Abstract
In less than a decade, half a dozen immune checkpoint inhibitors have been approved and are currently revolutionising the treatment of many cancer (sub)types. With the clinical evaluation of novel delivery approaches (e.g. oncolytic viruses, cancer vaccines, natural killer cell-mediated cytotoxicity) and combination therapies (e.g. chemo/radio-immunotherapy) as well as the emergence of novel promising targets (e.g. TIGIT, LAG-3, TIM-3), the ‘immunotherapy tsunami’ is not about to end anytime soon. However, this enthusiasm in the field is somewhat tempered by both the relatively low percentage (<15%) of patients who display an effective anti-cancer immune response and the inability to accurately identify them. Recently, several existing or acquired features/parameters have been shown to impact the efficacy of immune checkpoint inhibitors. In the present review, we critically discuss current knowledge regarding predictive biomarkers for checkpoint inhibitor-based immunotherapy, highlight the missing/unclear links and emphasise the importance of characterising each neoplasm and its microenvironment in order to better guide the course of treatment.
Similar content being viewed by others
Background
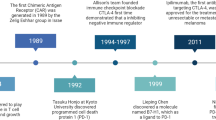
Cancer immunotherapy, aimed at harnessing the host immune system to attack tumour cells, is an old therapeutic concept, dating back to the late 19th century.1 Thanks to substantial advances in molecular immunology, non-specific approaches using bacterial toxins or certain cytokines (e.g. interferon (IFN)-α, interleukin (IL)-2) have now been replaced by targeted immunotherapies, and, in 2011, the cytotoxic T-lymphocyte-associated protein 4 (CTLA-4)-blocking antibody, ipilimumab, became the first immune checkpoint inhibitor approved by the Food and Drug Administration (FDA) for the treatment of advanced/metastatic melanoma.2 CTLA-4 is one of multiple immune checkpoints, key regulators that prevent the immune system from arbitrarily attacking its own host cells, and that are therefore essential for self-tolerance. As such, immune checkpoints are often exploited by cancer cells in order to avoid T-cell attack. A few years later, monoclonal antibodies targeting the programmed cell death protein 1 (PD-1) (e.g. pembrolizumab and nivolumab) or its ligand (PD-L1) (e.g. atezolizumab and avelumab), components of another immune checkpoint, have also been shown to be effective for patients diagnosed with diverse solid or liquid neoplasms. Despite the relatively short period of time since their FDA approvals, immune checkpoint inhibitors have been integrated into the clinical practice guidelines of many tumour (sub)types, either alone or in combination with chemotherapeutic agents, radiation therapy or inhibitors of other negative co-receptors (Fig. 1). The growing interest in immunotherapy is such that hundreds of clinical trials are currently ongoing worldwide, and many ‘next-generation’ agents targeting other well-established immune-regulatory proteins such as lymphocyte-activation gene 3 (LAG-3), T-cell immunoglobulin and ITIM domain (TIGIT) and T-cell immunoglobulin and mucin-domain containing-3 (TIM-3) are being tested in the context of various neoplasms.3 Although the estimated percentage of cancer patients who are eligible for treatment with immune checkpoint inhibitors has been multiplied by 30 over a 9-year period (from 1.54% in 2011 (95% confidence interval (CI), 1.51–1.57%) to 43.63% in 2018 (95% CI, 43.51–43.75%), the current response rate does not exceed 15% for most cancer subtypes.4 This low figure indicates that multiple factors impact host anti-tumour immune responses and, as a consequence, predicting a patient’s response to immune checkpoint inhibitors is challenging and clearly currently suboptimal. Most of the time, response prediction is based only on the expression, in 3–5 mm tumour biopsies, of PD-L1, assessed by immunohistochemistry, using a very low cut-off value (>1% positive cells), which is certainly not sufficient to select precisely the large majority of patients. Indeed, it is well known that not all PD-L1-positive neoplasms are successfully treated by immunotherapy and, conversely, the negative expression of this membrane protein is not automatically associated with an absence of therapeutic response (for a meta-analysis, see ref. 5). Moreover, for unclear reasons, some patients develop high-grade adverse events such as encephalitis, myocarditis or pneumonitis (especially in case of combination therapy) that still cannot be predicted either.
Inhibitory (red dot) and stimulatory (green dot) proteins which are currently being targeted by FDA-approved monoclonal antibodies or next-generation immunotherapeutic drugs (ongoing clinical trials) are illustrated (non-exhaustive list). A dotted line connects the currently approved drugs with their respective targets. These monoclonal antibodies are approved alone (regular), in combination (italics) or both (italics and underlined) depending on the cancer (sub)type. CD cluster of differentiation, GITR glucocorticoid-induced TNFR-related protein, HCC hepatocellular carcinoma, HNSCC head and neck squamous cell carcinoma, ICOS inducible T-cell costimulator, LAG-3 lymphocyte-activation gene 3, NSCLC non-small cell lung carcinoma, PMBCL primary mediastinal large B-cell lymphoma, RCC renal cell carcinoma, SCLC small cell lung carcinoma, SIRPα signal-regulatory protein α, TIGIT T-cell immunoglobulin and ITIM domain, TIM-3 T-cell immunoglobulin and mucin-domain containing-3, VISTA V-domain Ig suppressor of T-cell activation. Data have been retrieved from https://www.cancerresearch.org/scientists/immuno-oncology-landscape/pd-1-pd-l1-landscape (March 2021).
Several parameters that affect a patient’s response to immunotherapy, and that could be used as biomarkers/predictors, have been recently identified (tumour mutational burden (TMB), the degree of mismatch repair deficiency (MMR-D) and microsatellite instability (MSI), altered IFN-γ signalling, the extent/density of tumour-infiltrating immune cells, epigenetic modifications, intestinal microbiota, etc.) (Fig. 2). Might it be possible for these variables to be evaluated by oncologists, pathologists or geneticists and, of course, to be used in routine diagnostic and prognostic assessment? Do they all strongly influence the treatment response in humans or are some of them only predictive in preclinical models? Do they have synergistic effects? Focusing specifically on immune checkpoint inhibitors, we provide an overview of the recent literature, critically discuss the available data, highlight the missing/unknown links and indicate the most promising targets/strategies for better identifying and stratifying patients who might benefit from these therapies.
A high tumour mutational burden (TMB) results in the appearance of neoantigens. These latter are detected by antigen-presenting cells (e.g. dendritic cells, macrophages), which are responsible for the rise of tumour-neoantigen-specific CD8+ T-cell clones. In turn, tumour-infiltrating lymphocytes (TILs) indirectly increase the inflammation-driven expression of PD-L1 on both cancer and immune cells through the secretion of interferon (IFN)-γ. In parallel, cancer-associated fibroblasts might produce transforming growth factor (TGF)-β, which promotes a tolerogenic anti-tumour immune response through various mechanisms. Epigenetic modifications have been shown to influence several factors including the infiltration/phenotype of immune cells encountered within the tumour microenvironment, the TMB and PD-L1 expression. HLA and the intestinal microbiota are host-dependent features that are being increasingly studied in the context of cancer immunotherapy. Finally, the predictive value of many peripheral blood-based biomarkers has been assessed over the past few years.
PD-L1 expression
Early on in the use of anti-PD-1 and anti-PD-L1 agents, a rational presumption emerged that the membrane expression of PD-L1 by tumour cells could constitute a robust predictive biomarker for patient response. This was supported by several clinical studies [e.g. CheckMate 037 and 067 (melanoma), CheckMate 057 and 078 (non-small cell lung cancer, NSCLC), CheckMate 141 (head and neck carcinoma)] in which patients displayed a better response to nivolumab when tumoral PD-L1 immunoreactivity had been detected previously.5,6,7,8,9,10 However, nivolumab also showed benefit in patients bearing PD-L1-negative tumours, thereby already excluding PD-L1 expression as a discriminatory biomarker between responders and non-responders. However, it remains questionable whether or not these PD-L1-negative tumours are truly negative, or if the tiny/punch biopsies missed a positive area. Moreover, the primary lesion may not express PD-L1 while metastases display a strong/diffuse immunoreactivity. In a comprehensive evaluation of the primary studies associated with 45 FDA drug approvals from 2011 to 2019 across 15 different tumour types, PD-L1 expression was found to be predictive in only 28.9% of cases.11
The PD-1/PD-L1 axis undoubtedly plays an important role in damping immunity. In order to better understand the mechanisms underlying the efficacy of anti-PD-L1 treatment (or lack thereof), it seems important to differentiate two different types of PD-L1 expression, and the modulation of involved pathways, in tumour cells. One pathway that can influence PD-L1 expression is ‘inflammation-driven’, localised at sites of IFN-γ-mediated immune attack, and is therefore associated with T-cell infiltration. Interestingly, a link between epithelial-to-mesenchymal transition (EMT) (widely considered to promote tumour invasion) and the inflammation-mediated expression of PD-L1 was reported in the context of various solid cancers with an epithelial origin. Indeed, induced by inflammatory molecules such as tumour necrosis factor α (TNFα) and transforming growth factor β (TGFβ), the key EMT-inducing transcription factors SNAIL and ZEB1 have been shown to regulate PD-L1 mRNA levels through several mechanisms that involve DNA methylation of the promoter for PD-L1 gene (CD274) and microRNAs (most notably miR-200).12 The second pathway controlling PD-L1 induction/maintenance is ‘oncogene-driven’, constitutive, independent of inflammation and neither associated with a specific immune response nor T-cell tumour infiltration.13 In light of the findings of Herbst et al.14 that atezolizumab confers the best results in patients with pre-existing T-cell-mediated immunity that can be re-boosted by the treatment, it could be hypothesised that inflammation-driven PD-L1 expression would be a better indicator of durable response. In accordance with this assumption, Teng et al.15 proposed a classification of tumours based on the presence or absence of both tumour-infiltrating lymphocytes (TILs) and PD-L1 expression, with the idea that the efficacy of immune checkpoint inhibitors relies on immune cell infiltration within the tumour microenvironment (TME).
For PD-L1 expression to provide exploitable and reliable information, several pitfalls need to be overcome. The expression of PD-L1 induced by IFN-γ is both spatially and temporally highly labile, making it a dynamic biomarker which could easily be missed by a small biopsy. Moreover, the routine immunohistochemical detection of PD-L1 presents multiple unresolved/controversial issues. First, the cut-off score for determining staining positivity is still the subject of debate. Depending on the cancer subtype, the anti-PD-1/PD-L1 drug to be administered and the PD-L1 assay used, the cut-off might vary from 1% to 50% positive cells. Although several pooled analyses showed that, when a cut-off of 1% was used, patients bearing a PD-L1-positive cancer had a better overall response rate to anti-PD-L1 monoclonal antibodies compared to their counterparts with PD-L1-negative tumours, the odds ratio was relatively modest (<2.5).16 These findings and others correlating PD-L1 expression with the efficacy of treatment argue for increasing the cut-off value.17 Moreover, the different detection antibodies, variability in tissue processing and the absence of standardisation regarding the scoring methods used to quantify PD-L1 expression (the tumour proportion score (TPS) versus the combined positive score (CPS)) also stand in the way of a reproducible assay.13 Whereas the TPS only evaluates PD-L1 expression on cancer cells, the CPS takes into account both immune and tumour PD-L1-positive cells. This latter distinction between the cell types exhibiting membrane PD-L1 expression is of importance, especially as a stronger correlation was reported between treatment response and PD-L1 expression on immune cells than that on tumour cells.14
Already approved as a companion testing in some tumour types (e.g. breast cancer, NSCLC and bladder cancer),11 the limitations of this biomarker as a predictive tool clearly need to be addressed. In collaboration with the field of medical imaging, recent efforts to use 18-fluorodeoxyglucose PET/CT markers to non-invasively assess increased PD-L1 expression have been made. Interestingly, an inverse correlation between the metabolic-to-morphological volume ratio and PD-L1 positivity was noticed.18 Furthermore, as mentioned above, risk management frameworks are being proposed to reduce suboptimal patient selection for immunotherapy approaches based on the presence/absence of TILs and PD-L1 assessment.19
Tumour mutational burden
For some time now, the mutational burden of a tumour, which is representative of the number of non-synonymous mutations, has been regarded as a promising biomarker to predict the efficacy of immune checkpoint blockade. As the immunogenicity of a peptide can be affected by a single amino acid change,20 new T-cell epitopes are inevitably generated by various genetic alterations that occur during carcinogenesis. Traditionally referred to as tumour-specific antigens (just like any epitope derived from carcinogenic viruses (e.g. human papillomavirus)),21,22 and more likely to be detected in patients with a high TMB, these neoantigens contribute to the immune recognition of malignant cells and to the subsequent induction of anti-tumour responses, which can be boosted by the use of immune checkpoint inhibitors.23,24 More immunogenic than other classes of tumour antigen (e.g. cancer testis (MAGE-A1, MAGE-A3, NY-ESO-1, etc.) and differentiation antigens (MART-1, gp100, etc.)), these latter also have the advantage of not being expressed in healthy embryonic or adult tissues.22 Supporting the utility of TMB in precision oncology, data from many clinical trials have clearly demonstrated that patients whose tumours display a high mutational burden were associated with longer progression-free survivals following treatment with anti-PD-1/PD-L1 and/or anti-CTLA-4 antibodies.23,25,26,27 Interestingly, the predictive value of TMB was reported across most cancer subtypes.28,29,30 These results were further reinforced by a seminal study taking 27 different tumour types into consideration and reporting that 55% of the difference in the objective response rate across cancers was attributable to TMB.31
Detailed information relating to the TMB was originally obtained by whole exome sequencing using tumour biopsy samples. Although this approach undeniably gives the most unbiased and accurate determination of the mutational burden, it is hardly achievable in everyday clinic practice because of its relative complexity and cost.32 Aimed at expanding genomic testing to more patients and, therefore, making TMB assessment more clinically practicable, test panels such as MSK-IMPACT (which analyses 468 genes) or FoundationOne CDx (324 genes) have been recently approved by the FDA. Moreover, non-invasive strategies have been also explored, such as blood-based assays using circulating tumour DNA (ctDNA; see below). Despite its great potential, however, the use of ctDNA requires further investigation.33
Surprisingly, and very interestingly, the TMB does not correlate with PD-L1 expression on cancer cells, although both have a predictive capacity.34,35 Whether this absence of a correlation is artefactual (as a result of the imprecise assessment of tumour heterogeneity using needle biopsy specimens) or related to the aberrant inflammation-independent regulation of PD-L1 (through the activation of hypoxia-inducible factor 1α, epidermal growth factor receptor and/or mitogen-activated protein kinase (MAPK) signalling pathways) is still unclear, but the combined use of both factors would certainly result in greater predictive power. If the TMB evaluation was more broadly performed across the patient population, an evaluation of the risk/benefit ratio regarding the sampling would become essential. Indeed, performing a biopsy on a melanoma or a lung cancer involves very different levels of invasiveness and risk. In this respect, the size and/or number of samples should be carefully considered to meet the amount of DNA required for the assessment of the TMB in addition to any other predictive factor (e.g. PD-L1 expression) that we would want to measure, while taking also into consideration potential drawbacks for the patient. Furthermore, before its implementation in clinical practice, several challenges (e.g. determination of reliable sequencing panel sizes, definition of robust predictive cut-off values which might vary between cancer subtypes, establishment of clear, universally used standard operating procedures) still need to be overcome.36,37 Despite these requirements for standardisation, taking into account the accumulating data and recent improvements in technology, TMB seems on the verge of becoming a cornerstone in the decision of the immunotherapy course of treatment.
Mismatch repair deficiency and microsatellite instability
The DNA MMR pathway maintains genomic stability by repairing any mismatched bases during DNA replication and, thus, prevents genetic alterations.38 During cancer progression, a defect/deficiency in this machinery leads to DNA hypermutability and subsequent MSI. The MSI status is commonly determined by assessing the extent of instability in a ‘reference panel’ comprising some nearly monomorphic mononucleotide loci such as BAT-25, BAT-26, NR-21 and NR-24.39 Where MMR deficiency associated with the cancer predisposition condition Lynch syndrome is suspected, germline mutations in MSH2, MSH6, PMS2 and MLH1 genes (all of which encode proteins involved in MMR) are first generally evaluated by immunohistochemistry before being further validated by sequencing.40 Tumours displaying a MSI-high status are associated with a higher average number of somatic mutations (and, ultimately, neoantigens) than MMR-proficient cancers, and over 40% of neoplasms with a MSI-high status have been shown to respond positively to immune checkpoint blockade, resulting in prolonged progression-free survival.41 However, with the exception of colorectal and endometrial carcinomas, unfortunately only a small subset of tumours are MSI-high,42 which makes this diagnostic/predictive approach inappropriate for most patients. Moreover, a seminal study analysing 100,000 human cancer genomes reported that a higher percentage of malignancies are TMB-high rather than MSI-high.43 As an example, mutations in the DNA polymerase ε (POLE) gene, identified in 5–10% of most tumour subtypes and associated with exceptional responses to immune checkpoint inhibitors, have been shown to disrupt the proofreading function of the protein, leading to hypermutated phenotypes in the absence of MSI.44,45 Finally, although most MSI-high tumours are also TMB-high, intriguingly, ~18% of MSI-high neoplasms do not exhibit an elevated TMB.46 Whether these patients are less responsive to immunotherapy is still unknown but it would certainly be interesting to further investigate the issue. Altogether, MMR deficiency (and its associated MSI) could be regarded as an interesting biomarker to predict the response to immune checkpoint inhibitors but, compared with the potential universal utility of TMB, its potential use seems quite limited (and might not always be informative on its own).
Human leucocyte antigen
Encoded by highly polymorphic genes, human leucocyte antigen (HLA) allows the presentation of both self- and foreign antigens to T-cell receptors, thereby playing a pivotal role in initiating immunotolerance, helper or cytotoxic T-cell responses. As extensively studied in the last decades, immune escape can occur when this presentation is compromised. In the context of immunotherapy, recent data reported that impaired HLA class I (HLA-I) antigen processing led to acquired resistance to immune checkpoint inhibitors and evasion of tumour cell death by avoiding CD8+ T-cell recognition.47 In a seminal paper determining the HLA-I genotype of over 1,500 patients with metastatic cancer treated with immunotherapy, Chowell et al.48 showed that patients displaying a maximal heterozygosity at HLA-I loci A, B and C were more likely to display improved overall survival compared with those who were homozygous for at least one locus. The study also highlighted the extended survival of patients with the HLA-B44 supertype as well as the poor outcome associated with either the somatic loss of heterozygosity at HLA-I or the HLA-B62 supertype. Using a novel computational tool, HLA loss of heterozygosity was found to occur in 40% of early-stage NSCLC, suggesting that this event could influence a substantial proportion of treated metastatic cancer patients.49 HLA-I evolutionary divergence, calculated after quantifying physiochemical sequence divergence between HLA-I alleles, has also been shown to affect the efficacy of immune checkpoint inhibitors.50 Absence of the membrane expression of major histocompatibility complex (MHC) class I has been observed in almost half (43%) of melanoma specimens, and importantly, the transcriptional repression of HLA-A, HLA-B, HLA-C, and β2 microglobulin (B2M) predicted resistance to ipilimumab. Surprisingly, no predictive value was reported for anti-PD-1 blockade using nivolumab.51
As outlined above, different parameters (loss of heterozygosity, evolutionary divergence, mRNA/protein level), assessed by technological approaches such as exome sequencing, immunohistochemistry and real-time PCR, have been determined to study the influence of HLA on the response of cancer patients to immune checkpoint inhibitors. To be used in routine, a standardisation/harmonisation is absolutely required, which could potentially be conducted in a study comparing all these variables/approaches using a same cohort of patients. Moreover, it is important to notice that divergent results have been recently highlighted, which slightly tempers the enthusiasm for HLA status (or even precludes the direct clinical use of this parameter). Indeed, in patients with NSCLC, Negrao et al.52 reported that both PD-L1 expression and TMB were stronger predictors of response to immunotherapy than the HLA-I genotype which, actually, did not correlate with survival.
IFN-γ signalling
The interest for IFN-γ (and its associated signalling pathway) in the context of cancer immunotherapy originated from its implication in PD-L1 and PD-L2 upregulation.53,54 These findings led to several ongoing clinical assays investigating various combination therapies (e.g. nivolumab plus IFN-γ (NCT02614456), pembrolizumab and recombinant IL-12 (NCT03030378), pembrolizumab with intratumoral delivery of tavokinogene telseplasmid (an IL-12-encoding plasmid; NCT03567720)) that are aimed at enhancing presence/production of IFN-γ within the TME. Regarding the use of IFN-γ as a biomarker to predict the response of patients to immune checkpoint inhibitors, genomic defects in several type 2 IFN-related genes were reported to be responsible for acquired resistance to pembrolizumab in patients with advanced melanoma.55 Copy number alterations, including loss of IFNGR1, IRF-1, JAK2 and IFNGR2, and amplification of several IFN-γ pathway inhibitors (e.g. SOCS1 and PIAS4) were detected in non-responders to CTLA-4 therapy as well.56 In addition, loss-of-function mutations of JAK1/2, which encode two tyrosine kinases essential for transducing the IFN-γ signal intracellularly, were also reported.57 In parallel with genomic analyses, the mRNA level of a dozen IFN-stimulated genes (e.g. GZMA, CXCL9, CXCL10, PRF1) was determined in both metastatic melanoma and NSCLC patients, and a high pre-existing IFN-γ transcriptome signature has been shown to be associated with an improved response to anti-PD-1 blockade.34,51 Finally, the predictive value of some alternative strategies, such as the analysis of IFN-γ secretion, has also recently been assessed (mainly in pre-clinical models). For example, mass cytometry (CyTOF) has notably been used to measure the amounts of IFN-γ among immune cells.58
However, it is important to notice that the role of IFN-γ in anti-tumour immunity is complex and that its positive effect in mediating a durable response to cancer immunotherapy remains unclear/controversial.59 Indeed, combination therapy using PD-1 and CTLA-4 inhibitors in the context of TMB-low tumours has been shown to lead to an excessive production of IFN-γ and therapeutic resistance.58 Furthermore, Brown et al.60 also showed that IFN-γ-induced indoleamine 2,3-dioxygenase (IDO) expression provided adaptive resistance to immune checkpoint inhibitors in hepatocellular carcinoma patients. Despite these results, and further illustrating the complex role played by the IFN-γ–IDO–kynurenine pathway in treatment outcome, the addition of an IDO inhibitor (epacadostat) to pembrolizumab failed to confer any benefit in melanoma patients (ECHO-301/KEYNOTE-252).61 Therefore, the use of IFN-γ secretion and/or IFN-γ signalling components as potential biomarkers to predict the outcome of immunotherapy-treated patients undoubtedly needs further investigation and is not yet clinically applicable.
Immune infiltration
Immune checkpoint inhibitors are, in essence, designed to reinvigorate an inhibited or exhausted anti-cancer immune response. With this in mind, it seems natural that immune cells infiltrating the TME would play an important role not only in tumour control prior to treatment, but also in the response to immune checkpoint blockade. Indeed, an association between the density of pre-existing CD8+ T cells located at the invasive tumour margin and the response to anti-PD-1 treatment (pembrolizumab) has been especially demonstrated in patients with metastatic melanoma.62 Interestingly, the same cells were also thought to be responsible for the expression of PD-L1 on tumour cells through IFN-γ release.13 Moreover, a lack of response to atezolizumab in patients with urothelial cancer was associated with the presence in fibroblasts of a gene signature indicative of TGF-β signalling and with CD8+ T-cell exclusion.63 Blocking the TGF-β signalling pathway in stromal cells facilitated T-cell penetration into tumours and ultimately restored anti-tumour immunity in a mouse model. The predictive role of TILs in the response to anti-PD-1 has also been investigated in cohorts of patients with advanced NSCLC. Similar to the data reported in melanoma patients, in addition to the absolute number of TILs, a correlation between the CD8+/CD4+ ratio and the response to anti-PD-1 treatment (pembrolizumab or nivolumab) was highlighted.64
Easily detected by immunohistochemistry (or even using a simple haematoxylin and eosin staining), the challenge remains both to harmonise the quantification of TILs and to develop a validated method for clinical practice. Almost 15 years ago, Galon et al.65 developed the Immunoscore methodology in the context of colorectal cancer in an effort to quantify the in situ immune infiltrate. Technically, immunohistochemical experiments are first performed and both CD3+ and CD8+ T-cell populations are then automatically quantified at the centre of the tumour as well as at the invasive margin (the analysis spans 360 µm into the cancer region and 360µm into the surrounding healthy tissue) by a dedicated software (Immunoscore Analyzer, HalioDx). A score, ranging from I0 for low cell densities to I4 for high densities, is finally assigned, with a higher score being correlated with longer patient survival.66,67 The power of this method, which takes into account the two essential elements of density and localisation of TILs, is currently being validated in 23 international pathology expert centres.68 The added value of the Immunoscore as a prognostic marker for patients with colorectal cancer is now well-documented and its prognostic significance in other cancer types is currently being investigated.69,70 With further development, an immunoscore-like method to determine the quantity and ‘quality’ (e.g. CD8+/CD3+ ratio) of TILs as well as their potential exclusion within the collagen-rich peritumoral stroma could also be a precious tool to better stratify patients who would benefit from immune checkpoint blockade treatment. However, this still needs to be rigorously tested in a prospective clinical trial setting.
The importance of immune infiltration is further supported by the proven efficacy of combining cytotoxic chemotherapy or radiation therapy with immune checkpoint inhibitors.71,72 Indeed, the former treatments, by killing tumour cells, induce the release of both tumour-associated antigens and damage-associated molecular pattern molecules (e.g. calreticulin, HMGB1, ATP), which promote an immune response via the activation/maturation of (plasmacytoid) dendritic cells and the expansion/trafficking of CD8+ T cells. Chemoradiotherapy also shapes the TME towards a responsive state by depleting immunosuppressive cells such as regulatory T (TREG) cells and myeloid-derived suppressor cells (MDSCs).73,74 Patients with NSCLC are benefiting from the chemotherapy plus anti-PD-1 combination (carboplatin, pemetrexed and pembrolizumab) since it was accepted by the FDA in 2018.75 The now well-recognised ability of cytotoxic drugs to enhance PD-L1 expression in different types of cancer supports the great potential of combined treatments, in particular the numerous chemotherapy-immunotherapy combinations currently being tested.76,77 Other strategies (e.g. oncolytic virotherapy) that similarly aim to sensitise the TME before the administration of immune checkpoint inhibitors are also under investigation and would, in an ideal world, result in the generation of a personalised biomarker-based therapy tailored to each patient.78
In summary, combination therapies that integrate immune checkpoint inhibitors and chemotherapeutic agents are likely to constitute the future of cancer immunotherapy, and initiatives like Immunoscore could be regarded as a helpful tool in the prediction of treatment response (or the need for neoadjuvant chemoradiotherapy to first restore an immune-reactive microenvironment). If validated/adapted to each cancer type and universally accepted as a prognostic marker, this strategy could also be combined with other biomarkers to further improve patient selection.
Epigenetic modifications
Epigenetic changes such as promoter hypermethylation and histone modifications are well-known to affect the structure of chromatin and, ultimately, the gene expression profile, without altering the DNA sequence. Evidence accumulated over the past few years suggests that the chromatin state might also influence the response to immune checkpoint blockade. Indeed, using whole-genome bisulphite DNA sequencing, a genome-wide method that enables the study of cytosine methylation using small amounts of DNA, Ghoneim et al.79 showed that de novo DNA methylation programs promote T-cell exhaustion and, consequently, inhibit both antigen-specific CD8+ T-cell rejuvenation and clonal diversity following anti-PD-1 blockade. According to the authors, these epigenetic modifications repress key genes involved in effector functions, proliferation and tissue homing of exhausted T cells, which leads to the absence of a durable response to immune checkpoint inhibitors. Moreover, it has been reported that exhausted T cells have an epigenetic profile which differs from that of effector and memory T cells and, importantly, they would be minimally remodelled after anti-PD-1 therapy.80 Besides these global approaches, the potential predictive value of some proteins involved in the epigenetic mechanisms of gene regulation has also been highlighted. Thus, the expression levels of both DNA methyltransferase 1 (DNMT1) and enhancer of zeste homolog 2 (EZH2) have been found to correlate negatively with the extent of CD8+ T-cell infiltration within the TME and inhibition of EZH2 improved the response to anti-CTLA-4 therapy.81,82 Mechanistically, Xiao et al.83 showed that EZH2 directly enhances the H3K27me3 level of PD-L1/CD274 and interferon regulatory factor 1 (IRF-1) promoters, suppressing their expressions. The status (wild-type versus mutated) of ten-eleven translocation methylcytosine dioxygenase 1 (TET1) and AT-rich interaction domain 1A (ARID1A) genes, which are involved in DNA demethylation and chromatin remodelling, respectively, could also be regarded as promising biomarkers to predict patient response. Accordingly, patients with tumours containing TET1 and/or ARID1A mutations, and who have been treated by immunotherapy, have been reported to display both longer progression-free and overall survivals than their wild-type counterpart.84,85 Notably, MSI-high and TMB-high statuses are frequently observed in the ARID1A-mutated cancer group, which might potentially explain the results in these patients.85
In addition to their utility as predictive biomarkers, epigenetic modifications also represent promising therapeutic targets to improve the efficiency of immune checkpoint inhibitors. Using various mouse models of cancer, several research groups have recently shown that DNA methyltransferase inhibitors as well as drugs thwarting histone modifications (histone deacetylase inhibitors such as valproic acid, panabinostat or entinostat) improved anti-PD-1 and anti-CTLA-4 immunotherapies.86,87 However, these data still remain experimental, poorly explained and warrant clinical validation.
The intestinal microbiome
The symbiotic bacteria that colonise our digestive system have been found to influence many physiological processes, including immunity88 and, less than 5 years ago, preclinical studies established a link between gut microbiota and the response to immunotherapy. Indeed, cancer growth, spontaneous anti-tumour immunity and efficacy of both anti-CTLA-4 and anti-PD-L1 monoclonal antibodies differed in mice depending on the composition of the gut microbiota.89,90 These differences disappeared with cohousing or faecal transfer. Interestingly, the bacterial species associated with enhanced response to anti-CTLA-4 were not the same as the ones associated with enhanced efficacy of anti-PD-L1 therapy. These preclinical data suggested that the inter-individual heterogeneity of anti-tumour immunity (and hence the efficacy of immune checkpoint inhibitors) could partially rely on gut microbes, which could be manipulated for therapeutic purposes. Accordingly, when germ-free or antibiotic-treated mice were recolonised using patient stool during faecal microbiota transplantation experiments, a causal relationship between gut microbiota and sensitivity to immunotherapeutic drugs was confirmed. Stool from responders restored sensitivity to PD-1 blockade, whereas stool originating from non-responders conferred resistance to this agent.91 Interestingly, a human proof-of-concept study similarly reported the positive impact of faecal microbiota transplants in anti-PD-1-refractory melanoma patients.92
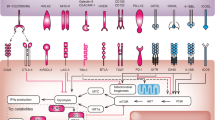
Several studies confirmed the so far suspected complexity of the part played by the colonic bacterial flora in the response to immune checkpoint inhibitors. Indeed, the baseline microbiota had an influence on the efficacy of both anti-CTLA-4 and anti-PD-L1 treatments. However, whereas the Bacteroides genus had previously been associated with an increased response to anti-CTLA-4 in mice,89 it was associated with a poor response in humans, and other bacteria strains such as Faecalibacterium prausnitzii were instead highlighted as favourable.93 Moreover, another prospective study focusing on the efficacy of anti-PD-1 antibodies showed that, among others, Bifidobacterium longum and Collinsella aerofaciens were enhanced in responders, while Ruminococcus obeum and Roseburia intestinalis were more abundant in non-responders.94 A study focusing on NSCLC patients highlighted that, in addition to Bifidobacterium longum, Alistipes putredinis and Prevotella copri were also enriched in responders to the anti-PD-1 antibody nivolumab in comparison with non-responders (Fig. 3).95 Of note, it is important to mention that gut microbiota profiles associated with a good response to treatment might also be associated with the occurrence of immune-related adverse effects (e.g. enterocolitis).93
Bifidobacterium longum, Collinsella aerofaciens, Alistipes putredinis and Prevotella copri have been shown to be associated with a more favourable response to anti-PD-L1 therapy. Patients enriched for Faecalibacterium prausnitzii responded better to anti-CTLA-4 therapy. Although the reasons are still unknown, some bacterial species (e.g. Roseburia intestinalis, Ruminococcus obeum) or families (e.g. Bacteroides) are associated with a poor response to anti-CTLA-4 or anti-PD-L1 therapy.
Discrepancies among different studies can be explained by several factors, including differences in experimental models (as well as the translation from mice to humans), tumour type and intra/inter-population variability of microbial composition. Furthermore, various metagenomic approaches (16S rRNA sequencing, used to find operational taxonomic units (OTUs), metagenomic shotgun sequencing, or both) and bioinformatic methods were applied, showing an urgent need for standardisation of the analytical approaches currently used.96,97 Despite all these variations and although the exact mechanism by which the response to immune checkpoint inhibitors is influenced remains unknown, constantly highlighting the predictive value of certain bacterial strains is likely to indicate their interest and relevance.
As described by Matson et al.,94 a ratio of ‘beneficial’ OTUs to ‘non-beneficial’ OTUs would be the strongest predictor of clinical response to immune checkpoint inhibitors, but would probably need to be adapted depending on the cancer type and/or the therapy considered. The integration of such a ratio into a multiparameter setting alongside tumour genomics and immune infiltration would undoubtedly strengthen its potential. We are probably several years and a considerable amount of research away from a fully developed tool, but the convenience and accessibility of sampling make it an information goldmine that needs to be exploited.
Peripheral blood biomarkers
Regardless of the disease or treatment, the interest in blood biomarkers is historical and is mainly based on the high cost/time efficiency, reduced invasiveness and high patient acceptance in comparison with existing tissue-derived biomarkers. During the past few years, various haematological parameters have been tested for their ability to predict a response to immunotherapy, with varying degrees of success and reproducibility (Fig. 4).
Blood cell subpopulations
Several peripheral blood cell subpopulations have been investigated as predictive biomarkers. Patients with metastatic cutaneous and uveal melanoma who have elevated relative counts of both eosinophils and lymphocytes have been shown to display a more favourable outcome after receiving anti-PD-1 monotherapy (pembrolizumab or nivolumab).98,99 A significantly better survival following ipilimumab treatment was also reported in patients with low absolute monocyte and MDSC counts and high eosinophil, lymphocyte and TREG cell frequencies.100 In a recent meta-analysis including 6,461 patients with renal cell carcinoma, the negative predictive value of a high neutrophil/lymphocyte ratio prior to treatment with immune checkpoint inhibitors was highlighted.101 Finally, the proportion of circulating exhausted T cells, characterised by a low proliferative capacity and the weak expression of several surface markers such as CD57 and killer cell lectin-like receptor subfamily G member 1 (KLRG1), has been correlated with patients’ response to immune checkpoint inhibitors and, hence, their clinical outcome.102
Soluble proteins
Some circulating/soluble proteins could also be regarded as interesting biomarkers. For example, elevated levels of lactate dehydrogenase (LDH) and C-reactive protein (CRP) have been associated with poor overall survival following anti-PD-1 therapy.99 In metastatic melanoma patients treated with anti-PD-1 or anti-CTLA-4 therapy, the level of angiopoietin-2 (ANGPT2) correlated with a reduced survival as well.103 Interestingly, two recent articles have reported the concentration of serum CD73 as a novel predictive peripheral blood biomarker inversely associated with patient response to nivolumab or pembrolizumab.104,105 Finally, the negative prognostic impact of both soluble natural killer group 2 member D (NKG2D) ligands (ULBP1-3, MICA and MICB) and CD25 for advanced melanoma patients treated with ipilimumab has been recognised for a few years.106,107,108
In addition to its expression on the membrane of tumour cells, immune and other cells (e.g. blood/lymphatic endothelial cells), several extracellular forms of PD-L1 have been reported. Although a few conflicting results exist, most studies report that high levels of exosomal or freely soluble PD-L1 in peripheral blood are associated with disease progression in patients with various cancers (for a review, see ref. 109,110) In addition to being a prognostic biomarker, the predictive value of exosomal PD-L1 in determining the response to the anti-PD-1 antibody pembrolizumab was also recently highlighted in patients with melanoma.111 However, intriguingly, soluble PD-L1 does not seem to be a surrogate marker for the tumour PD-L1 status.
Circulating tumour DNA
Tumour-derived DNA within the blood (ctDNA) could also provide interesting information. Not only is its low (or undetectable) level during treatment associated with a better prognosis,112 but the ability to sequence ctDNA prior to therapy enables the TMB to be determined and/or actionable genetic alterations to be detected.33 Such a strategy is the subject of ongoing Phase 2/3 clinical trials [e.g. Blood First Assay Screening Trial (BFAST)].
Combining parameters
Prelaj et al.113 have described a novel score (EPSILoN), which could help to identify patients with NSCLC who are most likely to benefit from second-line immunotherapy. This prognostic score combines several clinical (smoking and liver metastases) and blood parameters (LDH level and neutrophil/lymphocyte ratio). However, similar to all other approaches using peripheral blood, further investigations are needed to confirm these results in larger cohorts and in the context of other neoplasms.
Resistance to PD-1/PD-L1 blockade and potential biomarkers
Early in the clinical use of immune checkpoint inhibitors, it appeared that a substantial proportion of patients fails to benefit at all from any immunotherapeutic approach. Besides this primary resistance, and similar to what is commonly experienced with cytotoxic/cytostatic drugs and targeted therapies, there is a growing subset of initial responders who progressively become refractory to PD-1/PD-L1 blockade therapy (acquired resistance). Accumulating evidence suggests that both types of resistance are multifactorial, with underlying mechanisms that mostly overlap and that are antagonistic to those described in responders.114,115
Loss of immune checkpoint expression and/or (neo)antigen expression/presentation
The most obvious mechanism of resistance to immunotherapy is attributed to the primary or acquired absence of expression of targeted immune checkpoint(s) on the cell surface of cancer/immune cells. In addition, as previously discussed, a lack of neoantigen expression (low TMB) and/or presentation (disruption in the HLA machinery) can result in inadequate tumour recognition by immune cells and, ultimately, in drug resistance. Several strategies are under investigation to increase the immunogenicity of neoplasms that are refractory to checkpoint blockade (e.g. pretreatment with chemotherapy, intralesional injection of IL-2 in combination with hypofractionated radiotherapy (NCT03474497), use of a mRNA-based cancer vaccine targeting several tumour-associated antigens (NCT04526899)).
The presence of immunosuppressive cells
By altering anti-tumour immune responses via multiple mechanisms,116 both MDSC and TREG cells encountered within the cancer microenvironment are also thought to actively participate to the primary resistance to PD-1/PD-L1 blockade therapies. Accordingly, several preclinical studies have demonstrated an improved effectiveness of anti-PD-1/PD-L1 monotherapy following the depletion of these cell types.117,118,119 In order to better determine the non-responders, these two immunosuppressive cell populations could be easily detected by immunohistochemistry using anti-Ly-6C/Ly-6G (Gr-1) and anti-Foxp3 antibody, respectively, and then quantified using computerised image analysis.120
T-cell exclusion and resistance to IFN-γ
In both primary and acquired resistant tumours, malignant cells can develop an insensitivity to CD8+ T-cell-secreted IFN-γ through various genomic defects occurring in several genes involved in the IFN signalling pathway (see above). By inducing the overexpression of broad-spectrum immunosuppressive cytokines such as IL-8 and vascular endothelial growth factor (VEGF), activating mutations in key components of Wnt–β-catenin signalling pathway or MAPK cascade have been shown to promote both CD8+ T-cell and dendritic cell exclusions in syngeneic mouse models of melanoma and triple negative breast cancers.121,122,123 Aimed at overcoming this latter mechanism of resistance, which is frequently identified in patients with melanoma, NSCLC and colorectal cancer, several ongoing clinical trials are evaluating the efficacy of combining an anti-PD-1 antibody with inhibitors of the MAPK component BRAF/MEK (dabrafenib/trametinib) or VEGF receptor 2 (lenvatinib) inhibitors in the refractory advanced melanoma setting (e.g. NCT04305041, NCT02967692). If these explorative approaches prove to be efficient in re-inducing a positive response to immune checkpoint blockade, the mutational status of genes involved in these proto-oncogenic signalling pathways could be systematically assessed by exome sequencing.
Compensatory upregulation of alternative immune checkpoints
Finally, the expression of alternative inhibitory receptors including TIM-3 and the novel immune checkpoint VISTA (V-domain Ig suppressor of T-cell activation) has been shown to be strongly upregulated in humans and mice in cases of acquired resistance to anti-PD-1 or anti-CTLA-4 therapy.124,125,126 The restored responsiveness to anti-PD-1 treatment with the addition of an anti-TIM-3 blocking antibody has provided a scientific rationale for testing different treatment regimens combining PD-1/PD-L1 blockade with monoclonal antibodies that target other co-existing immune checkpoints in patients with refractory cancer [e.g. anti-CTLA-4 (NCT02694822), anti-B7-H3 (NCT02475213)].
Concluding remarks and discussion
The response to immune checkpoint blockade and its prediction are doubtless complex and multifactorial processes. The deeper we dig, the more evident it becomes that a single biomarker will never be able to capture the complexity of the tumour and/or its microenvironment and, obviously, to precisely predict the response to immunotherapy. A combination of multiple biomarkers will inevitably be necessary, as demonstrated by an increased performance of the combination of PD-L1 immunohistochemistry and T-cell infiltration or TMB assessment, compared with the three parameters alone.127 At this time, and based on the current state of knowledge, combining the assessment of immune infiltration and TMB with the more traditional analysis of tumour PD-L1 expression seems to be the most achievable compromise, as it presents the best risk-time-cost/benefit ratio. Indeed, both PD-L1 expression and CD3+/CD8+ T-cell infiltration can be assessed on the same slide (or on serial tissue sections), and TMB can be determined using DNA extracted from the same biopsy, which represents a considerable advantage. Using these three accessible biomarkers, the use of a workflow for better determining potential responder patients could be considered (Fig. 5). Of note, this review aims to present a global picture of the immunotherapeutic landscape, and we cannot exclude that this predictor combination might be more relevant in a specific cancer (sub)type or with a particular drug/monoclonal antibody in comparison with another. Additionally, as addressed above, each predictive biomarker still has its own challenges/pitfalls that need to be overcome before being used in a routine clinical setting. Once these challenges have been overcome, a standardisation of procedures as well as a universal harmonisation of biomarker measurements will be undoubtedly required. Of course, the field of cancer immunotherapy is moving so fast that, besides the biomarkers discussed in this article, other approaches such as integrative multi-omics analysis and algorithms/computing science (artificial intelligence) could also prove to be informative in the coming years.128,129 As an example, by extracting and analysing numerous features from CT scans (e.g. tumour heterogeneity, neovascularisation), an innovative machine-learning-based radiomics method for identifying patients at high risk of hyperprogression following immune checkpoint blockade has been recently described.128
The collaboration between oncologists ①, pathologists (2a, evaluations of both PD-L1 expression and intratumoral immune cell infiltration) and geneticists [2b, assessment of tumour mutational burden (TMB)] would lead to the establishment of a score that would guide the course of treatment (3a). Patients with a favourable/high score would be treated with immune checkpoint inhibitors, while patients considered as having a low probability of an efficient/durable response would undergo an alternative treatment (e.g. radiotherapy, chemotherapy) alone or prior to immunotherapy (after re-evaluation).
Instead of looking at the different aforementioned biomarkers separately, they could be considered as individual chain links that constitute one and the same chain of events. As appropriately explained by Linette and Carreno,130 these events could be simplified the following way: a high mutational burden gives rise to neoantigens, which will, in turn, be detected by our immune system. Neoantigen-specific lymphocytes are designed to control tumour growth and their actions are reinforced by checkpoint inhibitor treatments. Moreover, the accumulation of TILs contributes, by producing IFN-γ, to the upregulation of PD-L1 on both cancer and tumour-infiltrating immune cells (e.g. antigen-presenting cells). Given the parallel ‘oncogene-dependent’ regulation of PD-L1, the effect of this effector cytokine is supposed to be more important on immune cells encountered within the TME than on malignant cells.
It is, however, important to take into account that all these biomarkers are time- and space-dependent and, therefore, highly labile. The more downstream we attempt to detect a specific biomarker, the more we risk missing important information. This is why measuring several elements of this chain before treatment would enable a more representative picture of the tumour and its microenvironment to be created, allowing its potential ability to respond to immunotherapy to be determined. In this respect, considering that chemotherapy or radiotherapy is increasingly used prior to immune checkpoint inhibitors, the biomarkers should probably be assessed both before and after pretreatment, as the latter is likely to reshape the tumour and will therefore have an impact—hopefully positive—on factors such as the extent of intratumoral T-cell infiltration and PD-L1 expression.
Alongside the determination of ‘classical’ immune checkpoints (PD-1, PD-L1 and CTLA-4), the assessment of other proteins will soon become indispensable in light of the next, arising generation of immunotherapy agents.3 Hence, the establishment of a panel of different antibodies that can be used for the systematic determination of several proteins would be greatly appreciated. Moreover, as mentioned above, the expression of other immunosuppressive proteins such as IDO and the immune checkpoints TIM-3 and VISTA constitutes a compensatory mechanism that can undermine the efficacy of immune checkpoint inhibitors.124,125,126 Therefore, the expression of these emerging proteins could also provide useful predictive information even before the clinical approval of their inhibitors as treatment.
References
Dobosz, P. & Dzieciatkowski, T. The intriguing history of cancer immunotherapy. Front. Immunol. 10, 2965 (2019).
Hodi, F. S. et al. Improved survival with ipilimumab in patients with metastatic melanoma. N. Engl. J. Med. 363, 711–723 (2010).
Mazzarella, L. et al. The evolving landscape of ‘next-generation’ immune checkpoint inhibitors: a review. Eur. J. Cancer 117, 14–31 (2019).
Haslam, A. & Prasad, V. Estimation of the percentage of US patients with cancer who are eligible for and respond to checkpoint inhibitor immunotherapy drugs. JAMA Netw. Open 2, e192535 (2019).
Liu, X. et al. Association of PD-L1 expression status with the efficacy of PD-1/PD-L1 inhibitors and overall survival in solid tumours: a systematic review and meta-analysis. Int. J. Cancer 147, 116–127 (2020).
Weber, J. S. et al. Nivolumab versus chemotherapy in patients with advanced melanoma who progressed after anti-CTLA-4 treatment (CheckMate 037): a randomised, controlled, open-label, phase 3 trial. Lancet Oncol. 16, 375–384 (2015).
Hodi, F. S. et al. Nivolumab plus ipilimumab or nivolumab alone versus ipilimumab alone in advanced melanoma (CheckMate 067): 4-year outcomes of a multicentre, randomised, phase 3 trial. Lancet Oncol. 19, 1480–1492 (2018).
Horn, L. et al. Nivolumab versus docetaxel in previously treated patients with advanced non-small-cell lung cancer: two-year outcomes from two randomized, open-label, phase iii trials (CheckMate 017 and CheckMate 057). J. Clin. Oncol. 35, 3924–3933 (2017).
Wu, Y. L. et al. Nivolumab versus docetaxel in a predominantly chinese patient population with previously treated advanced NSCLC: CheckMate 078 randomized phase III clinical trial. J. Thorac. Oncol. 14, 867–875 (2019).
Ferris, R. L. et al. Nivolumab vs investigator’s choice in recurrent or metastatic squamous cell carcinoma of the head and neck: 2-year long-term survival update of CheckMate 141 with analyses by tumor PD-L1 expression. Oral Oncol. 81, 45–51 (2018).
Davis, A. A. & Patel, V. G. The role of PD-L1 expression as a predictive biomarker: an analysis of all US Food and Drug Administration (FDA) approvals of immune checkpoint inhibitors. J, Immunother, Cancer 7, 278 (2019).
Jiang, Y. & Zhan, H. Communication between EMT and PD-L1 signaling: new insights into tumor immune evasion. Cancer Lett. 468, 72–81 (2020).
Patel, S. P. & Kurzrock, R. PD-L1 expression as a predictive biomarker in cancer immunotherapy. Mol. Cancer Ther. 14, 847–856 (2015).
Herbst, R. S. et al. Predictive correlates of response to the anti-PD-L1 antibody MPDL3280A in cancer patients. Nature 515, 563–567 (2014).
Teng, M. W., Ngiow, S. F., Ribas, A. & Smyth, M. J. Classifying cancers based on T-cell infiltration and PD-L1. Cancer Res. 75, 2139–2145 (2015).
Passiglia, F. et al. PD-L1 expression as predictive biomarker in patients with NSCLC: a pooled analysis. Oncotarget 7, 19738–19747 (2016).
Xu, Y. et al. The association of PD-L1 expression with the efficacy of anti-PD-1/PD-L1 immunotherapy and survival of non-small cell lung cancer patients: a meta-analysis of randomized controlled trials. Transl. Lung Cancer Res. 8, 413–428 (2019).
Jreige, M. et al. (18)F-FDG PET metabolic-to-morphological volume ratio predicts PD-L1 tumour expression and response to PD-1 blockade in non-small-cell lung cancer. Eur. J. Nucl. Med. Mol. Imaging 46, 1859–1868 (2019).
Gonzalez-Ericsson, P. I. et al. The path to a better biomarker: application of a risk management framework for the implementation of PD-L1 and TILs as immuno-oncology biomarkers in breast cancer clinical trials and daily practice. J. Pathol. 250, 667–684 (2020).
Allen, P. M. et al. Identification of the T-cell and Ia contact residues of a T-cell antigenic epitope. Nature 327, 713–715 (1987).
Ward, J. P., Gubin, M. M. & Schreiber, R. D. The role of neoantigens in naturally occurring and therapeutically induced immune responses to cancer. Adv. Immunol. 130, 25–74 (2016).
Smith, C. C. et al. Alternative tumour-specific antigens. Nat. Rev. Cancer 19, 465–478 (2019).
Rizvi, N. A. et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science 348, 124–128 (2015).
Rooney, M. S., Shukla, S. A., Wu, C. J., Getz, G. & Hacohen, N. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell 160, 48–61 (2015).
Snyder, A. et al. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N. Engl. J. Med. 371, 2189–2199 (2014).
Hellmann, M. D. et al. Nivolumab plus ipilimumab in lung cancer with a high tumor mutational burden. N. Engl. J. Med. 378, 2093–2104 (2018).
Rosenberg, J. E. et al. Atezolizumab in patients with locally advanced and metastatic urothelial carcinoma who have progressed following treatment with platinum-based chemotherapy: a single-arm, multicentre, phase 2 trial. Lancet 387, 1909–1920 (2016).
Wu, Y. et al. The predictive value of tumor mutation burden on efficacy of immune checkpoint inhibitors in cancers: a systematic review and meta-analysis. Front. Oncol. 9, 1161 (2019).
Samstein, R. M. et al. Tumor mutational load predicts survival after immunotherapy across multiple cancer types. Nat. Genet. 51, 202–206 (2019).
Marabelle, A. et al. Association of tumour mutational burden with outcomes in patients with advanced solid tumours treated with pembrolizumab: prospective biomarker analysis of the multicohort, open-label, phase 2 KEYNOTE-158 study. Lancet Oncol. 21, 1353–1365 (2020).
Yarchoan, M., Hopkins, A. & Jaffee, E. M. Tumor mutational burden and response rate to PD-1 inhibition. N. Engl. J. Med. 377, 2500–2501 (2017).
Chan, T. A. et al. Development of tumor mutation burden as an immunotherapy biomarker: utility for the oncology clinic. Ann. Oncol. 30, 44–56 (2019).
Gandara, D. R. et al. Blood-based tumor mutational burden as a predictor of clinical benefit in non-small-cell lung cancer patients treated with atezolizumab. Nat. Med. 24, 1441–1448 (2018).
Yu, H. et al. Correlation of PD-L1 expression with tumor mutation burden and gene signatures for prognosis in early-stage squamous cell lung carcinoma. J. Thorac. Oncol. 14, 25–36 (2019).
Rizvi, H. et al. Molecular determinants of response to anti-programmed cell death (PD)-1 and anti-programmed death-ligand 1 (PD-L1) blockade in patients with non-small-cell lung cancer profiled with targeted next-generation sequencing. J. Clin. Oncol. 36, 633–641 (2018).
Sholl, L. M. et al. The promises and challenges of tumor mutation burden as an immunotherapy biomarker: a perspective from the international association for the study of lung cancer pathology committee. J. Thorac. Oncol. 15, 1409–1424 (2020).
Jardim, D. L., Goodman, A., de Melo Gagliato, D., Kurzrock, R. The challenges of tumor mutational burden as an immunotherapy biomarker. Cancer Cell 39, 154–173 (2021).
Li, G. M. Mechanisms and functions of DNA mismatch repair. Cell Res. 18, 85–98 (2008).
Buhard, O. et al. Multipopulation analysis of polymorphisms in five mononucleotide repeats used to determine the microsatellite instability status of human tumors. J. Clin. Oncol. 24, 241–251 (2006).
Le, D. T. et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science 357, 409–413 (2017).
Le, D. T. et al. PD-1 blockade in tumors with mismatch-repair deficiency. N. Engl. J. Med. 372, 2509–2520 (2015).
Cortes-Ciriano, I., Lee, S., Park, W. Y., Kim, T. M. & Park, P. J. A molecular portrait of microsatellite instability across multiple cancers. Nat. Commun. 8, 15180 (2017).
Chalmers, Z. R. et al. Analysis of 100,000 human cancer genomes reveals the landscape of tumor mutational burden. Genome Med. 9, 34 (2017).
Mehnert, J. M. et al. Immune activation and response to pembrolizumab in POLE-mutant endometrial cancer. J. Clin. Invest. 126, 2334–2340 (2016).
Gong, J., Wang, C., Lee, P. P., Chu, P. & Fakih, M. Response to PD-1 blockade in microsatellite stable metastatic colorectal cancer harboring a POLE mutation. J. Natl. Compr. Cancer Netw. 15, 142–147 (2017).
Goodman, A. M., Sokol, E. S., Frampton, G. M., Lippman, S. M. & Kurzrock, R. Microsatellite-stable tumors with high mutational burden benefit from immunotherapy. Cancer Immunol. Res. 7, 1570–1573 (2019).
Gettinger, S. et al. Impaired HLA class I antigen processing and presentation as a mechanism of acquired resistance to immune checkpoint inhibitors in lung cancer. Cancer Discov. 7, 1420–1435 (2017).
Chowell, D. et al. Patient HLA class I genotype influences cancer response to checkpoint blockade immunotherapy. Science 359, 582–587 (2018).
McGranahan, N. et al. Allele-specific HLA loss and immune escape in lung cancer evolution. Cell 171, 1259–1271.e1211 (2017).
Chowell, D. et al. Evolutionary divergence of HLA class I genotype impacts efficacy of cancer immunotherapy. Nat. Med. 25, 1715–1720 (2019).
Rodig, S. J. et al. MHC proteins confer differential sensitivity to CTLA-4 and PD-1 blockade in untreated metastatic melanoma. Sci. Transl. Med. 10, eaar3342 (2018).
Negrao, M. V. et al. PD-L1 expression, tumor mutational burden, and cancer gene mutations are stronger predictors of benefit from immune checkpoint blockade than HLA class I genotype in non-small cell lung cancer. J. Thorac. Oncol. 14, 1021–1031 (2019).
Spranger, S. et al. Up-regulation of PD-L1, IDO, and T(regs) in the melanoma tumor microenvironment is driven by CD8(+) T cells. Sci. Transl. Med. 5, 200ra116 (2013).
Garcia-Diaz, A. et al. Interferon receptor signaling pathways regulating PD-L1 and PD-L2 expression. Cell Rep. 19, 1189–1201 (2017).
Zaretsky, J. M. et al. Mutations associated with acquired resistance to PD-1 blockade in melanoma. N. Engl. J. Med. 375, 819–829 (2016).
Gao, J. et al. Loss of IFN-gamma pathway genes in tumor cells as a mechanism of resistance to anti-CTLA-4 therapy. Cell 167, 397–404 e399 (2016).
Shin, D. S. et al. Primary resistance to PD-1 blockade mediated by JAK1/2 mutations. Cancer Discov. 7, 188–201 (2017).
Pai, C. S. et al. Clonal deletion of tumor-specific T cells by interferon-gamma confers therapeutic resistance to combination immune checkpoint blockade. Immunity 50, 477–492 e478 (2019).
Ni, L. & Lu, J. Interferon gamma in cancer immunotherapy. Cancer Med. 7, 4509–4516 (2018).
Brown, Z. J. et al. Indoleamine 2,3-dioxygenase provides adaptive resistance to immune checkpoint inhibitors in hepatocellular carcinoma. Cancer Immunol. Immunother. 67, 1305–1315 (2018).
Long, G. V. et al. Epacadostat plus pembrolizumab versus placebo plus pembrolizumab in patients with unresectable or metastatic melanoma (ECHO-301/KEYNOTE-252): a phase 3, randomised, double-blind study. Lancet Oncol. 20, 1083–1097 (2019).
Tumeh, P. C. et al. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature 515, 568–571 (2014).
Mariathasan, S. et al. TGFbeta attenuates tumour response to PD-L1 blockade by contributing to exclusion of T cells. Nature 554, 544–548 (2018).
Uryvaev, A., Passhak, M., Hershkovits, D., Sabo, E. & Bar-Sela, G. The role of tumor-infiltrating lymphocytes (TILs) as a predictive biomarker of response to anti-PD1 therapy in patients with metastatic non-small cell lung cancer or metastatic melanoma. Med. Oncol. 35, 25 (2018).
Galon, J. et al. Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science 313, 1960–1964 (2006).
Mlecnik, B. et al. Comprehensive intrametastatic immune quantification and major impact of immunoscore on survival. J. Natl Cancer Inst. 110, https://doi.org/10.1093/jnci/djx123 (2018).
Kirilovsky, A. et al. Rational bases for the use of the Immunoscore in routine clinical settings as a prognostic and predictive biomarker in cancer patients. Int Immunol 28, 373–382 (2016).
Galon, J. et al. Cancer classification using the Immunoscore: a worldwide task force. J. Transl. Med. 10, 205 (2012).
Angell, H. & Galon, J. From the immune contexture to the Immunoscore: the role of prognostic and predictive immune markers in cancer. Curr. Opin. Immunol. 25, 261–267 (2013).
Galon, J. et al. Towards the introduction of the ‘Immunoscore’ in the classification of malignant tumours. J. Pathol. 232, 199–209 (2014).
Emens, L. A. & Middleton, G. The interplay of immunotherapy and chemotherapy: harnessing potential synergies. Cancer Immunol. Res. 3, 436–443 (2015).
Park, B., Yee, C. & Lee, K. M. The effect of radiation on the immune response to cancers. Int. J. Mol. Sci. 15, 927–943 (2014).
Ghiringhelli, F. et al. Metronomic cyclophosphamide regimen selectively depletes CD4+CD25+ regulatory T cells and restores T and NK effector functions in end stage cancer patients. Cancer Immunol. Immunother. 56, 641–648 (2007).
Suzuki, E., Kapoor, V., Jassar, A. S., Kaiser, L. R. & Albelda, S. M. Gemcitabine selectively eliminates splenic Gr-1+/CD11b+ myeloid suppressor cells in tumor-bearing animals and enhances antitumor immune activity. Clin. Cancer Res. 11, 6713–6721 (2005).
Murciano-Goroff, Y. R., Warner, A. B. & Wolchok, J. D. The future of cancer immunotherapy: microenvironment-targeting combinations. Cell Res. 30, 507–519 (2020).
Van Der Kraak, L. et al. 5-Fluorouracil upregulates cell surface B7-H1 (PD-L1) expression in gastrointestinal cancers. J. Immunother. Cancer 4, 65 (2016).
McDaniel, A. S. et al. Expression of PDL1 (B7-H1) before and after neoadjuvant chemotherapy in urothelial carcinoma. Eur. Urol. Focus 1, 265–268 (2016).
Zemek, R. M. et al. Sensitizing the tumor microenvironment to immune checkpoint therapy. Front. Immunol. 11, 223 (2020).
Ghoneim, H. E. et al. De novo epigenetic programs inhibit PD-1 blockade-mediated T cell rejuvenation. Cell 170, 142–157 e119 (2017).
Pauken, K. E. et al. Epigenetic stability of exhausted T cells limits durability of reinvigoration by PD-1 blockade. Science 354, 1160–1165 (2016).
Peng, D. et al. Epigenetic silencing of TH1-type chemokines shapes tumour immunity and immunotherapy. Nature 527, 249–253 (2015).
Goswami, S. et al. Modulation of EZH2 expression in T cells improves efficacy of anti-CTLA-4 therapy. J. Clin. Invest. 128, 3813–3818 (2018).
Xiao, G. et al. EZH2 negatively regulates PD-L1 expression in hepatocellular carcinoma. J. Immunother. Cancer 7, 300 (2019).
Wu, H. X. et al. Alteration in TET1 as potential biomarker for immune checkpoint blockade in multiple cancers. J. Immunother. Cancer 7, 264 (2019).
Okamura, R. et al. ARID1A alterations function as a biomarker for longer progression-free survival after anti-PD-1/PD-L1 immunotherapy. J. Immunother. Cancer 8, e000438 (2020).
Chiappinelli, K. B., Zahnow, C. A., Ahuja, N. & Baylin, S. B. Combining epigenetic and immunotherapy to combat cancer. Cancer Res. 76, 1683–1689 (2016).
Terranova-Barberio, M. et al. HDAC inhibition potentiates immunotherapy in triple negative breast cancer. Oncotarget 8, 114156–114172 (2017).
Geva-Zatorsky, N. et al. Mining the human gut microbiota for immunomodulatory organisms. Cell 168, 928–943 e911 (2017).
Vetizou, M. et al. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science 350, 1079–1084 (2015).
Sivan, A. et al. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science 350, 1084–1089 (2015).
Routy, B. et al. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science 359, 91–97 (2018).
Baruch, E. N. et al. Fecal microbiota transplant promotes response in immunotherapy-refractory melanoma patients. Science 371, 602–609 (2021).
Chaput, N. et al. Baseline gut microbiota predicts clinical response and colitis in metastatic melanoma patients treated with ipilimumab. Ann. Oncol. 28, 1368–1379 (2017).
Matson, V. et al. The commensal microbiome is associated with anti-PD-1 efficacy in metastatic melanoma patients. Science 359, 104–108 (2018).
Jin, Y. et al. The diversity of gut microbiome is associated with favorable responses to anti-programmed death 1 immunotherapy in Chinese patients with NSCLC. J. Thorac. Oncol. 14, 1378–1389 (2019).
Zitvogel, L., Ma, Y., Raoult, D., Kroemer, G. & Gajewski, T. F. The microbiome in cancer immunotherapy: diagnostic tools and therapeutic strategies. Science 359, 1366–1370 (2018).
Inamura, K. Roles of microbiota in response to cancer immunotherapy. Semin. Cancer Biol. 65, 164–175 (2020).
Weide, B. et al. Baseline biomarkers for outcome of melanoma patients treated with pembrolizumab. Clin. Cancer Res. 22, 5487–5496 (2016).
Heppt, M. V. et al. Prognostic factors and outcomes in metastatic uveal melanoma treated with programmed cell death-1 or combined PD-1/cytotoxic T-lymphocyte antigen-4 inhibition. Eur. J. Cancer 82, 56–65 (2017).
Martens, A. et al. Baseline peripheral blood biomarkers associated with clinical outcome of advanced melanoma patients treated with ipilimumab. Clin. Cancer Res. 22, 2908–2918 (2016).
Shao, Y. et al. Prognostic value of pretreatment neutrophil-to-lymphocyte ratio in renal cell carcinoma: a systematic review and meta-analysis. BMC Urol. 20, 90 (2020).
Huang, A. C. et al. T-cell invigoration to tumour burden ratio associated with anti-PD-1 response. Nature 545, 60–65 (2017).
Wu, X. et al. Angiopoietin-2 as a biomarker and target for immune checkpoint therapy. Cancer Immunol. Res. 5, 17–28 (2017).
Turiello, R. et al. Serum CD73 is a prognostic factor in patients with metastatic melanoma and is associated with response to anti-PD-1 therapy. J. Immunother. Cancer 8, e001689 (2020).
Morello, S. et al. Soluble CD73 as biomarker in patients with metastatic melanoma patients treated with nivolumab. J. Transl. Med. 15, 244 (2017).
Hannani, D. et al. Anticancer immunotherapy by CTLA-4 blockade: obligatory contribution of IL-2 receptors and negative prognostic impact of soluble CD25. Cell Res. 25, 208–224 (2015).
Maccalli, C. et al. Soluble NKG2D ligands are biomarkers associated with the clinical outcome to immune checkpoint blockade therapy of metastatic melanoma patients. Oncoimmunology 6, e1323618 (2017).
Maccalli, C. et al. Immunological markers and clinical outcome of advanced melanoma patients receiving ipilimumab plus fotemustine in the NIBIT-M1 study. Oncoimmunology 5, e1071007 (2016).
Daassi, D., Mahoney, K. M. & Freeman, G. J. The importance of exosomal PDL1 in tumour immune evasion. Nat. Rev. Immunol. 20, 209–215 (2020).
Yin, Z. et al. Mechanisms underlying low-clinical responses to PD-1/PD-L1 blocking antibodies in immunotherapy of cancer: a key role of exosomal PD-L1. J. Immunother. Cancer 9, e001698 (2021).
Chen, G. et al. Exosomal PD-L1 contributes to immunosuppression and is associated with anti-PD-1 response. Nature 560, 382–386 (2018).
Lee, J. H. et al. Circulating tumour DNA predicts response to anti-PD1 antibodies in metastatic melanoma. Ann. Oncol. 28, 1130–1136 (2017).
Prelaj, A. et al. EPSILoN: a prognostic score using clinical and blood biomarkers in advanced non-small-cell lung cancer treated with immunotherapy. Clin. Lung Cancer 21, 365–377 e365 (2020).
Nowicki, T. S., Hu-Lieskovan, S. & Ribas, A. Mechanisms of resistance to PD-1 and PD-L1 blockade. Cancer J. 24, 47–53 (2018).
Sharma, P., Hu-Lieskovan, S., Wargo, J. A. & Ribas, A. Primary, adaptive, and acquired resistance to cancer immunotherapy. Cell 168, 707–723 (2017).
Groth, C. et al. Immunosuppression mediated by myeloid-derived suppressor cells (MDSCs) during tumour progression. Br. J. Cancer 120, 16–25 (2019).
Highfill, S. L. et al. Disruption of CXCR2-mediated MDSC tumor trafficking enhances anti-PD1 efficacy. Sci. Transl. Med. 6, 237ra267 (2014).
Arce Vargas, F. et al. Fc-optimized anti-CD25 depletes tumor-infiltrating regulatory T cells and synergizes with PD-1 blockade to eradicate established tumors. Immunity 46, 577–586 (2017).
Taylor, N. A. et al. Treg depletion potentiates checkpoint inhibition in claudin-low breast cancer. J. Clin. Invest. 127, 3472–3483 (2017).
Bankhead, P. et al. QuPath: open source software for digital pathology image analysis. Sci. Rep. 7, 16878 (2017).
Spranger, S., Bao, R. & Gajewski, T. F. Melanoma-intrinsic beta-catenin signalling prevents anti-tumour immunity. Nature 523, 231–235 (2015).
Loi, S. et al. RAS/MAPK activation is associated with reduced tumor-infiltrating lymphocytes in triple-negative breast cancer: therapeutic cooperation between MEK and PD-1/PD-L1 immune checkpoint inhibitors. Clin. Cancer Res. 22, 1499–1509 (2016).
Liu, C. et al. BRAF inhibition increases tumor infiltration by T cells and enhances the antitumor activity of adoptive immunotherapy in mice. Clin. Cancer Res. 19, 393–403 (2013).
Koyama, S. et al. Adaptive resistance to therapeutic PD-1 blockade is associated with upregulation of alternative immune checkpoints. Nat. Commun. 7, 10501 (2016).
Gao, J. et al. VISTA is an inhibitory immune checkpoint that is increased after ipilimumab therapy in patients with prostate cancer. Nat. Med. 23, 551–555 (2017).
Kakavand, H. et al. Negative immune checkpoint regulation by VISTA: a mechanism of acquired resistance to anti-PD-1 therapy in metastatic melanoma patients. Mod. Pathol. 30, 1666–1676 (2017).
Lu, S. et al. Comparison of biomarker modalities for predicting response to PD-1/PD-L1 checkpoint blockade: a systematic review and meta-analysis. JAMA Oncol. 5, 1195–1204 (2019).
Vaidya, P. et al. Novel, non-invasive imaging approach to identify patients with advanced non-small cell lung cancer at risk of hyperprogressive disease with immune checkpoint blockade. J. Immunother. Cancer 8, e001343 (2020).
Mo, Q., Li, R., Adeegbe, D. O., Peng, G. & Chan, K. S. Integrative multi-omics analysis of muscle-invasive bladder cancer identifies prognostic biomarkers for frontline chemotherapy and immunotherapy. Commun. Biol. 3, 784 (2020).
Linette, G. P. & Carreno, B. M. Tumor-infiltrating lymphocytes in the checkpoint inhibitor era. Curr. Hematol. Malig. Rep. 14, 286–291 (2019).
Acknowledgements
The authors thank their colleagues at the University of Liege for their helpful discussions. C.P. and M.A. are Televie fellows.
Author information
Authors and Affiliations
Contributions
C.P., M.A. and M.H. reviewed the literature and wrote the manuscript. C.P., M.A. and M.H. generated the figures. G.J., P.D. and P.H. provided edits and approved the final version.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent to publish
Not applicable.
Data availability
Not applicable.
Competing interests
C.P., M.A., P.D., P.H. and M.H. have no conflict of interest to declare. G.J. reports grants, personal fees and/or non-financial support from Novartis, Roche, Pfizer, Lilly, Amgen, Bristol-Myers Squibb, Astra-Zeneca, Daiichi Sankyo, Abbvie, Medimmune and MerckKGaA (not directly related to the submitted work).
Funding information
M.H. is a Research Associate at the Belgian Fund for Scientific Research (FNRS). This work was supported in part by the FNRS (MIS F.4520.20; CDR/OL J.0111.20), the University of Liege (Crédits Sectoriels de Recherche en Sciences de la Santé 2018-2020), the Télévie (PDR Televie 7.8507.19) and the Leon Fredericq Foundation.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Pilard, C., Ancion, M., Delvenne, P. et al. Cancer immunotherapy: it’s time to better predict patients’ response. Br J Cancer 125, 927–938 (2021). https://doi.org/10.1038/s41416-021-01413-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41416-021-01413-x
This article is cited by
-
Improved automated one-pot two-step radiosynthesis of (S)-[18F]FETrp, a radiotracer for PET imaging of indoleamine 2,3-dioxygenase 1 (IDO1)
EJNMMI Radiopharmacy and Chemistry (2024)
-
A whirl of radiomics-based biomarkers in cancer immunotherapy, why is large scale validation still lacking?
npj Precision Oncology (2024)
-
NF1 mutations as biomarker of response to immune checkpoint blockades for lung adenocarcinoma patients
npj Precision Oncology (2024)
-
Current approaches in glioblastoma multiforme immunotherapy
Clinical and Translational Oncology (2024)
-
Longitudinal plasma proteomic analysis identifies biomarkers and combinational targets for anti-PD1-resistant cancer patients
Cancer Immunology, Immunotherapy (2024)