthogonal tissue- and plasma-based methodologies. Clin Cancer Res. 2018;24:3539–49.
Vosoughi A, Zhang T, Shohdy KS, Vlachostergios PJ, Wilkes DC, Bhinder B, et al. Common germline-somatic variant interactions in advanced urothelial cancer. Nat Commun. 2020;11:6195.
Rhodes DR, Kalyana-Sundaram S, Mahavisno V, Varambally R, Yu J, Briggs BB, et al. Oncomine 3.0: Genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia. 2007;9:166–80.
Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, et al. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6:1–6.
Huang L, Fernandes H, Zia H, Tavassoli P, Rennert H, Pisapia D, et al. The cancer precision medicine knowledge base for structured clinical-grade mutations and interpretations. J Am Med Inform Assoc. 2017;24:513–9.
Chakravarty D, Gao J, Phillips S, Kundra R, Zhang H, Wang J, et al. OncoKB: a precision oncology knowledge base. JCO Precis Oncol. 2017;1:1–16.
Iyer G, Hanrahan AJ, Milowsky MI, Al-Ahmadie H, Scott SN, Janakiraman M, et al. Genome sequencing identifies a basis for everolimus sensitivity. Science 2012;338:221–221.
Loriot Y, Necchi A, Park SH, Garcia-Donas J, Huddart R, Burgess E, et al. Erdafitinib in locally advanced or metastatic urothelial carcinoma. N. Engl J Med. 2019;381:338–48.
Findlay GM, Daza RM, Martin B, Zhang MD, Leith AP, Gasperini M, et al. Accurate classification of BRCA1 variants with saturation genome editing. Nature. 2018;562:217–22.
Bick AG, Weinstock JS, Nandakumar SK, Fulco CP, Bao EL, Zekavat SM, et al. Inherited causes of clonal haematopoiesis in 97,691 whole genomes. Nature. 2020;586:763–8.
Nassar AH, Mouw KW, Jegede O, Shinagare AB, Kim J, Liu C-J, et al. A model combining clinical and genomic factors to predict response to PD-1/PD-L1 blockade in advanced urothelial carcinoma. Br J Cancer. 2020;122:555–63.
Khaki AR, Li A, Diamantopoulos LN, Miller NJ, Carril-Ajuria L, Castellano D, et al. A new prognostic model in patients with advanced urothelial carcinoma treated with first-line immune checkpoint inhibitors. Eur Urol Oncol. 2021;4:464–72.
Shen J, Ju Z, Zhao W, Wang L, Peng Y, Ge Z, et al. ARID1A deficiency promotes mutability and potentiates therapeutic antitumor immunity unleashed by immune checkpoint blockade. Nat Med. 2018;24:556–62.
Lee HW, Sa JK, Gualberto A, Scholz C, Sung HH, Jeong BC, et al. A phase II trial of tipifarnib for patients with previously treated, metastatic urothelial carcinoma harboring hras mutations. Clin Cancer Res. 2020;26:5113–9.
Bajorin DF, Dodd PM, Mazumdar M, Fazzari M, McCaffrey JA, Scher HI, et al. Long-term survival in metastatic transitional-cell carcinoma and prognostic factors predicting outcome of therapy. J Clin Oncol. 1999;17:3173–81.
Apolo AB, Ostrovnaya I, Halabi S, Iasonos A, Philips GK, Rosenberg JE, et al. Prognostic model for predicting survival of patients with metastatic urothelial cancer treated with cisplatin-based chemotherapy. J Natl Cancer Inst. 2013;105:499–503.
Bratman SV, Yang SYC, Iafolla MAJ, Liu Z, Hansen AR, Bedard PL, et al. Personalized circulating tumor DNA analysis as a predictive biomarker in solid tumor patients treated with pembrolizumab. Nat Cancer. 2020;1:873–81.
Birkenkamp-Demtröder K, Christensen E, Nordentoft I, Knudsen M, Taber A, Høyer S, et al. Monitoring treatment response and metastatic relapse in advanced bladder cancer by liquid biopsy analysis. Eur Urol. 2018;73:535–40.
Avanzini S, Kurtz DM, Chabon JJ, Moding EJ, Hori SS, Gambhir SS, et al. A mathematical model of ctDNA shedding predicts tumor detection size. Sci Adv. 2020;6:1–10.
Acknowledgements
BMF was supported by the Department of Defense CDMRP grant (CA160212), a STARR Cancer Consortium grant (I14-0047), and the Gellert Family-John P. Leonard, MD Research Scholarship in Hematology and Medical Oncology. This work was also supported by a Conquer Cancer Foundation Long Term International Fellowship Award (KSS) and the Englander Institute for Precision Medicine at WCM (OE, BMF). Guardant performed sequencing and initial mutational testing of ctDNA samples. We thank Duy Nguyen for his assistance with editing the text.
Funding
Not applicable.
Author information
Authors and Affiliations
Contributions
Initiation and design of the study: KSS, KSP, RN, PG, NJV and BMF. Subject enrollment, sample, and clinical data collection: KSS, DV, YC, JT, STT, AMM, CNS, DMN, JMM, OE, GPS, PV, NJV and BMF. Sample sequencing: KSP and RN. Statistical, and bioinformatic analyses: KSS and OE. Supervision of research: NV and BMF. Writing of the first draft of the manuscript: KSS, OE, GPV, PG, NV and BMF. All authors contributed to the writing and editing of the revised manuscript and approved the manuscript.
Corresponding authors
Ethics declarations
Competing interests
KSS, DMV, YC and JT: No competing interests. AMM: Honoraria—ASCO, Consulting or advisory role—EISAi; Exelixis; Janssen. KSP and RN: Employment—Guardant Health Inc. CNS: Consulting or advisory role—Astellas Pharma; AstraZeneca; Bayer; Genzyme; Immunomedics; Incyte; Medscape; Merck; MSD; Pfizer; Roche; UroToday. STT: Consulting or advisory role—Abbvie; Amgen; Astellas Pharma; Bayer; Clovis Oncology; Dendreon; Endocyte; Genentech; Immunomedics; Janssen; Karyopharm Therapeutics; Medivation; Pfizer; QED Therapeutics; Sanofi; Tolmar, Research funding—Abbvie (Inst); Amgen (Inst); Astellas Pharma (Inst); AstraZeneca (Inst); AVEO (Inst); Bayer (Inst); Boehringer Ingelheim (Inst); Bristol-Myers Squibb (Inst); Clovis Oncology (Inst); Dendreon (Inst); Endocyte (Inst); Exelixis (Inst); Genentech (Inst); Immunomedics (Inst); Inovio Pharmaceuticals (Inst); Janssen (Inst); Karyopharm Therapeutics (Inst); Lilly (Inst); Medivation (Inst); Merck (Inst); Millennium (Inst); Newlink Genetics (Inst); Novartis (Inst); Progenics (Inst); Rexahn Pharmaceuticals (Inst); Sanofi (Inst); Stem CentRx (Inst), Travel, Accommodations, expenses—Amgen; Immunomedics; Sanofi. DMN: Consulting or advisory role—Roche/Genentech, Research funding—Boehringer Ingelheim (Inst); Novartis (Inst); Zenith Epigenetics (Inst). JMM: Research funding—Personal genome diagnostics, Travel, accommodations, expenses—Personal genome diagnostics. OE: Stock and other ownership interests—OneThree Biotech; Owkin; Volastra Therapeutics. GPS: Honoraria—UpToDate, Consulting or advisory role—Astellas Pharma; AstraZeneca; Bicycle Therapeutics; Bristol-Myers Squibb; Eisai; EMD Serono; Exelixis; Genentech; Janssen; Merck; Pfizer; Seattle Genetics, Speakers’ Bureau - Medscape; Onclive; Physicans’ Education Resource; Research to practice, research funding—AstraZeneca (Inst); Janssen (Inst); Sanofi (Inst), Travel, Accommodations, expenses—Bristol-Myers Squibb, Other relationship—Astellas Pharma; AstraZeneca; Bavarian Nordic; Bristol-Myers Squibb; Debiopharm Group; Elsevier; QED Therapeutics. PG: Consulting or advisory role—AstraZeneca; Bayer; Bristol-Myers Squibb; Clovis Oncology; Driver, Inc; EMD Serono; Exelixis; Foundation Medicine; Genzyme; GlaxoSmithKline; HERON; Janssen; Merck; Mirati Therapeutics; Pfizer; QED Therapeutics; Roche; Seattle Genetics, Research funding—Bavarian Nordic (Inst); Bristol-Myers Squibb (Inst); Clovis Oncology (Inst); Debiopharm Group (Inst); Immunomedics (Inst); Pfizer (Inst). NJV: Employment—US Oncology, Stock and Other Ownership interests—Caris Life Sciences, Honoraria—Novartis; Pfizer; UpToDate, Consulting or Advisory role—Astellas Pharma, AstraZeneca; Bayer; Boehringer Ingelheim; Caris Life Sciences; Clovis Oncology; Corvus Pharmaceuticals; Eisai; Genentech/Roche; Janssen Oncology; Merck; Modra Pharmaceuticals; Pfizer; Tolero Pharmaceuticals, Speakers’ Bureau—Bayer; Bristol-Myers Squibb; Clovis Oncology; Genentech/Roche; Sanofi; Seattle Genetics/Astellas, Research funding—Endocyte (Inst); Merck (Inst); Suzhou Kintor Pharmaceuticals (Inst); US Oncology (Inst), Expert testimony—Novartis, Travel, accommodations, expenses—AstraZeneca/MedImmune; Bayer/Onyx; Exelixis; Genentech/Roche; Pfizer; Sanofi/Aventis; US Oncology. BMF: Honoraria—Digital Science Press, Consulting or advisory role—QED therapeutics, Immunomedics, Merck, Seattle Genetics. Patent royalties Immunomedics/Gilead Research funding—Eli Lilly.
Ethics approval and consent to participate
The study was approved by the Western Institutional Review Board (Protocol No. 20152817).
Consent to publish
Not applicable.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Shohdy, K.S., Villamar, D.M., Cao, Y. et al. Serial ctDNA analysis predicts clinical progression in patients with advanced urothelial carcinoma. Br J Cancer 126, 430–439 (2022). https://doi.org/10.1038/s41416-021-01648-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41416-021-01648-8
This article is cited by
-
Blood-based liquid biopsy: insights into early detection, prediction, and treatment monitoring of bladder cancer
Cellular & Molecular Biology Letters (2023)
-
Circulating and urinary tumour DNA in urothelial carcinoma — upper tract, lower tract and metastatic disease
Nature Reviews Urology (2023)
-
Minimal residual disease in solid tumors: an overview
Frontiers of Medicine (2023)
-
Patient-specific targeted analysis of circulating tumour DNA in plasma is feasible and may be a potential biomarker in UTUC
World Journal of Urology (2023)