Abstract
Background
The mechanisms underlying metastasis of colorectal cancer (CRC) remain unclear. C14orf159 is a mitochondrial matrix protein converting d-glutamate to 5-oxo-d-proline. Other metabolic functions of C14orf159, especially on mitochondrial metabolism, and its contribution to CRC metastasis, are not elucidated.
Methods
Metabolome analysis by gas chromatography-mass spectrometry, RNA-sequencing analysis, flow cytometry, migration and invasion assay, sphere-formation assay using C14orf159-knockout and -stable expressing cells, immunohistochemistry of C14orf159 in human CRC specimens, and xenograft experiments using Balb/c nude mice were conducted.
Results
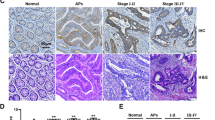
C14orf159 maintained the mitochondrial membrane potential of human CRC cells, and its involvement in amino acid and glutathione metabolism was demonstrated. In human CRC specimens, a decrease in C14orf159 expression at the invasive front of the tumour and in metastasis was determined. C14orf159 was also shown to attenuate the migration, invasion, and spheroid growth of CRC cells in vitro and colorectal tumour growth and metastasis in vivo. Mechanistically, C14orf159 reduced the expression of genes involved in CRC metastasis, including members of the Wnt and MMP family, by maintaining the mitochondrial membrane potential.
Conclusions
Our findings link mitochondrial membrane potential to Wnt/β-catenin signalling and reveal a previously unrecognised function of the mitochondrial matrix protein C14orf159 as a suppressor of CRC metastasis.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 24 print issues and online access
$259.00 per year
only $10.79 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The data that support the findings of this study are available from the corresponding authors upon reasonable request. RNA-sequence data that support the findings of this study have been deposited in GEO with the accession code GSE167947. The previously published data of colorectal cancer cells that have been reanalysed here are available in GEO with the accession code GSE131418 [37]. Data in Fig. 2a are publicly available online at www.oncomine.org [36].
References
Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–49, https://doi.org/10.3322/caac.21660.
Biller LH, Schrag D. Diagnosis and treatment of metastatic colorectal cancer: a review. J Am Med Assoc. 2021;325:669–85.
Riihimaki M, Hemminki A, Sundquist J, Hemminki K. Patterns of metastasis in colon and rectal cancer. Sci Rep. 2016;6:1–9.
Lord AC, Knijn N, Brown G, Nagtegaal ID. Pathways of spread in rectal cancer: a reappraisal of the true routes to distant metastatic disease. Eur J Cancer. 2020;128:1–6.
Langley RR, Fidler IJ. The seed and soil hypothesis revisited-The role of tumor-stroma interactions in metastasis to different organs. Int J Cancer. 2011;128:2527–35.
Tauriello DVF, Calon A, Lonardo E, Batlle E. Determinants of metastatic competency in colorectal cancer. Mol Oncol. 2017;11:97–119.
Faubert B, Solmonson A, DeBerardinis RJ. Metabolic reprogramming and cancer progression. Science. 2020;368:eaaw5473.
Vander Heiden MG, DeBerardinis RJ. Understanding the intersections between metabolism and cancer biology. Cell. 2017;168:657–69.
Schild T, Low V, Blenis J, Gomes AP. Unique metabolic adaptations dictate distal organ-specific metastatic colonization. Cancer Cell. 2018;33:347–54.
Ohshima K, Morii E. Metabolic reprogramming of cancer cells during tumor progression and metastasis. Metabolites. 2021;11:1–23.
Elia I, Doglioni G, Fendt SM. Metabolic hallmarks of metastasis formation. Trends Cell Biol. 2018;28:673–84.
Knott SRV, Wagenblast E, Khan S, Kim SY, Soto M, Wagner M, et al. Asparagine bioavailability governs metastasis in a model of breast cancer. Nature. 2018;554:378–81.
Sullivan MR, Mattaini KR, Dennstedt EA, Nguyen AA, Sivanand S, Reilly MF, et al. Increased serine synthesis provides an advantage for tumors arising in tissues where serine levels are limiting. Cell Metab. 2019;29:1410–21.
Mcdonald OG, Li X, Saunders T, Tryggvadottir R, Mentch SJ, Warmoes MO, et al. Epigenomic reprogramming during pancreatic cancer progression links anabolic glucose metabolism to distant metastasis. Nat Genet. 2017;49:367–76.
Ubellacker JM, Tasdogan A, Ramesh V, Shen B, Mitchell EC, Martin-Sandoval MS, et al. Lymph protects metastasizing melanoma cells from ferroptosis. Nature. 2020;585:113–8.
Bu P, Chen KY, Xiang K, Johnson C, Crown SB, Rakhilin N, et al. Aldolase B-mediated fructose metabolism drives metabolic reprogramming of colon cancer liver metastasis. Cell Metab. 2018;27:1249–62.
Denisenko TV, Gorbunova AS, Zhivotovsky B. Mitochondrial involvement in migration, invasion and metastasis. Front Cell Dev Biol. 2019;7, https://doi.org/10.3389/fcell.2019.00355.
Chuang CH, Dorsch M, Dujardin P, Silas S, Ueffing K, Holken JM, et al. Altered mitochondria functionality defines a metastatic cell state in lung cancer and creates an exploitable vulnerability. Cancer Res. 2021;81:567–579.
Zong WX, Rabinowitz JD, White E. Mitochondria and cancer. Mol Cell. 2016;61:667–76.
Tomlinson IPM, Alam NA, Rowan AJ, Barclay E, Jaeger EEM, Kelsell D, et al. Germline mutations in FH predispose to dominantly inherited uterine fibroids, skin leiomyomata and papillary renal cell cancer the multiple leiomyoma consortium. Nat Genet. 2002;30:406–10.
Letouzé E, Martinelli C, Loriot C, Burnichon N, Abermil N, Ottolenghi C, et al. SDH mutations establish a hypermethylator phenotype in paraganglioma. Cancer Cell. 2013;23:739–52.
Killian JK, Kim SY, Miettinen M, Smith C, Merino M, Tsokos M, et al. Succinate dehydrogenase mutation underlies global epigenomic divergence in gastrointestinal stromal tumor. Cancer Discov. 2013;3:648–57.
Yan H, Parsons DW, Jin G, McLendon R, Rasheed BA, Yuan W, et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360:765–73.
Kosmider O, Gelsi-Boyer V, Slama L, Dreyfus F, Beyne-Rauzy O, Quesnel B, et al. Mutations of IDH1 and IDH2 genes in early and accelerated phases of myelodysplastic syndromes and MDS/myeloproliferative neoplasms. Leukemia. 2010;24:1094–6.
Losman JA, Kaelin WG. What a difference a hydroxyl makes: mutant IDH, (R)-2-hydroxyglutarate, and cancer. Genes Dev. 2013;27:836–52.
Newman AC, Maddocks ODK. One-carbon metabolism in cancer. Br J Cancer. 2017;116:1499–504.
Yang L, Venneti S, Nagrath D. Glutaminolysis: a hallmark of cancer metabolism. Annu Rev Biomed Eng. 2017;19:163–94.
Ariyoshi M, Katane M, Hamase K, Miyoshi Y, Nakane M, Hoshino A, et al. D-Glutamate is metabolized in the heart mitochondria. Sci Rep. 2017;7:43911.
Ohshima K, Nojima S, Tahara S, Kurashige M, Kawasaki K, Hori Y, et al. Serine racemase enhances growth of colorectal cancer by producing pyruvate from serine. Nat Metab. 2020;2:81–96.
Zhu YM, Li Q, Gao XZ, Meng X, Sun LL, Shi Y, et al. C14orf159 suppresses gastric cancer cells’ invasion and proliferation by inactivating ERK signaling. Cancer Manag Res. 2019;11:1717–23.
Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–5.
Ohshima K, Nojima S, Tahara S, Kurashige M, Hori Y, Hagiwara K, et al. Argininosuccinate synthase 1-deficiency enhances the cell sensitivity to arginine through decreased DEPTOR expression in endometrial cancer. Sci Rep. 2017;7:45504.
Spandidos A, Wang X, Wang H, Seed B. PrimerBank: a resource of human and mouse PCR primer pairs for gene expression detection and quantification. Nucleic Acids Res. 2009;38:D792–D799.
Irino Y, Toh R, Nagao M, Mori T, Honjo T, Shinohara M, et al. 2-Aminobutyric acid modulates glutathione homeostasis in the myocardium. Sci Rep. 2016;6:36749.
Lai Z, Tsugawa H, Wohlgemuth G, Mehta S, Mueller M, Zheng Y, et al. Identifying metabolites by integrating metabolome databases with mass spectrometry cheminformatics. Nat Methods. 2018;15:53–56.
Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, et al. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia 2004;6:1–6.
Kamal Y, Schmit SL, Hoehn HJ, Amos CI, Frost HR. Transcriptomic differences between primary colorectal adenocarcinomas and distant metastases reveal metastatic colorectal cancer subtypes. Cancer Res. 2019;79:4227–41.
Hughes CS, Postovit LM, Lajoie GA. Matrigel: a complex protein mixture required for optimal growth of cell culture. Proteomics. 2010;10:1886–90.
Bafico A, Liu G, Goldin L, Harris V, Aaronson SA. An autocrine mechanism for constitutive Wnt pathway activation in human cancer cells. Cancer Cell. 2004;6:497–506.
Voloshanenko O, Erdmann G, Dubash TD, Augustin I, Metzig M, Moffa G, et al. Wnt secretion is required to maintain high levels of Wnt activity in colon cancer cells. Nat Commun. 2013;4:2610.
Wang X, Moon J, Dodge ME, Pan X, Zhang L, Hanson JM, et al. The development of highly potent inhibitors for porcupine. J Med Chem. 2013;56:2700–4.
David Sanin AE, Matsushita M, Klein Geltink RI, Ching-Cheng Huang S, Pearce EL, Pearce EJ, et al. Mitochondrial membrane potential regulates nuclear gene expression in macrophages exposed to prostaglandin E2. Immunity. 2018;49:1021–.e6.
Kurniawan H, Franchina DG, Guerra L, Bonetti L, Soriano Baguet L, Grusdat M, et al. Glutathione restricts serine metabolism to preserve regulatory T cell function. Cell Metab. 2020;31:920–.e7.
Chandel NS. Evolution of mitochondria as signaling organelles. Cell Metab. 2015;22:204–6.
Kim J, DeBerardinis RJ. Mechanisms and implications of metabolic heterogeneity in cancer. Cell Metab. 2019;30:434–46.
Tasdogan A, Faubert B, Ramesh V, Ubellacker JM, Shen B, Solmonson A, et al. Metabolic heterogeneity confers differences in melanoma metastatic potential. Nature. 2020;577:115–20.
Rossi M, Doglioni G, Bornes L, Broekaert D, Planque M, Fernández-García J, et al. Heterogeneity in PHGDH protein expression potentiates cancer cell dissemination and metastasis. bioRxiv [Preprint]. [cited 2021 Jan 26]. Available from: https://doi.org/10.1101/2021.01.24.427949.
Momcilovic M, Jones A, Bailey ST, Waldmann CM, Li R, Lee JT, et al. In vivo imaging of mitochondrial membrane potential in non-small-cell lung cancer. Nature. 2019;575:380–4.
Dekker E, Tanis PJ, Vleugels JLA, Kasi PM, Wallace MB. Colorectal cancer. Lancet. 2019;394:1467–80.
Massagué J, Obenauf A. Metastatic colonization by circulating tumour cells. Nature. 2016;529:298–306.
Massagué J, Obenauf A. Surviving at a distance: organ-specific Metastasis. Trends Cancer. 2015;1:76–91.
Acknowledgements
We thank Mr. Masaharu Kohara, Ms. Etsuko Maeno and Ms. Takako Sawamura from the Department of Pathology, Osaka University Graduate School of Medicine, and this study was supported by Eiji Oiki, Tomoaki Mizuno and Center for Medical Research and Education, Osaka University Graduate School of Medicine.
Funding
This work was supported by JSPS KAKENHI Grant Number 20K16220 (KO), 19H03452 (EM) and the Takeda Science Foundation (KO).
Author information
Authors and Affiliations
Contributions
KO carried out most of the experiments, analysed data and managed clinical information. RO performed immunohistochemistry of C14orf159 in human colorectal adenocarcinoma specimens. DO performed RNA-seq and analysed the data. MS performed metabolome analysis by GC–MS. SN double-checked the tissue samples. KO designed the project and wrote the manuscript, and EM checked the manuscript and supervised the project.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
All experimental protocols involving human specimens were approved by the Ethical Review Board of the Graduate School of Medicine, Osaka University (No. 15234-2), and were performed in accordance with the committee guidelines and regulations. All patients provided informed consent. The study was performed in accordance with the Declaration of Helsinki. The protocols used for all animal experiments in this study were approved by the Animal Research Committee of Osaka University, Japan (No. 30050002).
Consent to publish
Not applicable.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Ohshima, K., Oi, R., Okuzaki, D. et al. Mitochondrial matrix protein C14orf159 attenuates colorectal cancer metastasis by suppressing Wnt/β-catenin signalling. Br J Cancer 125, 1699–1711 (2021). https://doi.org/10.1038/s41416-021-01582-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41416-021-01582-9