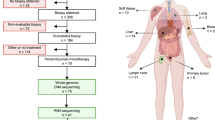
Abstract
Background
FGFR3-altered urothelial cancer (UC) correlates with a non-T cell-inflamed phenotype and has therefore been postulated to be less responsive to immune checkpoint blockade (ICB). Preclinical work suggests FGFR3 signalling may suppress pathways such as interferon signalling that alter immune microenvironment composition. However, correlative studies examining clinical trials have been conflicting as to whether FGFR altered tumours have equivalent response and survival to ICB in patients with metastatic UC. These findings have yet to be validated in real world data, therefore we evaluated clinical outcomes of patients with FGFR3-altered metastatic UC treated with ICB and investigate the underlying immunogenomic mechanisms of response and resistance.
Methods
103 patients with metastatic UC treated with ICB at a single academic medical center from 2014 to 2018 were identified. Clinical annotation for demographics and cancer outcomes, as well as somatic DNA and RNA sequencing, were performed. Objective response rate to ICB, progression-free survival, and overall survival was compared between patients with FGFR3-alterations and those without. RNA expression, including molecular subtyping and T cell receptor clonality, was also compared between FGFR3-altered and non-altered patients.
Results
Our findings from this dataset confirm that FGFR3-altered (n = 17) and wild type (n = 86) bladder cancers are equally responsive to ICB (12 vs 19%, p = 0.73). Moreover, we demonstrate that despite being less inflamed, FGFR3-altered tumours have equivalent T cell receptor (TCR) diversity and that the balance of a CD8 T cell gene expression signature to immune suppressive features is an important determinant of ICB response.
Conclusions
Our work in a real world dataset validates prior observations from clinical trials but also extends this prior work to demonstrate that FGFR3-altered and wild type tumours have equivalent TCR diversity and that the balance of effector T cell to immune suppression signals are an important determinant of ICB response.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 24 print issues and online access
$259.00 per year
only $10.79 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
DNA sequencing data are available upon request. RNAseq data have been deposited to Gene Expression Omnibus under accession ID GSE176307.
References
Rosenberg JE, O’Donnell PH, Balar AV, McGregor BA, Heath EI, Yu EY, et al. Pivotal trial of enfortumab vedotin in urothelial carcinoma after platinum and anti-programmed death 1/programmed death ligand 1 therapy. J Clin Oncol. 2019;37:2592–600.
Loriot Y, Necchi A, Park SH, Garcia-Donas J, Huddart R, Burgess E, et al. Erdafitinib in locally advanced or metastatic urothelial carcinoma. N Engl J Med. 2019;381:338–48.
Balar AV, Galsky MD, Rosenberg JE, Powles T, Petrylak DP, Bellmunt J, et al. Atezolizumab as first-line treatment in cisplatin-ineligible patients with locally advanced and metastatic urothelial carcinoma: a single-arm, multicentre, phase 2 trial. Lancet. 2017;389:67–76.
Rosenberg JE, Hoffman-Censits J, Powles T, Heijden MS van der, Balar AV, Necchi A, et al. Atezolizumab in patients with locally advanced and metastatic urothelial carcinoma who have progressed following treatment with platinum-based chemotherapy: a single-arm, multicentre, phase 2 trial. Lancet. 2016;387:1909–20.
Sharma P, Retz M, Siefker-Radtke A, Baron A, Necchi A, Bedke J, et al. Nivolumab in metastatic urothelial carcinoma after platinum therapy (CheckMate 275): a multicentre, single-arm, phase 2. Lancet Oncol. 2017;18:312–22.
Bellmunt J, Wit R, de, Vaughn DJ, Fradet Y, Lee J-L, Fong L, et al. Pembrolizumab as second-line therapy for advanced urothelial carcinoma. N Engl J Med. 2017;376:1015–26.
Apolo AB, Infante JR, Balmanoukian A, Patel MR, Wang D, Kelly K, et al. Avelumab, an anti-programmed death-ligand 1 antibody, in patients with refractory metastatic urothelial carcinoma: results from a multicenter, phase Ib study. J Clin Oncol. 2017;35;2117–24.
Helsten T, Elkin S, Arthur E, Tomson BN, Carter J, Kurzrock R.The FGFR landscape in cancer: analysis of 4,853 tumors by next-generation Sequencing.Clin Cancer Res. 2016;22:259–67.
Robertson AG, Kim J, Al-Ahmadie H, Bellmunt J, Guo G, Cherniack AD, et al. Comprehensive molecular characterization of muscle-invasive bladder. Cancer Cell. 2017;171:540–556.e25.
Robinson BD, Vlachostergios PJ, Bhinder B, Liu W, Li K, Moss TJ, et al. Upper tract urothelial carcinoma has a luminal-papillary T-cell depleted contexture and activated FGFR3 signaling. Nat Commun. 2019;10:2977.
Audenet F, Isharwal S, Cha EK, Donoghue MTA, Drill EN, Ostrovnaya I, et al. Clonal relatedness and mutational differences between upper tract and bladder urothelial carcinoma. Clin Cancer Res. 2019;25:967–76.
Sfakianos JP, Cha EK, Iyer G, Scott SN, Zabor EC, Shah RH, et al. Genomic characterization of upper tract urothelial carcinoma. Eur Urol. 2015;68:970–7.
Kardos J, Chai S, Mose LE, Selitsky SR, Krishnan B, Saito R, et al. Claudin-low bladder tumors are immune infiltrated and actively immune suppressed. JCI insight. 2016;1:e85902.
Kamoun A, Reyniès A de, Allory Y, Sjödahl G, Robertson AG, Seiler R, et al. A consensus molecular classification of muscle-invasive bladder cancer. Eur Urol. 2019;:420–33.
Saito R, Smith CC, Utsumi T, Bixby LM, Kardos J, Wobker SE, et al. Molecular subtype-specific immunocompetent models of high-grade urothelial carcinoma reveal differential neoantigen expression and response to immunotherapy. Cancer Res. 2018;78:3954–68. 15
Wang L, Gong Y, Saci A, Szabo PM, Martini A, Necchi A, et al. Fibroblast growth factor receptor 3 alterations and response to PD-1/PD-L1 blockade in patients with metastatic urothelial cancer. Eur Urol. 2019;76:599–603.
Necchi A, Anichini A, Raggi D, Briganti A, Massa S, Lucianò R, et al. Pembrolizumab as neoadjuvant therapy before radical cystectomy in patients with muscle-invasive urothelial bladder carcinoma (PURE-01): an open-label, single-arm, phase II study. J Clin Oncol. 2018;36:3353–60.
Necchi A, Raggi D, Giannatempo P, Marandino L, Farè E, Gallina A, et al. Can patients with muscle-invasive bladder cancer and fibroblast growth factor receptor-3 alterations still be considered for neoadjuvant pembrolizumab? A comprehensive assessment from the updated results of the PURE-01 study. Euro Urol Oncol. 2020; https://doi.org/10.1016/j.euo.2020.04.005.
Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. 1
Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinforma. 2011;12:323.
Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550.
Damrauer JS, Hoadley KA, Chism DD, Fan C, Tiganelli CJ, Wobker SE, et al. Intrinsic subtypes of high-grade bladder cancer reflect the hallmarks of breast cancer biology. Proc Natl Acad. Sci USA 2014;111:3110–5.
Network CGAR. Comprehensive molecular characterization of urothelial bladder carcinoma. Nature. 2014;507:315–22. 20
Dabney AR. Classification of microarrays to nearest centroids. Bioinformatics. 2005;21:4148–54.
Bolotin DA, Poslavsky S, Mitrophanov I, Shugay M, Mamedov IZ, Putintseva EV, et al. MiXCR: software for comprehensive adaptive immunity profiling. Nat Methods. 2015;12:380–1.
Lefranc M-P. IMGT, the International ImMunoGeneTics Information System. Cold Spring Harb Protoc. 2011;2011:595–603.
Michalik L, Desvergne B, Wahli W. Peroxisome-proliferator-activated receptors and cancers: complex stories. Nat. Rev. Cancer 2004;4:61–70.
Rebouissou S, Bernard-Pierrot I, Reyniès A, de, Lepage M-L, Krucker C, Chapeaublanc E, et al. EGFR as a potential therapeutic target for a subset of muscle-invasive bladder cancers presenting a basal-like phenotype. Sci Transl Med. 2014;6:244ra91–244ra91.
Sjödahl G, Lauss M, Lövgren K, Chebil G, Gudjonsson S, Veerla S, et al. A molecular taxonomy for urothelial carcinoma. Clin Cancer Res. 2012;18:3377–86.
Volkmer J-P, Sahoo D, Chin RK, Ho PL, Tang C, Kurtova AV, et al. Three differentiation states risk-stratify bladder cancer into distinct subtypes. Proc Natl Acad Sci USA 2012;109:2078–83.
Choi W, Porten S, Kim S, Willis D, Plimack ER, Hoffman-Censits J, et al. Identification of distinct basal and luminal subtypes of muscle-invasive bladder cancer with different sensitivities to frontline chemotherapy. Cancer Cell. 2014;25:152–65.
Wang L, Saci A, Szabo PM, Chasalow SD, Castillo-Martin M, Domingo-Domenech J, et al. EMT- and stroma-related gene expression and resistance to PD-1 blockade in urothelial cancer. Nat Commun. 2018;9:3503.
Mariathasan S, Turley SJ, Nickles D, Castiglioni A, Yuen K, Wang Y, et al. TGFβ attenuates tumour response to PD-L1 blockade by contributing to exclusion of T cells. Nature 2018;554:544–8.
Sweis RF, Spranger S, Bao R, Paner GP, Stadler WM, Steinberg G, et al. Molecular drivers of the non-t-cell-inflamed tumor microenvironment in urothelial bladder. Cancer Cancer Immunol. Res. 2016;4:563–8.
Necchi A, Raggi D, Gallina A, Madison R, Colecchia M, Lucianò R, et al. Updated results of PURE-01 with preliminary activity of neoadjuvant pembrolizumab in patients with muscle-invasive bladder carcinoma with variant histologies. Euro Urol. 2020;77:439–46.
Palakurthi S, Kuraguchi M, Zacharek SJ, Zudaire E, Huang W, Bonal DM, et al. The Combined Effect of FGFR Inhibition and PD-1 Blockade Promotes Tumor-Intrinsic Induction of Antitumor Immunity. Cancer Immunol. Res. 2019;7:1457–71.
Rochel N, Krucker C, Coutos-Thévenot L, Osz J, Zhang R, Guyon E, et al. Recurrent activating mutations of PPARγ associated with luminal bladder tumors. Nat Commun. 2019;10:253.
Korpal M, Puyang X, Wu ZJ, Seiler R, Furman C, Oo HZ, et al. Evasion of immunosurveillance by genomic alterations of PPARγ/RXRα in bladder cancer. Nat Commun. 2017;8:103.
Acknowledgements
We acknowledge the members of the Kim Lab for useful discussions, the UNC translational pathology laboratory (TPL) and the UNC Translational Genomics Facility (TGL) for their technical assistance.
Funding
This work was supported by the University Cancer Research Fund (UCRF) and NCI grant R01-CA241810 [WYK], a research collaboration agreement between Janssen Research & Development, LLC and GeneCentric Therapeutics, Inc. and a sponsored research agreement between GeneCentric Therapeutics, Inc and the University of North Carolina at Chapel Hill. TLR is supported by the National Cancer Institute K12 Career Development Award in Clinical Oncology (grant 5K12CA120780) and K08CA248967, as well as the Doris Duke Charitable Foundation (grant number 2015213).
Author information
Authors and Affiliations
Contributions
Tracy L Rose: Conceptualisation, methodology, validation, formal analysis, data curation, writing—original draft, writing—review and editing, visualisation, project administration. William H Weir: Formal analysis, data curation, writing—original draft, writing—review and editing, visualisation. Greg M Mayhew: Methodology, formal analysis, data curation, writing—review and editing. Yoichiro Shibata: Software, validation, formal analysis, data curation, writing—original draft. Patrick Eulitt: Data curation. Josh M Uronis: Project administration, Mi Zhou: Data curation, writing—original draft. Matthew Nielsen: Resources, writing— review and editing. Angela Smith: Writing—review and editing. Michael Woods: Writing—review and editing. Michele C Hayward: Data curation, project administration, writing—review and editing. Ashley H Salazar: Data curation. Matthew I Milowsky: Writing—review and editing. Sara E Wobker: Formal analysis, data curation. Katrina McGinty: Formal analysis, data curation. Michael V Millburn: Conceptualisation, supervision. Joel R Eisner: Writing—original draft, supervision, writing—review and editing, supervision. William Y Kim: Conceptualisation, methodology, validation, writing—original draft, writing—review and editing, supervision. Tracy L Rose was placed first in the co-first authorship spot because she conceived the project. William H. Weir was placed as second in the co-first authorship because he made substantial contributions to computational biology analysis, figure generation, and intellectual insights.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
This study was reviewed and approved by the Institutional Review Board at UNC with an approved IRB 18-1478. All patient data were de-identified at the time of data abstraction from the electronic medical record. The study was performed in accordance with the Declaration of Helsinki.
Consent to publish
Not applicable. No individual patient data are included in the publication.
Competing interests
This work was funded in part through a research collaboration agreement between Janssen Research & Development, LLC and GeneCentric Therapeutics, Inc. and a sponsored research agreement between GeneCentric Therapeutics, Inc and the University of North Carolina at Chapel Hill. TLR is supported by the Doris Duke Charitable Foundation (grant number 2015213) and the National Cancer Institute of the National Institutes of Health (1K08CA248967-01 Clinical Investigator Award). WYK and TLR receive research funding from GeneCentric Therapeutics and Merck. TLR receives research funding from Genentech/Hoffman-La Roche and Bristol-Myers Squibb. GMM, YS, MVM, JMU and JRE are employees of GeneCentric Therapeutics, Inc. and have stock interest in the company. GM a patent holder of the GeneCentric bladder cancer subtype classifier.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The original online version of this article was revised: Due to a figure error.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Rose, T.L., Weir, W.H., Mayhew, G.M. et al. Fibroblast growth factor receptor 3 alterations and response to immune checkpoint inhibition in metastatic urothelial cancer: a real world experience. Br J Cancer 125, 1251–1260 (2021). https://doi.org/10.1038/s41416-021-01488-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41416-021-01488-6
This article is cited by
-
Quantified pathway mutations associate epithelial-mesenchymal transition and immune escape with poor prognosis and immunotherapy resistance of head and neck squamous cell carcinoma
BMC Medical Genomics (2024)
-
Progress in systemic therapy for advanced-stage urothelial carcinoma
Nature Reviews Clinical Oncology (2024)
-
ZNF689 deficiency promotes intratumor heterogeneity and immunotherapy resistance in triple-negative breast cancer
Cell Research (2024)
-
High B7-H3 expression with low PD-L1 expression identifies armored-cold tumors in triple-negative breast cancer
npj Breast Cancer (2024)
-
FGFR-targeted therapeutics: clinical activity, mechanisms of resistance and new directions
Nature Reviews Clinical Oncology (2024)