Abstract
Background
Most patients with hormone receptor (HR)-positive, human epidermal growth factor receptor type 2 (HER2)-negative breast cancer can be cured by surgery and endocrine therapy, but a significant proportion suffer recurrences. Actinin-4 is associated with cancer invasion and metastasis, and its genetic alteration may be used for breast cancer prognostication.
Methods
The copy number of the actinin-4 (ACTN4) gene was determined by fluorescence in situ hybridisation (FISH) in two independent cohorts totalling 597 patients (336 from Japan and 261 from the USA) with HR-positive, HER2-negative, node-negative breast cancer.
Results
In the Japanese cohort, multivariate analysis revealed that a copy number increase (CNI) of ACTN4 was an independent factor associated with high risks of recurrence (P = 0.01; hazard ratio (HR), 2.95) and breast cancer death (P = 0.014; HR, 4.27). The prognostic significance of ACTN4 CNI was validated in the US cohort, where it was the sole prognostic factor significantly associated with high risks of recurrence (P = 0.04; HR, 2.73) and death (P = 0.016; HR, 4.01).
Conclusions
Copy number analysis of a single gene, ACTN4, can identify early-stage luminal breast cancer patients with a distinct outcome. Such high-risk patients may benefit from adjuvant chemotherapy.
Similar content being viewed by others
Background
In most developed countries, the breast cancer mortality rate has declined over the last few decades1 owing to the development of new screening, diagnostic and therapeutic modalities. However, breast cancer is a heterogeneous disease with a wide spectrum of biological behaviour, and a significant proportion of patients still suffer recurrences and die of the disease. On the basis of intrinsic molecular characteristics, breast cancer is classified into at least four subtypes: the HER2-enriched, basal-like, luminal A, and luminal B (HER2-positive and -negative) subtypes.2 This classification is closely associated with various pathological features, response to therapeutics and prognosis.
The luminal A subtype typically expresses a high level of oestrogen receptor (ER)/progesterone receptor (PgR) and is histologically low-grade, characterised by low cell proliferation. Luminal B tumours are ER-positive, but may have variable degrees of ER/PgR expression; histologically, they are high-grade and have high cell proliferation.3 Approximately 20% of luminal B tumours are HER2-positive and treated with anti-HER2 therapeutics, but patients with luminal B/HER2-negative tumours need to be accurately diagnosed and receive adjuvant chemotherapy. Luminal tumours were originally defined by gene expression profiling,4 but the distinction between luminal A- and B-like tumours is most commonly made by Ki-67 immunostaining. However, problems such as inter-observer or inter-institutional variation and lack of an optimal cut-off value make Ki-67 immunostaining unreliable.5,6 Several commercially available multi-gene assays including the 21-gene recurrence score (Oncotype DX) and 70-gene signature (MammaPrint) assays are well standardised and more accurate for predicting the outcome of axillary node-negative luminal breast tumours.7 However, these multi-gene assays are expensive,8 and the development of a more cost-effective single gene assay would be desirable.
We originally identified actinin-4 as an actin-binding protein closely associated with cell motility and cancer invasion.9 Forced expression of actinin-4 increased cell motility and promoted metastasis, whereas knockdown of actinin-4 suppressed the invasiveness of cancer cells.9,10,11,12,13 Amplification of the ACTN4 gene was detected in a small (8–16%) but substantial subset of stage I (node-negative) lung adenocarcinomas with a markedly unfavourable outcome.14 We and others have shown that amplification/overexpression of the ACTN4 gene/actinin-4 protein is a significant prognostic factor associated with poor outcome in patients with colorectal,15 pancreatic,11 ovarian,16,17 salivary gland18 and high-grade neuroendocrine pulmonary tumours.19
Luminal B breast cancer has a unique profile of gene copy number alterations.2 Gene amplifications and other gross chromosomal aberrations are frequently detected in luminal B tumours. We have hypothesised that an increase in the copy number of the ACTN4 gene might also identify luminal B-like/HER2-negative breast cancers with a high risk of recurrence. Here we report for the first time that ACTN4 can be used as a highly specific single genetic biomarker to identify high-risk patients with HR-positive, HER2-negative, node-negative invasive breast cancer.
Methods
Patients and tissue microarrays
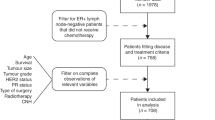
The use of human samples for this study was reviewed and approved by the Institutional Review Board of the National Cancer Center (Tokyo, Japan). We constructed tissue microarrays (TMAs) from formalin-fixed, paraffin-embedded tissue blocks that had been prepared for routine pathological diagnosis of 709 primary breast cancers surgically resected at the National Cancer Center Hospital (NCCH) (Tokyo, Japan) between 1996 and 2000 and pathologically diagnosed as node-negative, using a tissue microarrayer (Azumaya, Tokyo, Japan) as described previously.6 The status of ER, PpR, HER2 and Ki-67 was determined by immunohistochemistry and FISH, as described previously.6 Histological and nuclear grading was performed as described previously.6 Among the 709 cases, there were 369 HR-positive, HER2-negative invasive breast cancers (Fig. 1). Pathological staging was performed according to the Unio Internationalis Contra Cancrum (UICC) Tumor-Node-Metastasis (TNM) classification (7th edition, 2009).
We obtained a set of 10 TMAs containing 550 tissue cores of node-negative breast cancers (Breast Stage I Prognostic TMA) from the Cooperative Human Tissue Network (CHTN) through the Cancer Diagnosis Program (CDP) of the National Cancer Institute (NCI).20 The TMAs included 330 patients with HR-positive, HER2-negative, node-negative invasive breast cancer (Supplementary Fig. S1).
FISH
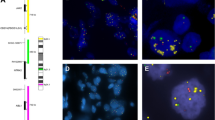
Bacterial artificial chromosome (BAC) clones containing the ACTN4 gene and centromere of chromosome 19 were isolated as described previously,14 and Good Manufacturing Practice (GMP)-grade FISH probes were prepared by Abnova (Taipei, Taiwan). TMAs were hybridised with the FISH probes at 37 °C for 48 h. The nuclei were counterstained with 4,6-diamidino-2-phenylindole (DAPI). The numbers of fluorescence signals of ACTN4 (red) and centromere (green) in the nuclei of 20 interphase tumour cells were counted by an investigator blinded to the clinical data. The FISH patterns with gene amplification (≥2.0-fold increase relative to the centromere) and high polysomy (≥5 mean copies per cell) were defined as having an ACTN4 copy number increase (CNI), and all others were defined as having a normal copy number (NCN) according to the standard Colorado criteria21 (Fig. 2).
Statistical analyses
The statistical significance of correlations was examined using Fisher’s exact test. Breast cancer-specific survival (BCSS) was measured as the interval from surgery to the date of breast cancer death or last follow-up. Disease-free survival (DFS) was defined as the length of time from surgery to the first detection of new lesions. BCSS and DFS were estimated by the Kaplan–Meier method and compared by log-rank test. Univariate and multivariate analyses were carried out using the Cox regression model. All statistical analyses were performed with EZR (Saitama Medical Center, Jichi Medical University, Saitama, Japan), a graphical user interface for R (The R Foundation for Statistical Computing, Vienna, Austria).22 Differences were considered to be significant at P < 0.05.
Results
Patient characteristics
As the TMAs had been used in our previous study,6 several tissue cores had worn out or peeled off from the slides. Among the 369 HR-positive, HER2-negative, node-negative invasive breast cancers in the Japanese cohort, 336 samples were evaluable by FISH. A total of 36 recurrences and 13 deaths from breast cancer occurred during the median follow-up period of 118.1 (range, 0.6–160.3) months. The age of the patients ranged from 27 to 85 years (median 57 years). Other clinicopathological characteristics of the patients are shown in Table 1.
Among the 336 cases, high polysomy was detected in 6 patients (1.7%) and gene amplification was evident in 17 patients (5.1%). A total of 23 patients (6.8%) were defined as having ACTN4 CNI, and the remaining 313 patients (93.2%) were defined as having NCN. ACTN4 gene status was not significantly associated with age, menopausal status, area of invasive tumour, histology, histological grade, lymphovascular invasion or Ki-67 status (Table 1).
Prognostic impact of ACTN4 CNI in breast cancers
ACTN4 CNI was significantly correlated (P = 0.0008, log-rank test) with poor DFS in the 336 patients with HR-positive, HER2-negative, lymph node-negative invasive breast cancer (Fig. 3a). The 10-year DFS rates for patients with ACTN4 NCN and CNI were 89.1% and 65.5%, respectively. Furthermore, BCSS of patients with CNI was significantly worse than that of patients without CNI (P = 0.0009, log-rank test) (Fig. 3b). The 10-year BCSS rates for patients with NCN and CNI were 96.5% and 79.1%, respectively. There were 249 stage I and 87 stage II HR-positive, HER2-negative, node-negative patients. ACTN4 CNI was associated with significantly poor DFS and BSS in both stages (Supplementary Figs. S2 and S3).
Kaplan–Meier estimate of DFS (a) and BCSS (b) of patients with HR-positive, HER2-negative, node-negative invasive breast cancer carrying (red) and not carrying (black) ACTN4 CNI. Differences between the curves were assessed using the log-rank test. This figure has originally appeared in an abstract of the 36th annual meeting of the Japan Society for Molecular Tumor Marker Research and is reproduced with permission from the society.
ACTN4 CNI was not significantly associated with poor outcome in patients with HER2-positive or triple-negative breast cancer (Supplementary Figs. S4 and S5).
Multivariate analysis
We calculated the hazard ratios (HR) of selected parameters including patient age, menopausal status, area of invasive tumour, histology, histological grade, nuclear grade, lymphovascular invasion, Ki-67 status, and ACTN4 copy number status for recurrence and death using univariate and multivariate Cox regression analyses. Univariate analysis revealed that histological grade (P = 0.006; HR, 2.52; 95% confidence interval [CI]: 1.3–4.86), nuclear grade (P = 0.005, HR, 2.57; 95% CI: 1.33–4.94), and ACTN4 FISH (P = 0.002, HR, 3.62; 95% CI: 1.59–8.27) were significantly correlated with recurrence of breast cancer. Histological and nuclear grades are mutually correlated. Multivariate analysis revealed that the copy number status of ACTN4 (P = 0.01, HR, 2.95; 95% CI: 1.27–6.83) was the sole independent risk factor for recurrence (Table 2).
Univariate analysis revealed that histological grade (P = 0.0004; HR, 10.16; 95% CI: 2.81–37.1), nuclear grade (P = 0.002; HR, 7.52; 95% CI: 2.07–27.34), and ACTN4 copy number status (P = 0.0035; HR, 5.78; 95% CI: 1.78–18.77) were significantly associated with breast cancer-associated death. Histological and nuclear grades are mutually correlated. Multivariate analysis revealed that ACTN4 copy number status (P = 0.014; HR, 4.27; 95% CI: 1.3–14.05) was the sole independent risk factor for death (Table 3).
Validation cohort
In order to validate the universality of the above results obtained by examining a Japanese cohort, we examined another patient cohort from a different country (the USA) using FISH. This US cohort included 261 tissue samples from patients with HR-positive, HER2-negative, node-negative invasive breast cancers. The clinicopathological characteristics of this US cohort and the correlations with ACTN4 CNI are shown in Supplementary Table S1.
ACTN4 CNI was significantly associated with histological (P = 0.024) and nuclear (P = 0.021) grades. Kaplan–Meier analysis revealed that patients with ACTN4 CNI had a significantly poorer outcome than those with ACTN4 NCN in terms of both DFS and BCSS (P = 0.035 and 0.009, respectively, log-rank test) (Fig. 4). The 10-year BCSS rates for patients with ACTN4 NCN and CNI were 97.2% and 80.9%, respectively. Univariate Cox regression analysis showed that ACTN4 CNI was the sole factor significantly associated with high risks of recurrence (P = 0.04; HR, 2.73; 95% CI 1.03–7.24) and breast cancer death (P = 0.016; HR, 4.01; 95% CI: 1.29–12.49) (Supplementary Tables S2 and S3).
Discussion
HR-positive, HER2-negative, node-negative invasive breast cancers comprise a heterogeneous population, and a significant proportion of patients suffer recurrences even after complete surgical resection of their primary tumours. Therefore, it is important to establish a method of identifying high-risk patients for whom adjuvant chemotherapy would be necessary. ER and PgR status is determined by IHC, HER2 status is determined by IHC and/or FISH,3 and the tumour proliferative fraction is most commonly assessed by Ki-67 immunostaining, but the 15th St. Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2017 (St. Gallen 2017) raised an issue of caution about the reproducibility of IHC for Ki67 and its use for clinical decision-making. We previously examined the prognostic impact of 3 different Ki-67 cut-off values (10%, 14 and 20%), but no absolute cut-off value was found to reproducibly stratify patients with node-negative luminal breast cancer.6 St. Gallen 2017 alternatively recommended the use of multi-gene molecular assays to obtain accurate prognostic information.3 These multi-gene assays were reproducible across institutions and had been successfully incorporated into several clinical trials of adjuvant chemotherapy.23,24,25 However, their cost and complexity preclude their worldwide application to routine oncological practice.
In the present study, we examined the feasibility of ACTN4 copy number status as a novel single-gene assay for stratification of node-negative luminal breast cancer. We first examined the genetic status of ACTN4 in 336 HR (ER/I)-positive, lymph node-negative breast cancers that had been resected in Japan. The prognostic significance was then subjected to independent validation in a large cohort of patients with node-negative (Stage I) luminal breast cancer treated in the USA. The result was highly reproducible across the two countries with different racial backgrounds. The 10-year DFS rates for patients with NCN and CNI of the ACTN4 gene were 89.1% and 65.5%, respectively, in Japan and 89.8% and 76.3%, respectively, in the USA. The rates of patients with ACTN4 CNI seem comparable to those of high-risk patients categorised by the 21-gene recurrence score (31 or higher).7 The NSABP B-20 prospective-retrospective study revealed that addition of chemotherapy to tamoxifen had therapeutic benefits for patients with node-negative, ER-positive breast cancer with a 21-gene recurrence score of ≥31,26 indicating that patients with node-negative luminal breast cancer carrying ACTN4 CNI might similarly benefit from postoperative adjuvant chemotherapy.
Metastasis is the major form of cancer recurrence. The actinin-4 protein is highly concentrated at the leading edges of actin-rich cell protrusions and directly regulates cell motility through remodelling of the actin cytoskeleton.15 It has been shown that the expression of actinin-4 in focally dedifferentiated cancer cells at the invasive front is significantly correlated with metastasis.15 Epithelial-mesenchymal transition (EMT) is an essential step in cancer metastasis.27 Actinin-4 overexpression changes cell morphology from epithelial-like to mesenchymal-like28 and promotes cancer cell migration and invasion through the expression of Snail and stabilised β-catenin.29 Therefore, molecular therapeutics targeting actinin-4 would likely inhibit cancer metastasis and cure patients with a high risk of recurrence. We recently revealed that a small-molecule Traf2 and Nck-interacting kinase (TNIK) inhibitor, NCB-0846,30 suppressed the EMT and metastasis of non-small cell lung cancer cells through post-translational modification of actinin-4 (unpublished observation). This compound could be applicable for prevention of recurrence in breast cancer patients with ACTN4 CNI.
ER has been considered one of the major therapeutic targets in HR-positive breast cancer. In fact, adjuvant administration of tamoxifen to HR-positive breast cancer patients reduces the rates of recurrence and mortality.31 Oestrogen binds to ER in the cytoplasm and translocates into the nucleus, where it binds to DNA sequences and promotes mammary cell differentiation and proliferation. In the nucleus, actinin-4 is known to function as a co-activator of several transcriptional factors, independently from its cytoplasmic actin-binding function.32,33 Khurana et al. showed that overexpression of ACTN4 activated ERα-mediated transcription activity, whereas knockdown of ACTN4 inhibited oestrogen-mediated cancer cell proliferation.34 ER signalling may be activated in patients with ACTN4 CNI and promote the aggressiveness of HR-positive tumours. Consistently, ACTN4 CNI was not significantly associated with poor outcome in patients with HER2-positive or triple-negative breast cancer (Supplementary Figs. S4 and S5).
This study had a few limitations. The first is that it was retrospective in design. However, this study was carefully performed in accordance with the Reporting Recommendations for Tumor Marker Prognostic Studies (REMARK),35 and we have provided diagrams (Fig. 1 and Supplementary Fig. S1) to clearly indicate the numbers of individuals included at different stages, and incorporated into univariate and multivariable analyses. Second, the treatment of patients in the Japanese cohort was not based on a pre-fixed protocol but determined empirically by their attending physicians. Purely for this reason, we did not analyse the effects of different therapeutics in patients with and without ACTN4 CNI. Third, we re-used TMAs that had been prepared for a previous study,6 and therefore 33 tissue cores (out of 369) were missing and not evaluable (Fig. 1). However, a significant number of tissue cores were found to be intact, and this random case exclusion was unlikely to have affected the overall results.
We conclude that ACTN4 copy number analysis is a cost-effective single gene assay and can be potentially applicable for selection of high-risk patients with HR-positive, HER2-negative, lymph node-negative invasive breast cancer. Its prognostic impact is significant and reproducible, warranting further independent validation and clinical trials by other investigators.
References
DeSantis, C., Ma, J., Bryan, L. & Jemal, A. Breast cancer statistics, 2013. CA Cancer J. Clin. 64, 52–62 (2014).
Ades, F., Zardavas, D., Bozovic-Spasojevic, I., Pugliano, L., Fumagalli, D., de Azambuja, E. et al. Luminal B breast cancer: molecular characterization, clinical management, and future perspectives. J. Clin. Oncol. 32, 2794–2803 (2014).
Curigliano, G., Burstein, H. J., E, P. W., Gnant, M., Dubsky, P., Loibl, S. et al. De-escalating and escalating treatments for early-stage breast cancer: the St. Gallen International Expert Consensus Conference on the Primary Therapy of Early Breast Cancer 2017. Ann. Oncol. 28, 1700–1712 (2017).
Sorlie, T., Perou, C. M., Tibshirani, R., Aas, T., Geisler, S., Johnsen, H. et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc. Natl. Acad. Sci. USA 98, 10869–10874 (2001).
Dowsett, M., Nielsen, T. O., A’Hern, R., Bartlett, J., Coombes, R. C., Cuzick, J. et al. Assessment of Ki67 in breast cancer: recommendations from the International Ki67 in Breast Cancer working group. J. Natl. Cancer Inst. 103, 1656–1664 (2011).
Ono, M., Tsuda, H., Yunokawa, M., Yonemori, K., Shimizu, C., Tamura, K. et al. Prognostic impact of Ki-67 labeling indices with 3 different cutoff values, histological grade, and nuclear grade in hormone-receptor-positive, HER2-negative, node-negative invasive breast cancers. Breast Cancer 22, 141–152 (2015).
Paik, S., Shak, S., Tang, G., Kim, C., Baker, J., Cronin, M. et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N. Engl. J. Med 351, 2817–2826 (2004).
Wang, S. Y., Chen, T., Dang, W., Mougalian, S. S., Evans, S. B. & Gross, C. P. Incorporating tumor characteristics to maximize 21-gene assay utility: a cost-effectiveness analysis. J. Natl. Compr. Canc Netw. 17, 39–46 (2019).
Honda, K., Yamada, T., Endo, R., Ino, Y., Gotoh, M., Tsuda, H. et al. Actinin-4, a novel actin-bundling protein associated with cell motility and cancer invasion. J. Cell Biol. 140, 1383–1393 (1998).
Hayashida, Y., Honda, K., Idogawa, M., Ino, Y., Ono, M., Tsuchida, A. et al. E-cadherin regulates the association between β-catenin and actinin-4. Cancer Res 65, 8836–8845 (2005).
Kikuchi, S., Honda, K., Tsuda, H., Hiraoka, N., Imoto, I., Kosuge, T. et al. Expression and gene amplification of actinin-4 in invasive ductal carcinoma of the pancreas. Clin. Cancer Res 14, 5348–5356 (2008).
Barbolina, M. V., Adley, B. P., Kelly, D. L., Fought, A. J., Scholtens, D. M., Shea, L. D. et al. Motility-related actinin α-4 is associated with advanced and metastatic ovarian carcinoma. Lab Invest 88, 602–614 (2008).
Yamada, S., Yanamoto, S., Yoshida, H., Yoshitomi, I., Kawasaki, G., Mizuno, A. et al. RNAi-mediated down-regulation of α-actinin-4 decreases invasion potential in oral squamous cell carcinoma. Int J. Oral. Maxillofac. Surg. 39, 61–67 (2010).
Noro, R., Honda, K., Tsuta, K., Ishii, G., Maeshima, A. M., Miura, N. et al. Distinct outcome of stage I lung adenocarcinoma with ACTN4 cell motility gene amplification. Ann. Oncol. 24, 2594–2600 (2013).
Honda, K., Yamada, T., Hayashida, Y., Idogawa, M., Sato, S., Hasegawa, F. et al. Actinin-4 increases cell motility and promotes lymph node metastasis of colorectal cancer. Gastroenterology 128, 51–62 (2005).
Yamamoto, S., Tsuda, H., Honda, K., Kita, T., Takano, M., Tamai, S. et al. Actinin-4 expression in ovarian cancer: a novel prognostic indicator independent of clinical stage and histological type. Mod. Pathol. 20, 1278–1285 (2007).
Yamamoto, S., Tsuda, H., Honda, K., Onozato, K., Takano, M., Tamai, S. et al. Actinin-4 gene amplification in ovarian cancer: a candidate oncogene associated with poor patient prognosis and tumor chemoresistance. Mod. Pathol. 22, 499–507 (2009).
Watabe, Y., Mori, T., Yoshimoto, S., Nomura, T., Shibahara, T., Yamada, T. et al. Copy number increase of ACTN4 is a prognostic indicator in salivary gland carcinoma. Cancer Med 3, 613–622 (2014).
Miyanaga, A., Honda, K., Tsuta, K., Masuda, M., Yamaguchi, U., Fujii, G. et al. Diagnostic and prognostic significance of the alternatively spliced ACTN4 variant in high-grade neuroendocrine pulmonary tumours. Ann. Oncol. 24, 84–90 (2013).
Taube, S. E., Jacobson, J. W. & Lively, T. G. Cancer diagnostics: decision criteria for marker utilization in the clinic. Am. J. Pharmacogenomics 5, 357–364 (2005).
Cappuzzo, F., Marchetti, A., Skokan, M., Rossi, E., Gajapathy, S., Felicioni, L. et al. Increased MET gene copy number negatively affects survival of surgically resected non-small-cell lung cancer patients. J. Clin. Oncol. 27, 1667–1674 (2009).
Kanda, Y. Investigation of the freely available easy-to-use software ‘EZR’ for medical statistics. Bone Marrow Transpl. 48, 452–458 (2013).
Krop, I., Ismaila, N., Andre, F., Bast, R. C., Barlow, W., Collyar, D. E. et al. Use of biomarkers to guide decisions on adjuvant systemic therapy for women with early-stage invasive breast cancer: American Society of Clinical Oncology Clinical Practice Guideline Focused Update. J. Clin. Oncol. 35, 2838–2847 (2017).
Cardoso, F., van’t Veer, L. J., Bogaerts, J., Slaets, L., Viale, G., Delaloge, S. et al. 70-gene signature as an aid to treatment decisions in early-stage breast cancer. N. Engl. J. Med. 375, 717–729 (2016).
Sparano, J. A., Gray, R. J., Makower, D. F., Pritchard, K. I., Albain, K. S., Hayes, D. F. et al. Adjuvant chemotherapy guided by a 21-gene expression assay in breast cancer. N. Engl. J. Med 379, 111–121 (2018).
Paik, S., Tang, G., Shak, S., Kim, C., Baker, J., Kim, W. et al. Gene expression and benefit of chemotherapy in women with node-negative, estrogen receptor-positive breast cancer. J. Clin. Oncol. 24, 3726–3734 (2006).
Chaffer, C. L. & Weinberg, R. A. A perspective on cancer cell metastasis. Science 331, 1559–1564 (2011).
Wang, M. C., Chang, Y. H., Wu, C. C., Tyan, Y. C., Chang, H. C., Goan, Y. G. et al. α-Actinin 4 is associated with cancer cell motility and is a potential biomarker in non-small cell lung cancer. J. Thorac. Oncol. 10, 286–301 (2015).
An, H. T., Yoo, S. & Ko, J. α-Actinin-4 induces the epithelial-to-mesenchymal transition and tumorigenesis via regulation of Snail expression and β-catenin stabilization in cervical cancer. Oncogene 35, 5893–5904 (2016).
Masuda, M., Uno, Y., Ohbayashi, N., Ohata, H., Mimata, A., Kukimoto-Niino, M. et al. TNIK inhibition abrogates colorectal cancer stemness. Nat. Commun. 7, 12586 (2016).
Davies, C., Godwin, J., Gray, R., Clarke, M., Cutter, D., Darby, S. et al. Relevance of breast cancer hormone receptors and other factors to the efficacy of adjuvant tamoxifen: patient-level meta-analysis of randomised trials. Lancet 378, 771–784 (2011).
Aksenova, V., Turoverova, L., Khotin, M., Magnusson, K. E., Tulchinsky, E., Melino, G. et al. Actin-binding protein α-actinin 4 (ACTN4) is a transcriptional co-activator of RelA/p65 sub-unit of NF-kB. Oncotarget 4, 362–372 (2013).
Zhao, X., Hsu, K. S., Lim, J. H., Bruggeman, L. A. & Kao, H. Y. α-Actinin 4 potentiates nuclear factor kappa-light-chain-enhancer of activated B-cell (NF-kappaB) activity in podocytes independent of its cytoplasmic actin binding function. J. Biol. Chem. 290, 338–349 (2015).
Khurana, S., Chakraborty, S., Cheng, X., Su, Y. T. & Kao, H. Y. The actin-binding protein, actinin α4 (ACTN4), is a nuclear receptor coactivator that promotes proliferation of MCF-7 breast cancer cells. J. Biol. Chem. 286, 1850–1859 (2011).
McShane, L. M., Altman, D. G., Sauerbrei, W., Taube, S. E., Gion, M. & Clark, G. M. REporting recommendations for tumour MARKer prognostic studies (REMARK). Br. J. Cancer 93, 387–391 (2005).
Sugano, T., Yoshida, M., Masuda, M., Ono, M., Kakuya, T., Goto, N. et al. Distinct prognostic impact of ACTN4 gene copy number increase in hormone receptor-positive, HER2-negative, node-negative invasive breast cancers. Proc. 36th Annu. Meet. Jpn. Soc. Mol. Tumor Marker Res. 32, 23–24 (2017).
Acknowledgements
The provisional data of this study were orally presented in the 36th annual meeting of the Japan Society for Molecular Tumor Marker Research held on October 5, 2016 in Yokohama, Japan, and its meeting abstract36 is available online in the following Web site: https://www.jstage.jst.go.jp/article/jsmtmr/32/0/32_23/_article/-char/ja/.
Author information
Authors and Affiliations
Contributions
T.S., M.M. and T.Y. conceived the study concept. M.Y., T.S., M.O., T.K., and H.T. provided materials and clinical/pathological data. T.S. and M.Y. performed the experiments. All authors analysed and interpreted the data. S.T., M.M., and T.Y. drafted the paper. All authors approved the final version of the paper and provided their consent for publication.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The use of human samples for this study was reviewed and approved by the Institutional Review Board (IRB) of the National Cancer Center (Tokyo, Japan) [Authorisation numbers: 2014-381 (Japanese cohort) and 2016-017 (US cohort)] and conducted in accordance with the Declaration of Helsinki. The IRB waived the requirement for obtaining new informed consent to this study.
Consent to publish
No individually identifiable data are presented.
Data availability
The datasets generated and/or analysed during the present study are not publicly available, but anonymised versions may be available from the corresponding author on reasonable request.
Competing interests
The authors declare no competing interests.
Funding information
This study was supported by the National Cancer Center Research and Development Fund (26-A-13 and 26-A-5 to T. Yamada), the Acceleration Transformative Research for Medical Innovation (ACT-MS) program of the Japan Agency for Medical Research and Development (AMED) (16im0210804h0001 to T. Yamada), the Kobayashi Foundation for Cancer Research (to T. Yamada), a KAKENHI Grant-in-Aid for Challenging Exploratory Research (16K14627 to M. Masuda), a Grant-in Aid for Scientific Research (B) (17H03603 to M. Masuda) from the Japan Society for the Promotion of Science (JSPS), a Cancer Research Grant from the Foundation for Promotion of Cancer Research in Japan (to M. Masuda), and a Research Grant from the Princess Takamatsu Cancer Research Fund (to M. Masuda).
Additional information
Note This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution 4.0 International (CC BY 4.0).
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Sugano, T., Yoshida, M., Masuda, M. et al. Prognostic impact of ACTN4 gene copy number alteration in hormone receptor-positive, HER2-negative, node-negative invasive breast carcinoma. Br J Cancer 122, 1811–1817 (2020). https://doi.org/10.1038/s41416-020-0821-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41416-020-0821-y