Abstract
Background
Epidemiological studies provide strong evidence for a role of endogenous sex hormones in the aetiology of breast cancer. The aim of this analysis was to identify genetic variants that are associated with urinary sex-hormone levels and breast cancer risk.
Methods
We carried out a genome-wide association study of urinary oestrone-3-glucuronide and pregnanediol-3-glucuronide levels in 560 premenopausal women, with additional analysis of progesterone levels in 298 premenopausal women. To test for the association with breast cancer risk, we carried out follow-up genotyping in 90,916 cases and 89,893 controls from the Breast Cancer Association Consortium. All women were of European ancestry.
Results
For pregnanediol-3-glucuronide, there were no genome-wide significant associations; for oestrone-3-glucuronide, we identified a single peak mapping to the CYP3A locus, annotated by rs45446698. The minor rs45446698-C allele was associated with lower oestrone-3-glucuronide (−49.2%, 95% CI −56.1% to −41.1%, P = 3.1 × 10–18); in follow-up analyses, rs45446698-C was also associated with lower progesterone (−26.7%, 95% CI −39.4% to −11.6%, P = 0.001) and reduced risk of oestrogen and progesterone receptor-positive breast cancer (OR = 0.86, 95% CI 0.82–0.91, P = 6.9 × 10–8).
Conclusions
The CYP3A7*1C allele is associated with reduced risk of hormone receptor-positive breast cancer possibly mediated via an effect on the metabolism of endogenous sex hormones in premenopausal women.
Similar content being viewed by others
Background
Epidemiological studies provide strong evidence for a role of endogenous hormones in the aetiology of breast cancer.1,2 Pooled analyses of data from prospective studies estimated that a doubling of circulating oestradiol or oestrone was associated with a 30–50% increase in breast cancer risk in postmenopausal women and a 20–30% increase in breast cancer risk in premenopausal women; there was no evidence that premenopausal progesterone levels were associated with breast cancer risk.2,3 We have previously screened 642 SNPs tagging 42 genes involved in sex steroid synthesis or metabolism, and tested for the association with premenopausal urinary oestrone glucuronide and pregnanediol-3-glucuronide levels, measured in urine samples collected at pre-specified days of the woman’s menstrual cycle.4 Oestrone-3-glucuronide and pregnanediol-3-glucuronide are urinary metabolites of oestrogen and progesterone, respectively,5,6 that are used in the context of reproductive medicine to monitor ovarian activity.7 None of the variants that we tested was associated with urinary pregnanediol-3-glucuronide, but a rare haplotype, defined by two SNPs spanning the cytochrome P450 family 3 subfamily A (CYP3A) gene cluster, was associated with a highly significant 32% difference in urinary oestrone-3-glucuronide.4 Fine-scale mapping analyses identified the SNP rs45446698 as a putative causal variant at this locus; rs45446698 is one of seven highly correlated SNPs that cluster within the CYP3A7 promoter and comprise the CYP3A7*1C allele.8 A genome-wide association study (GWAS) of postmenopausal plasma oestradiol levels found no association at this locus.9 A subsequent GWAS of pre- and postmenopausal hormone levels similarly found no association with plasma oestradiol at this locus; they did however find associations at this locus with DHEAS and progesterone.10
The CYP3A genes (CYP3A5, CYP3A7 and CYP3A4) encode enzymes that metabolise a diverse range of substrates;11 in addition to a role in the oxidative metabolism of hormones, CYP3A enzymes metabolise ~50% of all clinically used drugs, including many of the agents used in treating cancer.12 CYP3A4, the major isoform in adults, is predominantly expressed in the liver, where it is the most abundant P450, accounting for 30% of total CYP450 protein. CYP3A7, the major isoform in the foetus, is generally silenced shortly after birth.13 In CYP3A7*1C carriers, a region within the foetal CYP3A7 promoter has been replaced with the equivalent region from the adult CYP3A4 gene;14 this results in adult expression of CYP3A7 in CYP3A7*1C carriers and may influence metabolism of endogenous hormones, exogenous hormones used in menopausal hormone treatment and clinically prescribed drugs, including agents used in treating cancer, in these individuals.12,15 In order to identify additional variants that are associated with premenopausal urinary hormone levels and to further characterise the associations at the CYP3A locus, we carried out a GWAS of urinary oestrone-3-glucuronide and pregnanediol-3-glucuronide levels, using mid-luteal-phase urine samples from women of European ancestry and followed up by testing for an association with breast cancer risk in cases and controls from the Breast Cancer Association Consortium (BCAC). To determine whether the CYP3A7*1C allele influences metabolism of exogenous hormones, we evaluated gene-environment interactions with menopausal hormone treatment for breast cancer risk, and to investigate whether adult expression of CYP3A7 impacts on agents used in treating cancer, we analysed associations with breast cancer-specific survival.
Methods
GWAS subjects
Generations Study
Full details of the Generations Study have been published previously.16 Briefly, the Generations Study is a cohort study of more than 110,000 women from the UK general population, who were recruited beginning in 2003 and from whom detailed questionnaires and blood samples have been collected to investigate risk factors for breast cancer.
British Breast Cancer Study
Full details of the British Breast Cancer Study have been published previously.17 Briefly, the British Breast Cancer Study is a national case–control study of breast cancer, in which cases of breast cancer were ascertained through the cancer registries of England and Scotland and through the National Cancer Research Network. Cases were asked to invite a healthy female first-degree relative with no history of cancer and a female friend or non-blood relative to participate in the study.
Mammography Oestrogens and Growth Factors study
Full details of the Mammography Oestrogens and Growth Factors study have been published previously.18 Briefly, this is an observational study nested within a trial of annual mammography screening in young women that was conducted in Britain.19 Approximately 54,000 women aged 39–41 years were randomly assigned to the intervention arm from 1991 to 1997 and offered annual mammograms until age 48 years. From 2000 to 2003, women in the intervention arm who were still participating in this trial were invited to participate in the Mammography Oestrogens and Growth Factors study; they were asked to provide a blood sample and complete a questionnaire detailing demographic, lifestyle and reproductive factors. More than 8000 women were enrolled in the study.
GWAS subjects were drawn from the Generations Study (N = 184), the British Breast Cancer Study (N = 284) and the Mammography Oestrogens and Growth Factors study (N = 109). To be eligible for the GWAS analysis of oestrone-3-glucuronide and pregnanediol-3-glucuronide levels, women had to be having regular menstrual cycles (i.e., their usual cycle length had to be between 21 and 35 days) and not using menopausal hormone therapy or oral contraceptives. All of the women included in this analysis reported being of European ancestry, and none had been diagnosed with breast cancer at the time of study recruitment.
Measurement of hormone levels
The protocol for collecting timed urine samples has been published previously.18 Briefly, a woman’s predicted date of ovulation was estimated from the date of the first day of her last menstrual period and her usual cycle length; ovulation was predicted to occur 14 days before the date of her next menstrual period. On this basis, women were asked to provide a series of early morning urine samples on pre-specified days of their cycle. For this analysis, the mid-luteal-phase sample, taken at 7 days after the predicted day of ovulation, was used. To confirm that ovulation had occurred, consistent with the predicted date of ovulation, pregnanediol-3-glucuronide was measured; to take account of the differences in volume in early morning urine samples from different women, we measured creatinine, a waste product of normal muscle and protein metabolism that is released at a constant rate by the body. Samples in which pregnanediol-3-glucuronide, adjusted for creatinine levels, was >0.3 µmol/mol, were taken forward for measurement of creatinine-adjusted oestrone-3-glucuronide. Pregnanediol-3-glucuronide and oestrone-3-glucuronide were analysed by commercial competitive ELISA Kits (Arbor Assays, Ann Arbor, USA) according to the manufacturer’s instructions. For pregnanediol-3-glucuronide, the lower limit of detection was determined as 0.64 nmol/l; intra- and inter-assay coefficients of variation were 3.7% and 5.2%, respectively. For oestrone-3-glucuronide, the lower limit of detection was determined as 19.6 pmol/l; intra- and inter-assay coefficients of variation were 3.5% and 5.9%, respectively. Creatinine was determined using the creatininase/creatinase-specific enzymatic method20 using a commercial kit (Alpha Laboratories Ltd. Eastleigh, UK) adapted for use on a Cobas Fara centrifugal analyser (Roche Diagnostics Ltd, Welwyn Garden City, UK). For within-run precision, the coefficient of variation was <3%, while for intra-batch precision, the coefficient of variation was <5%.
For 303 premenopausal women participating in the Generations Study (184 as above and an additional 119 for whom timed urine samples were accrued more recently), urinary progesterone levels were also measured using an “in house” ELISA. In all, 96-well plates (Greiner Bio-One GmbH, Frickenhausen, Germany) were coated with 100 µl of 5 µg/ml GAM (Arbor Assays, Ann Arbor, USA) in ELISA coating buffer (100 mM Na Bicarbonate, pH 9.6) covered and incubated in a fridge at 4 °C overnight. Before use, the plates were washed three times with wash buffer 0.05 M Tris/HCl and 0.05% Tween 20, pH 7.4 (Tween® 20, Sigma-Aldrich, Inc., St. Louis, MO, USA). Standards, samples and controls (20 µl per well) were added to each well, followed by 80 µl of progesterone 3-HRP conjugate (Astra Biotech GmbH, Berlin, Germany) at 1:10,000 in assay buffer (PBS pH 7.4 containing 0.1% BSA and 250 ng/ml Cortisol), followed by 50 μl of monoclonal progesterone Ab (Astra Biotech GmbH, Berlin, Germany) 1:50,000 in assay buffer. Plates were incubated at room temperature for 2 h on a microtitre plate shaker (IKA®, Schuttler MTS4, IKA Labortechnik, Staufen, Germany), then washed five times with assay wash buffer and 120 µl of substrate solution (3,3,5,5-tetramethylbenzidine, Millipore Corporation, Temecula, CA, USA) was added to each well. Plates were incubated at room temperature without shaking in the dark. After 20 min, the reaction was stopped by adding 80 µl of 2 N H2SO4 solution (Sigma-Aldrich Company Ltd., Dorset, UK). Finally, the plates were read on a plate reader at 450 nm. Standard curves were prepared with a total of eight different concentrations (16, 8, 4, 2, 1, 0.5, 0.25 and 0 ng/ml). Samples, standards and controls were included in duplicate. Inter- and intra-assay coefficients of variation were calculated from two controls of low and high progesterone in duplicate in each of eight assays. The inter-assay coefficients of variation for low and high pools, respectively, were 11.4 and 9.1%; the intra-assay coefficients of variation were 8.9 and 5.6%. The lower limit of detection was calculated at 0.1 ng/ml. Cross-reaction with other steroids was oestrone: 0.17%, oestradiol: 0.28%, oestriol: 0.18%, dehydroepiandrosterone: 0.02%, testosterone: 0.36%, dihydrotestosterone: 0.15%, 17α-hydroxyprogesterone: 2.9%, androstenedione: 0.14%, 11-deoxycortisol: 0.46%, corticosterone: 0.18%, cortisone: 0.04% and cortisol: 0.04%.
GWAS genotyping and quality control
DNA from 577 women was genotyped using Illumina Infinium OncoArray 500 K BeadChips. We excluded samples for which <95% of SNPs were successfully genotyped. Identity-by-descent analysis was used to identify closely related individuals enabling exclusion of first-degree relatives. We applied SmartPCA21 to our data and used phase II HapMap samples to identify individuals with non-Caucasian ancestry. The first two principal components for each individual were plotted, and k-means clustering was used to identify samples separated from the main Caucasian cluster. SNPs with call rates <95% were excluded, as were SNPs with minor allele frequency (MAF) < 2% and those whose genotype frequencies deviated from Hardy–Weinberg proportions at P < 1 × 10–05. Following QC, 487,659 SNPs were successfully genotyped in 560 samples (Generations Study: N = 179, British Breast Cancer Study: N = 278 and Mammography Oestrogens and Growth Factors study: N = 103). Genome-wide imputation was performed using 1KGP Phase 3 reference data. Haplotypes were pre-phased using SHAPEIT2.22 Imputation was performed using IMPUTE2.23 Imputed SNPs with INFO scores <0.8 and MAFs <2% were excluded from subsequent analyses. After QC, a set of 7,792,694 successfully imputed SNPs were available for association analysis.
Genotyping rs45446698 and sequencing of the CYP3A7*1C allele
For the 119 Generations Study women who were not included in the GWAS but for whom progesterone was subsequently measured, rs45446698 was genotyped by TaqMan (Thermo Fisher Scientific Ltd, UK). The call rate was 100% with 100% concordance between 12 duplicates. To confirm that rs45446698 tags the CYP3A7*1C allele, we sequenced this region in 31 women selected on the basis of their rs45446698 genotype (9 common homozygotes and 22 carriers). A 370-bp DNA region (chr7: 99 332 745-99 333 114; GRCh37/hg19) was amplified using Phusion High-Fidelity DNA Polymerase (New England Biolabs, UK) and primers CCATAGAGACAAGAGGAGA (forward) and CTGAGTCTTTTTTTCAGCAGC (reverse). The PCR product was purified using QIAquick Gel Extraction Kit (Qiagen) and Sanger sequenced using a commercially available service (Eurofins Genomics, Germany).
Statistical analysis of GWAS data
Tests of association between SNP genotypes and log-transformed creatinine-adjusted oestrone-3-glucuronide and pregnanediol-3-glucuronide adjusted for study were performed using linear regression in SNPTEST v2.5.24 Test statistic inflation was assessed visually using a QQ Plot (Supplementary Fig. S1) and formally by calculating the inflation factor, λ. There was no evidence of systematic test statistic inflation (λ = 1.01 for both oestrone-3-glucuronide and pregnanediol-3-glucuronide). For the single significant association (rs45446698), we used multivariate linear regression to adjust for potential confounders: age at menarche (<12, 12, 13, 14 and >14 years), age at collection of urine samples (<35, 35–40 and ≥40 years), body mass index (BMI: < 18.5, 18.5–<20.0, 20.0–<25.0, 25.0–<30.0 and ≥ 30.0 kg/m2) and parity (0, 1, 2 and ≥3 live births).
Follow-up genotyping of rs45446698
Genotype data for rs45446698 were generated as part of iCOGS25 and OncoArray.26 Full details of SNP selection, array design, genotyping and post-genotyping QC have been published.25,26 Participants genotyped in both collaborations were excluded from the iCOGS data sets with the exception of the GxE interaction analysis of menopausal hormone treatment, for which five studies (CPS-II, PBCS, UKBGS, MCCS and pKARMA) were excluded from OncoArray, rather than iCOGS, in order to maximise the number of studies with sufficient cases and controls for analysis. We excluded cases with breast tumours of unknown invasiveness, or in situ disease, and those for whom age at diagnosis was not known. After QC exclusions,26 the call rate for rs45446698 in OncoArray data was 99.66% and there was no evidence of deviation from Hardy–Weinberg equilibrium in controls (Supplementary Table S1). In iCOGS data, rs45446698 was imputed using 1KGP Phase 3 reference data (info score = 0.94); we used gene dosages (≤0.2 = 0, >0.8 and ≤1.2 = 1, >1.8 = 2) to call genotypes for 99.22% of samples.
Statistical analysis of rs45446698 and breast cancer risk
Due to the low MAF of rs45446698 (3.7%, 0.03% and 0.4% in individuals of European, Asian and African ancestry, respectively), we restricted our analyses to individuals of European ancestry and excluded studies with <50 cases or controls; there were 35 (iCOGS) and 56 (OncoArray) studies for the current case–control analysis (Supplementary Tables S1 and S2).
We combined heterozygote and rare homozygote genotypes and estimated carrier ORs using logistic regression, adjusted for 15 principal components25,26 and study. Stratum-specific carrier ORs were estimated for a set of pre-specified prognostic variables (oestrogen receptor (ER), progesterone receptor (PR), HER2, grade and stage). We excluded studies with <50 cases or controls in any individual stratum from stratified analyses. Interactions were assessed based on case-only models (ER, PR, Her2, stage and grade). In the subset of studies for which covariate data were available, we used multivariable logistic regression to adjust for reference age (defined as age at diagnosis for cases and age at interview for controls), age at menarche, BMI and parity (as above). Finally, we stratified our analyses on menopausal status at reference age. When menopausal status was missing, the reference age was used as a surrogate (<54 premenopausal and ≥54 postmenopausal). To select the reference age that most accurately captured menopausal status in this group of studies, we generated AUC curves based on women who had reported natural menopause with different reference age cut-offs (50–56 years); on this basis, a reference age of 54 was selected. P values were estimated using likelihood ratio tests with one degree of freedom. All P values reported, for all analyses, are two-sided. Statistical analyses were performed using STATA version 11.0 (StataCorp, College Station, TX, USA).
Statistical analysis of gene-environment interaction (GxE) with menopausal hormone treatment
Postmenopausal women from 13 (iCOGS) and 27 (OncoArray) studies provided the data on menopausal hormone treatment. Menopausal status and postmenopausal hormone use were derived as of the reference date (defined as date of diagnosis for cases and interview for controls); women with unknown age at reference date were excluded from this analysis. All analyses were conducted only in postmenopausal women. Carrier ORs for breast cancer risk were estimated using logistic regression stratified by current use of menopausal hormone treatment, oestrogen–progesterone therapy and oestrogen-only therapy, respectively. Analyses were adjusted for study, ten principal components, reference age, age at menarche, parity, BMI, former use of menopausal hormone treatment and use of any menopausal hormone treatment preparation other than the one of interest in analyses of current use of menopausal hormone treatment by type. To account for potential heterogeneity of the main effects of menopausal hormone treatment/oestrogen–progesterone therapy/oestrogen-only therapy by study design, we included an interaction term between the risk factor of interest and an indicator variable for study design (prospective cohorts/population-based case–control studies, non-population-based studies). Interactions between rs45446698 and current use of menopausal hormone treatment, oestrogen–progesterone therapy and oestrogen-only therapy were assessed using likelihood ratio tests, based on logistic regression models with and without interaction between rs45446698 and current use of menopausal hormone treatment, oestrogen–progesterone therapy and oestrogen-only therapy, respectively. Statistical analyses were performed using SAS 9.4 and R (version 3.4.4).
Statistical analysis of breast cancer-specific survival in cases
In total, 38 (iCOGS) and 63 (OncoArray) studies provided follow-up data for analysis of breast cancer-specific survival. Analysis of outcome was restricted to patients who were at least 18 years old at diagnosis and for whom vital status at, and date of the last follow-up were known. Patients ascertained for a second tumour were excluded. Time-to-event was calculated from the date of diagnosis. For prevalent cases with study entry after diagnosis, left truncation was applied, i.e., follow-up started at the date of study entry.27 Follow-up was right-censored at the date of death (death known to be due to breast cancer considered an event), the date the patient was last known to be alive if death did not occur or at 10 years after diagnosis, whichever came first. Follow-up was censored at 10 years due to limited data availability after this time. Hazard ratios (HR) for association of rs45446698 genotype with breast cancer-specific survival were estimated using Cox proportional hazards regression implemented in the R package survival (v. 2.43–3) stratified by country. iCOGS and OncoArray estimates were combined using an inverse-variance-weighted meta-analysis.
Results
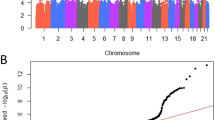
We tested 8,280,353 autosomal SNPs for association with luteal-phase creatinine-adjusted oestrone-3-glucuronide and pregnanediol-3-glucuronide in 560 premenopausal women. For oestrone-3-glucuronide, we identified a single peak mapping to the CYP3A locus at chromosome 7q22.1 (Fig. 1 and Supplementary Table S3); conditioning on any of the top SNPs, there were no additional independent signals. Four of the SNPs that were significant at P < 1 × 10−8 comprise part of the seven-SNP CYP3A7*1C allele,8,15 including the top, directly genotyped SNP, rs45446698 (Supplementary Table S3). The rare rs45446698-C allele (MAF = 0.035) was associated with a 49.2% reduction in luteal-phase oestrone-3-glucuronide (95% CI −56.1% to −41.1%, P = 3.1 × 10−18, Table 1) and explained 11.5% of the variation in oestrone-3-glucuronide in these premenopausal women. Since hormone levels may be influenced by both demographic and reproductive factors, we adjusted for age at urine collection, age at menarche, body mass index and parity; these adjustments did not alter the association (fully adjusted model: 44.8% reduction, 95% CI −53.3% to −34.8%, P = 2.1 × 10−12, Table 1).
For pregnanediol-3-glucuronide, there were no associations that were significant at a threshold of P < 1 × 10−8 (Supplementary Fig. S2). An association between the CYP3A locus and progesterone levels has been reported previously;10 accordingly, we measured progesterone in addition to pregnanediol-3-glucuronide in premenopausal women from the Generations Study. Progesterone was moderately correlated with pregnanediol-3-glucuronide (r = 0.37, P = 7.4 × 10−12), but while there was no association between the rs45446698-C allele and urinary pregnanediol-3-glucuronide levels (5.5% reduction, 95% CI −24.2% to +17.7%, P = 0.61) in this group of women, the rs45446698-C allele was associated with significantly lower luteal-phase urinary progesterone levels (26.7% reduction, 95% CI −39.4% to −11.6%, P = 0.001, Table 1). Adjusting these analyses for covariates, as above, did not alter the results (Table 1).
To test for the association between rs45446698 and breast cancer risk, we combined genotype data from 56 studies (OncoArray; Supplementary Table S1) with imputed data from 35 studies (iCOGS; Supplementary Table S2) in a total of 90,916 cases and 89,893 controls of European Ancestry. The rs45446698-C allele was associated with a reduction in breast cancer risk (OR = 0.94, 95% CI 0.91–0.98, P = 0.002, Table 2) with no evidence of heterogeneity between data sets (Phet = 0.58). There was no evidence that the reduction in breast cancer risk associated with being a rs45446698-C carrier differed according to Her2 status, tumour grade or stage (Supplementary Table S4). Stratifying by ER status, the association was limited to ER-positive (ER + ) breast cancers (OR = 0.91, 95% CI 0.87–0.96, P = 0.0002 and OR = 1.03, 95% CI 0.95–1.11, P = 0.50 for ER + and ER− cancers, respectively; Pint = 0.03, Table 2). Stratifying by ER and PR status, the association was limited to ER + /PR + cancers (ER + /PR + : OR = 0.86, 95% CI 0.82–0.91, P = 6.9 × 10–8; ER + /PR−: OR = 1.06, 95% CI 0.96–1.16, P = 0.25; Pint = 0.0001, Table 2). Adjusting for demographic and reproductive factors in the subset of studies for which these additional covariates were available did not alter this association (Supplementary Table S5). Defining reference age as age at diagnosis for cases and age at interview for controls and using this as a proxy for menopausal status (<54 or ≥54 years), we further stratified our analysis on menopausal status; there was little evidence that the association with ER + /PR + breast cancer differed by menopausal status (premenopausal OR = 0.94, 95% CI 0.84–1.06, P = 0.31, postmenopausal OR = 0.86, 95% CI 0.80–0.93, P = 0.0001, Phet = 0.28).
On the assumption that genetic variants that influence metabolism of endogenous hormones5 may also impact on metabolism of exogenous hormones, we investigated whether menopausal hormone treatment modified the association between rs45446698 genotype and ER + /PR + breast cancer risk in 17,831 postmenopausal breast cancer cases and 40,437 postmenopausal controls. The rs45446698-C carrier OR was lower (i.e., more protective) in current users of any menopausal hormone treatment but particularly in those who used combined oestrogen–progesterone therapy (current users: OR = 0.68, 95% CI 0.52–0.90, P = 0.007; never users: OR = 0.85, 95% CI 0.76–0.95, P = 0.005, Table 3). This difference was not, however, statistically significant (Pint = 0.15, Table 3).
Finally, to determine whether rs45446698 genotype could affect patient outcome by influencing metabolism of cytotoxic agents that are CYP3A substrates,15 we tested for the association between rs45446698 genotype and 10-year breast cancer-specific survival in 91,539 breast cancer cases from 71 studies for whom follow-up data were available. There was neither overall association between rs45446698 genotype and breast cancer-specific survival (HR = 0.99, 95% CI 0.91–1.09, P = 0.90, Table 4) nor was there any evidence of an association in analyses stratified by tumour characteristics (Supplementary Table S6). Stratifying by treatment regimen, we found no evidence that rs45446698 genotype influenced outcome in cases who were treated with a hormonal agent (i.e., tamoxifen or an aromatase inhibitor, Table 4). There was, however, some evidence that in cases who were treated with a taxane, carriers of the rs45446698-C allele had reduced breast cancer-specific survival compared with non-carriers (HR = 1.46, 95% CI 1.08–1.97, P = 0.01, Table 4).
Discussion
This present GWAS identified a single, highly significant association between the CYP3A7*1C allele (tagged by rs4546698) and premenopausal urinary oestrone-3-glucuronide. This finding alone is not novel; we have previously reported an association between the CYP3A7*1C allele, parent oestrogens and several oestrogen metabolites.5 What we have demonstrated for the first time is the extent to which this signal dominates the genetic architecture of hormone levels in premenopausal women of Northern European ancestry (Fig. 1; rs45446698 P = 3.1 × 10−18, all other signals P > 1 × 10–8) and we estimate that 11.5% of the variance in urinary oestrone-3-glucuronide levels is explained by this one allele.
Two previous GWAS of circulating oestrogen levels have been published, neither reported an association with the CYP3A locus.9,10 This lack of replication may be explained by our choice of study population. The first GWAS9 was conducted in postmenopausal women (N = 1623) participating in the Nurses’ Health Study and the Sisters in Breast Screening Study. The second was conducted within the Twins UK study (N = 2913) and included men as well as pre-, peri- and postmenopausal women. A strength of our GWAS is that all of the women were premenopausal and had regular menstrual cycles; circulating levels of oestrogens in premenopausal women are much higher compared with those in postmenopausal women.4,28 For each woman, we assayed a single urine sample taken in the mid-luteal phase of her cycle at exactly 7 days after her predicted day of ovulation. Thus, although our study is relatively small (N = 560), we may have had greater power to detect an association at the CYP3A locus than previous studies due to the very homogeneous premenopausal study population that we selected.
Our findings also demonstrate the potential significance of the choice of hormone or hormone metabolite; both of the previous GWAS assayed plasma oestradiol. In a targeted analysis of urinary oestrogen metabolites, we have previously shown that the association between the CYP3A7*1C allele and oestrone (45.3% lower levels in carriers, P = 0.0005) is more pronounced than the association with oestradiol (26.7% lower levels, P = 0.07) with the implication that measuring urinary oestrone-3-glucuronide (rather than plasma oestradiol) may have contributed to our positive findings. Similarly, by measuring pregnanediol-3-glucuronide and progesterone in premenopausal women from the Generations Study, we were able to demonstrate a significant association of rs45446698 with progesterone (27% reduction, P = 0.001) in the absence of an association with pregnanediol-3-glucuronide (6% reduction, P = 0.61).
The fact that we measured a urinary oestrogen metabolite (oestrone-3-glucuronide) rather than serum or plasma oestrogens (oestradiol or oestrone) limits the interpretation of our results in terms of a causal association. Estimates of the association between circulating oestrogens and breast cancer risk are based on measurements of hormone levels in plasma or serum,3 and in a recent study that measured luteal-phase serum oestrogens and urinary oestrogen metabolites in 249 premenopausal women,29 serum oestradiol and oestrone were only moderately correlated with urinary oestrone (serum oestradiol: r = 0.39, serum oestrone: r = 0.48). Our analysis of rs45446698 genotypes in 90,916 cases and 89,893 controls from BCAC, however, provides robust evidence of an association of the CYP3A7*1C allele with breast cancer risk overall (OR = 0.94, P = 0.002) and a more pronounced protective effect on ER + /PR + breast cancers (OR = 0.86, P = 6.9 × 10−8). The specificity of this association (comparing ER + /PR− with ER + /PR + cancers, Phet = 0.001) and our replication of Ruth and colleagues report of a signal at the CYP3A locus in their analysis of circulating progesterone levels10 raise the possibility that premenopausal progesterone levels might influence risk of ER + /PR + breast cancers. This would be in contrast to the findings from Key et al. who reported no evidence of an association between premenopausal progesterone levels and breast cancer risk overall and no heterogeneity in estimates stratified by PR status.3 However, the number of cases of PR + (N = 158) and PR− (N = 61) breast cancer was small, and this analysis may have lacked power to detect modest associations in subgroups of cancers. Alternatively, the association of rs45446698 genotype with ER + /PR + breast cancer risk, specifically, may be due to the fact that PR is a marker for an intact oestrogen signalling pathway30 confirming a direct link between the levels of oestrogen (or oestrogen signalling) and proliferation in this subgroup of cancers.
Our analysis of the CYP3A7*1C allele, menopausal hormone treatment and breast cancer risk was inconclusive; while the carrier ORs were consistent with a greater protective effect of this allele in women taking exogenous hormones, particularly oestrogen–progesterone therapy, none of the interactions was statistically significant. Overall, there were 14,119 ER + /PR + breast cancer cases and 32,418 controls for this subgroup analysis, but for what was, arguably, the most pertinent subgroup (i.e., current oestrogen–progesterone therapy use), the number of cases who were current users was relatively small (CYP3A7*1C carriers N = 107, non-carriers N = 1498) and power was limited to detect modest interactions. There are limitations to this analysis; we focussed on current menopausal hormone treatment use (adjusted for past use) as it is for current use that the association with breast cancer risk is the strongest,31 but we did not have information on dose, duration or the formulation that was used.
Finally, we found no association between CYP3A7*1C carrier status and survival in patients treated with tamoxifen, a known CYP3A substrate. This may reflect the fact that compared to CYP3A4, CYP3A7 is a poor metaboliser of tamoxifen,32 or that standard doses of tamoxifen achieve high levels of oestrogen receptor saturation.33 There was some evidence that breast cancer-specific survival was reduced in CYP3A7*1C carriers who were treated with a taxane, compared with non-carriers (P = 0.01); this may, however, be a chance finding given the number of comparisons that were tested.
In conclusion, we present strong evidence that the CYP3A7*1C allele impacts on the metabolism of endogenous hormones, which in turn, reduces the risk of hormone receptor-positive breast cancer in carriers. Optimal strategies for breast cancer prevention in women at high risk of breast cancer and in the general population are an area of active research. In this context, CYP3A7*1C carriers represent a naturally occurring cohort in which the effects of reduced exposure to endogenous oestrogens and progesterones throughout a woman’s premenopausal years can be further investigated. Our results regarding the impact of CYP3A7*1C carrier status on exogenous hormones and chemotherapeutic agents are preliminary but warrant further investigation, preferably in the setting of randomised trials.
References
Chen, W. Y. Exogenous and endogenous hormones and breast cancer. Best. Pr. Res Clin. Endocrinol. Metab. 22, 573–585 (2008).
Key, T., Appleby, P., Barnes, I., Reeves, G. & Endogenous, H., Breast Cancer Collaborative, G. Endogenous sex hormones and breast cancer in postmenopausal women: reanalysis of nine prospective studies. J. Natl Cancer Inst. 94, 606–616 (2002).
Key, T. J., Appleby, P. N., Reeves, G. K., Travis, R. C., Alberg, A. J., Barricarte, A. et al. Sex hormones and risk of breast cancer in premenopausal women: a collaborative reanalysis of individual participant data from seven prospective studies. Lancet Oncol. 14, 1009–1019 (2013).
Johnson, N., Walker, K., Gibson, L. J., Orr, N., Folkerd, E., Haynes, B. et al. CYP3A variation, premenopausal estrone levels, and breast cancer risk. J. Natl Cancer Inst. 104, 657–669 (2012).
Sood, D., Johnson, N., Jain, P., Siskos, A. P., Bennett, M., Gilham, C. et al. CYP3A7*1C allele is associated with reduced levels of 2-hydroxylation pathway oestrogen metabolites. Br. J. Cancer 116, 382–388 (2017).
Chang, E., Slaunwhite, W. R. Jr & Sandberg, A. A. Biliary and urinary metabolites of 4-C14-progesterone in human subjects. J. Clin. Endocrinol. Metab. 20, 1568–1575 (1960).
Blackwell, L. F., Cooke, D. G. & Brown, S. The use of estrone-3-glucuronide and pregnanediol-3-glucuronide excretion rates to navigate the continuum of ovarian activity. Front. Public Health 6, 153 (2018).
Kuehl, P., Zhang, J., Lin, Y., Lamba, J., Assem, M., Schuetz, J. et al. Sequence diversity in CYP3A promoters and characterization of the genetic basis of polymorphic CYP3A5 expression. Nat. Genet. 27, 383–391 (2001).
Prescott, J., Thompson, D. J., Kraft, P., Chanock, S. J., Audley, T., Brown, J. et al. Genome-wide association study of circulating estradiol, testosterone, and sex hormone-binding globulin in postmenopausal women. PLoS ONE 7, e37815 (2012).
Ruth, K. S., Campbell, P. J., Chew, S., Lim, E. M., Hadlow, N., Stuckey, B. G. et al. Genome-wide association study with 1000 genomes imputation identifies signals for nine sex hormone-related phenotypes. Eur. J. Hum. Genet. 24, 284–290 (2016).
Rodriguez-Antona, C. & Ingelman-Sundberg, M. Cytochrome P450 pharmacogenetics and cancer. Oncogene 25, 1679–1691 (2006).
Perera, M. A. The missing linkage: what pharmacogenetic associations are left to find in CYP3A? Expert Opin. Drug Metab. Toxicol. 6, 17–28 (2010).
Schuetz, J. D., Beach, D. L. & Guzelian, P. S. Selective expression of cytochrome P450 CYP3A mRNAs in embryonic and adult human liver. Pharmacogenetics 4, 11–20 (1994).
Gonzalez, F. J. The molecular biology of cytochrome P450s. Pharm. Rev. 40, 243–288 (1988).
Johnson, N., De Ieso, P., Migliorini, G., Orr, N., Broderick, P., Catovsky, D. et al. Cytochrome P450 allele CYP3A7*1C associates with adverse outcomes in chronic lymphocytic leukemia, breast, and lung cancer. Cancer Res. 76, 1485–1493 (2016).
Swerdlow, A. J., Jones, M. E., Schoemaker, M. J., Hemming, J., Thomas, D., Williamson, J. et al. The Breakthrough Generations Study: design of a long-term UK cohort study to investigate breast cancer aetiology. Br. J. Cancer 105, 911–917 (2011).
Johnson, N., Fletcher, O., Naceur-Lombardelli, C., dos Santos Silva, I., Ashworth, A. & Peto, J. Interaction between CHEK2*1100delC and other low-penetrance breast-cancer susceptibility genes: a familial study. Lancet 366, 1554–1557 (2005).
Walker, K., Fletcher, O., Johnson, N., Coupland, B., McCormack, V. A., Folkerd, E. et al. Premenopausal mammographic density in relation to cyclic variations in endogenous sex hormone levels, prolactin, and insulin-like growth factors. Cancer Res. 69, 6490–6499 (2009).
Moss, S. M., Cuckle, H., Evans, A., Johns, L., Waller, M., Bobrow, L. et al. Effect of mammographic screening from age 40 years on breast cancer mortality at 10 years’ follow-up: a randomised controlled trial. Lancet 368, 2053–2060 (2006).
Borner, U., Szasz, G., Bablok, W. & Busch, E. W. [A specific fully enzymatic method for creatinine: reference values in serum (author’s transl)]. J. Clin. Chem. Clin. Biochem. 17, 679–682 (1979).
Price, A. L., Patterson, N. J., Plenge, R. M., Weinblatt, M. E., Shadick, N. A. & Reich, D. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 38, 904–909 (2006).
Delaneau, O., Zagury, J. F. & Marchini, J. Improved whole-chromosome phasing for disease and population genetic studies. Nat. Methods 10, 5–6 (2013).
Howie, B. N., Donnelly, P. & Marchini, J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 5, e1000529 (2009).
Marchini, J., Howie, B., Myers, S., McVean, G. & Donnelly, P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat. Genet. 39, 906–913 (2007).
Michailidou, K., Hall, P., Gonzalez-Neira, A., Ghoussaini, M., Dennis, J., Milne, R. L. et al. Large-scale genotyping identifies 41 new loci associated with breast cancer risk. Nat. Genet. 45, 353–361 (2013).
Michailidou, K., Lindstrom, S., Dennis, J., Beesley, J., Hui, S., Kar, S. et al. Association analysis identifies 65 new breast cancer risk loci. Nature 551, 92–94 (2017).
Azzato, E. M., Greenberg, D., Shah, M., Blows, F., Driver, K. E., Caporaso, N. E. et al. Prevalent cases in observational studies of cancer survival: do they bias hazard ratio estimates? Br. J. Cancer 100, 1806–1811 (2009).
Mauriac, L. & Smith, I. Aromatase inhibitors in early breast cancer treatment. Semin Oncol. 30, 46–57 (2003).
Maskarinec, G., Beckford, F., Morimoto, Y., Franke, A. A. & Stanczyk, F. Z. Association of estrogen measurements in serum and urine of premenopausal women. Biomark. Med. 9, 417–424 (2015).
Wu, V. S., Kanaya, N., Lo, C., Mortimer, J. & Chen, S. From bench to bedside: what do we know about hormone receptor-positive and human epidermal growth factor receptor 2-positive breast cancer? J. Steroid Biochem. Mol. Biol. 153, 45–53 (2015).
Collaborative Group on Hormonal Factors in Breast, C. Type and timing of menopausal hormone therapy and breast cancer risk: individual participant meta-analysis of the worldwide epidemiological evidence. Lancet 394, 1159–1168 (2019).
Williams, J. A., Ring, B. J., Cantrell, V. E., Jones, D. R., Eckstein, J., Ruterbories, K. et al. Comparative metabolic capabilities of CYP3A4, CYP3A5, and CYP3A7. Drug Metab. Dispos. 30, 883–891 (2002).
Lash, T. L., Lien, E. A., Sorensen, H. T. & Hamilton-Dutoit, S. Genotype-guided tamoxifen therapy: time to pause for reflection? Lancet Oncol. 10, 825–833 (2009).
Acknowledgements
We thank all the individuals who took part in these studies and all the researchers, clinicians, technicians and administrative staff who have enabled this work to be carried out. The COGS study would not have been possible without the contributions of the following: Kyriaki Michailidou, (BCAC), Andrew Berchuck (OCAC), Rosalind A. Eeles, Ali Amin Al Olama, Zsofia Kote-Jarai, Sara Benlloch (PRACTICAL), Antonis Antoniou, Lesley McGuffog and Ken Offit (CIMBA), Andrew Lee, and Ed Dicks, Craig Luccarini and the staff of the Centre for Genetic Epidemiology Laboratory, the staff of the CNIO genotyping unit, Daniel C. Tessier, Francois Bacot, Daniel Vincent, Sylvie LaBoissière and Frederic Robidoux and the staff of the McGill University and Génome Québec Innovation Centre, the staff of the Copenhagen DNA laboratory, and Julie M. Cunningham, Sharon A. Windebank, Christopher A. Hilker, Jeffrey Meyer and the staff of Mayo Clinic Genotyping Core Facility. ABCFS thank Maggie Angelakos, Judi Maskiell, Gillian Dite. ABCS thanks the Blood bank Sanquin, The Netherlands. ABCTB Investigators: Christine Clarke, Deborah Marsh, Rodney Scott, Robert Baxter, Desmond Yip, Jane Carpenter, Alison Davies, Nirmala Pathmanathan, Peter Simpson, Dinny Graham, Mythily Sachchithananthan. Samples are made available to researchers on a non-exclusive basis. BBCS thanks Eileen Williams, Elaine Ryder-Mills, Kara Sargus. BCEES thanks Allyson Thomson, Christobel Saunders, Terry Slevin, BreastScreen Western Australia, Elizabeth Wylie, Rachel Lloyd. The BCINIS study would not have been possible without the contributions of Dr. K. Landsman, Dr. N. Gronich, Dr. A. Flugelman, Dr. W. Saliba, Dr. E. Liani, Dr. I. Cohen, Dr. S. Kalet, Dr. V. Friedman, Dr. O. Barnet of the NICCC in Haifa, and all the contributing family medicine, surgery, pathology and oncology teams in all medical institutes in Northern Israel. BIGGS thanks Niall McInerney, Gabrielle Colleran, Andrew Rowan, Angela Jones. The BREOGAN study would not have been possible without the contributions of the following: Manuela Gago-Dominguez, Jose Esteban Castelao, Angel Carracedo, Victor Muñoz Garzón, Alejandro Novo Domínguez, Maria Elena Martinez, Sara Miranda Ponte, Carmen Redondo Marey, Maite Peña Fernández, Manuel Enguix Castelo, Maria Torres, Manuel Calaza (BREOGAN), José Antúnez, Máximo Fraga and the staff of the Department of Pathology and Biobank of the University Hospital Complex of Santiago-CHUS, Instituto de Investigación Sanitaria de Santiago, IDIS, Xerencia de Xestion Integrada de Santiago-SERGAS; Joaquín González-Carreró and the staff of the Department of Pathology and Biobank of University Hospital Complex of Vigo, Instituto de Investigacion Biomedica Galicia Sur, SERGAS, Vigo, Spain. BSUCH thanks Peter Bugert, Medical Faculty Mannheim. CBCS thanks study participants, co-investigators, collaborators and staff of the Canadian Breast Cancer Study, and project coordinators Agnes Lai and Celine Morissette. CCGP thanks Styliani Apostolaki, Anna Margiolaki, Georgios Nintos, Maria Perraki, Georgia Saloustrou, Georgia Sevastaki, Konstantinos Pompodakis. CGPS thanks staff and participants of the Copenhagen General Population Study. For the excellent technical assistance: Dorthe Uldall Andersen, Maria Birna Arnadottir, Anne Bank, Dorthe Kjeldgård Hansen. The Danish Cancer Biobank is acknowledged for providing infrastructure for the collection of blood samples for the cases. CNIO-BCS thanks Guillermo Pita, Charo Alonso, Nuria Álvarez, Pilar Zamora, Primitiva Menendez, the Human Genotyping-CEGEN Unit (CNIO). Investigators from the CPS-II cohort thank the participants and Study Management Group for their invaluable contributions to this research. They also acknowledge the contribution to this study from central cancer registries supported through the Centers for Disease Control and Prevention National Program of Cancer Registries, as well as cancer registries supported by the National Cancer Institute Surveillance Epidemiology and End Results program. The authors would like to thank the California Teachers Study Steering Committee that is responsible for the formation and maintenance of the Study within which this research was conducted. A full list of California Teachers Study team members is available at https://www.calteachersstudy.org/team. We thank the participants and the investigators of EPIC (European Prospective Investigation into Cancer and Nutrition). ESTHER thanks Hartwig Ziegler, Sonja Wolf, Volker Hermann, Christa Stegmaier, Katja Butterbach. FHRISK thanks NIHR for funding. GC-HBOC thanks Stefanie Engert, Heide Hellebrand, Sandra Kröber and LIFE - Leipzig Research Centre for Civilization Diseases (Markus Loeffler, Joachim Thiery, Matthias Nüchter, Ronny Baber). The GENICA Network: Dr. Margarete Fischer-Bosch-Institute of Clinical Pharmacology, Stuttgart, and University of Tübingen, Germany [HB, Wing-Yee Lo, Reiner Hoppe], Department of Internal Medicine, Evangelische Kliniken Bonn gGmbH, Johanniter Krankenhaus, Bonn, Germany [Yon-Dschun Ko, Christian Baisch], Institute of Pathology, University of Bonn, Germany [Hans-Peter Fischer], Molecular Genetics of Breast Cancer, Deutsches Krebsforschungszentrum (DKFZ), Heidelberg, Germany [Ute Hamann], Institute for Prevention and Occupational Medicine of the German Social Accident Insurance, Institute of the Ruhr University Bochum (IPA), Bochum, Germany [Thomas Brüning, Beate Pesch, Sylvia Rabstein, Anne Lotz]; and Institute of Occupational Medicine and Maritime Medicine, University Medical Center Hamburg-Eppendorf, Germany [Volker Harth]. HABCS thanks Michael Bremer. HEBCS thanks Sofia Khan, Johanna Kiiski, Kristiina Aittomäki, Rainer Fagerholm, Kirsimari Aaltonen, Karl von Smitten, Irja Erkkilä. HMBCS thanks Peter Hillemanns, Hans Christiansen and Johann H. Karstens. HUBCS thanks Shamil Gantsev. KARMA and SASBAC thank the Swedish Medical Research Counsel. KBCP thanks Eija Myöhänen, Helena Kemiläinen. kConFab/AOCS wish to thank Heather Thorne, Eveline Niedermayr, all the kConFab research nurses and staff, the heads and staff of the Family Cancer Clinics, and the Clinical Follow Up Study (which has received funding from the NHMRC, the National Breast Cancer Foundation, Cancer Australia, and the National Institute of Health (USA)) for their contributions to this resource, and the many families who contribute to kConFab. LMBC thanks Gilian Peuteman, Thomas Van Brussel, EvyVanderheyden and Kathleen Corthouts. MABCS thanks Milena Jakimovska (RCGEB “Georgi D. Efremov”), Emilija Lazarova (University Clinic of Radiotherapy and Oncology), Katerina Kubelka-Sabit, Mitko Karadjozov (Adzibadem-Sistina Hospital), Andrej Arsovski and Liljana Stojanovska (Re-Medika Hospital) for their contributions and commitment to this study. MARIE thanks Petra Seibold, Dieter Flesch-Janys, Judith Heinz, Nadia Obi, Alina Vrieling, Sabine Behrens, Ursula Eilber, Muhabbet Celik, Til Olchers and Stefan Nickels. MBCSG (Milan Breast Cancer Study Group): Paolo Radice, Bernard Peissel, Jacopo Azzollini, Dario Zimbalatti, Daniela Zaffaroni, Bernardo Bonanni, Irene Feroce, Mariarosaria Calvello, Aliana Guerrieri Gonzaga, Monica Marabelli, Davide Bondavalli and the personnel of the Cogentech Cancer Genetic Test Laboratory. The MCCS was made possible by the contribution of many people, including the original investigators, the teams that recruited the participants and continue working on follow-up, and the many thousands of Melbourne residents who continue to participate in the study. We thank the coordinators, the research staff and especially the MMHS participants for their continued collaboration on research studies in breast cancer. MTLGEBCS would like to thank Martine Tranchant (CHU de Québec – Université Laval Research Center), Marie-France Valois, Annie Turgeon and Lea Heguy (McGill University Health Center, Royal Victoria Hospital; McGill University) for DNA extraction, sample management and skilful technical assistance. J.S. is Chair holder of the Canada Research Chair in Oncogenetics. The following are NBCS Collaborators: Anne-Lise Børresen-Dale, Grethe I. Grenaker Alnæs, Kristine K. Sahlberg (PhD), Lars Ottestad (MD), Rolf Kåresen (Prof. Em.), Ellen Schlichting (MD), Marit Muri Holmen (MD), Toril Sauer (MD), Vilde Haakensen (MD), Olav Engebråten (MD), Bjørn Naume (MD), Alexander Fosså (MD), Cecile E. Kiserud (MD), Kristin V. Reinertsen (MD), Åslaug Helland (MD), Margit Riis (MD), Jürgen Geisler (MD) and OSBREAC. NBHS thank study participants and research staff for their contributions and commitment to the studies. For NHS and NHS2 the study protocol was approved by the institutional review boards of the Brigham and Women’s Hospital and Harvard T.H. Chan School of Public Health, and those of participating registries as required. We would like to thank the participants and staff of the NHS and NHS2 for their valuable contributions as well as the following state cancer registries for their help: AL, AZ, AR, CA, CO, CT, DE, FL, GA, ID, IL, IN, IA, KY, LA, ME, MD, MA, MI, NE, NH, NJ, NY, NC, ND, OH, OK, OR, PA, RI, SC, TN, TX, VA, WA, WY. The authors assume full responsibility for analyses and interpretation of these data. OBCS thanks Arja Jukkola-Vuorinen, Mervi Grip, Saila Kauppila, Meeri Otsukka, Leena Keskitalo and Kari Mononen for their contributions to this study. The OFBCR thanks Teresa Selander, Nayana Weerasooriya and Steve Gallinger. ORIGO thanks E. Krol-Warmerdam, and J. Blom for patient accrual, administering questionnaires, and managing clinical information. The LUMC survival data were retrieved from the Leiden hospital-based cancer registry system (ONCDOC) with the help of Dr. J. Molenaar. PBCS thanks Louise Brinton, Mark Sherman, Neonila Szeszenia-Dabrowska, Beata Peplonska, Witold Zatonski, Pei Chao, Michael Stagner. PROCAS thanks NIHR for funding. The RBCS thanks Corine M. Beaufort, Jannet Blom, Renée Broeren—Foekens, Saskia Pelders, Wendy J.C. Prager—van der Smissen, Kirsten Ruigrok – Ritstier, Anita M.A.C. Trapman—Jansen, Michelle van der Vlugt – Daane, Vanja de Weerd, and the Erasmus MC Family Cancer Clinic. SBCS thanks Sue Higham, Helen Cramp, Dan Connley, Ian Brock, Sabapathy Balasubramanian and Malcolm W.R. Reed. We thank the SEARCH and EPIC teams. SZBCS thanks Ewa Putresza. UCIBCS thanks Irene Masunaka. UKBGS thanks Breast Cancer Now and the Institute of Cancer Research for support and funding of the Generations Study, and the study participants, study staff, and the doctors, nurses and other health care providers and health information sources who have contributed to the study. We acknowledge NHS funding to the Royal Marsden/ICR NIHR Biomedical Research Centre.
NBCS collaborators
Anne-Lise Børresen-Dale86,129, Grethe I. Grenaker Alnæs129, Kristine K. Sahlberg129,130, Lars Ottestad129, Rolf Kåresen86,131, Ellen Schlichting131, Marit Muri Holmen132, Toril Sauer86,133, Vilde Haakensen129, Olav Engebråten86,134,135, Bjørn Naume86,134, Alexander Fosså134,136, Cecile E. Kiserud134,136, Kristin V. Reinertsen134,136, Åslaug Helland129,134, Margit Riis131, Jürgen Geisler86,137, Osbreac138
AOCS group
David D. L. Bowtell139,140,141, Anna deFazio142,143,144, Penelope M Webb145, Georgia Chenevix-Trench145
ABCTB investigators
Christine Clarke146, Deborah Marsh147, Rodney Scott148,149, Robert Baxter150, Desmond Yip151,152, Jane Carpenter153, Alison Davis154,155, Nirmala Pathmanathan156,157, Peter Simpson158, Dinny Graham146, Mythily Sachchithananthan146
kConFab investigators
David Amor159, Lesley Andrews160, Yoland Antill161, Rosemary Balleine162, Jonathan Beesley163, Ian Bennett164, Michael Bogwitz165, Leon Botes166, Meagan Brennan167, Melissa Brown168, Michael Buckley169, Jo Burke170, Phyllis Butow171, Liz Caldon172, Ian Campbell173, Deepa Chauhan174, Manisha Chauhan175, Georgia Chenevix-Trench176, Alice Christian177, Paul Cohen178, Alison Colley179, Ashley Crook180, James Cui181, Margaret Cummings182, Sarah-Jane Dawson183, Anna DeFazio184, Martin Delatycki185, Rebecca Dickson186, Joanne Dixon187, Ted Edkins188, Stacey Edwards189, Gelareh Farshid190, Andrew Fellows191, Georgina Fenton192, Michael Field193, James Flanagan194, Peter Fong195, Laura Forrest196, Stephen Fox197, Juliet French198, Michael Friedlander199, Clara Gaff200, Mike Gattas201, Peter George202, Sian Greening203, Marion Harris204, Stewart Hart205, Nick Hayward206, John Hopper207, Cass Hoskins208, Clare Hunt209, Paul James210, Mark Jenkins211, Alexa Kidd212, Judy Kirk213, Jessica Koehler160, James Kollias214, Sunil Lakhani215, Mitchell Lawrence216, Geoff Lindeman217, Lara Lipton218, Liz Lobb219, Graham Mann220, Deborah Marsh221, Sue Anne McLachlan222, Bettina Meiser160, Roger Milne223, Sophie Nightingale224, Shona O’Connell225, Sarah O’Sullivan226, David Gallego Ortega227, Nick Pachter228, Briony Patterson229, Amy Pearn230, Kelly Phillips231, Ellen Pieper232, Edwina Rickard233, Bridget Robinson234, Mona Saleh235, Elizabeth Salisbury236, Christobel Saunders237, Jodi Saunus238, Rodney Scott239, Clare Scott240, Adrienne Sexton241, Andrew Shelling242, Peter Simpson243, Melissa Southey244, Amanda Spurdle245, Jessica Taylor246, Renea Taylor247, Heather Thorne248, Alison Trainer249, Kathy Tucker250, Jane Visvader251, Logan Walker252, Rachael Williams253, Ingrid Winship254, Mary Ann Young255
Author information
Authors and Affiliations
Consortia
Contributions
N.J., K.T., M.E.J., M.J.S., C. G., A.F.H., J.P., I.dS.S. and A.J.S. generated the data for the GWAS reported in this paper. N.J., S.M., A.M., P.M.K., J.C-C., M.K.S., N.O. and O.F. analysed the data relating to this paper and drafted the initial paper. D.F.E. coordinated the BCAC and led the iCOGS and OncoArray genotyping. P.H. led the COGS collaboration, J.S. led the OncoArray collaboration. M.K.B., Q.W. and J.D. coordinated the BCAC database. The remaining authors led individual studies and contributed to the design of the study, data collection and revising the paper.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Collection of blood samples, urine samples and questionnaire information was undertaken with written informed consent and relevant ethical review board approval in accordance with the tenets of the Declaration of Helsinki (Supplementary Table S7).
Consent to publish
Not applicable.
Data availability
GWAS data and the complete dataset for follow-up genotyping will not be made publicly available due to restraints imposed by the ethics committees of individual studies; requests for data can be made to the corresponding author (GWAS data) or the Data Access Coordination Committee (follow-up genotyping data) of BCAC (http://bcac.ccge.medschl.cam.ac.uk/). Summary results for all variants genotyped by BCAC (including rs45446698) are available at http://bcac.ccge.medschl.cam.ac.uk/.
Competing interests
M.W.B. conducts research funded by Amgen, Novartis and Pfizer. P.A.F. conducts research funded by Amgen, Novartis and Pfizer. He received honoraria from Roche, Novartis and Pfizer. A.W.K.’s institution has received research funding from Myriad Genetics for an unrelated project (funding dates 2017–2019). P.H., .P.D.P.P., O.F., K.C. and A.C. are members of the Editorial Board of BJC. The remaining authors declare no competing interests.
Funding information
This work was supported by Programme Grants from Breast Cancer Now as part of Programme Funding to the Breast Cancer Now Toby Robins Research Centre to O.F. BCAC is funded by Cancer Research UK (C1287/A16563, C1287/A10118), the European Union’s Horizon 2020 Research and Innovation Programme (grant numbers 634935 and 633784 for BRIDGES and B-CAST respectively) and by the European Community’s Seventh Framework Programme under grant agreement number 223175 (grant number HEALTH-F2-2009-223175) (COGS). The EU Horizon 2020 Research and Innovation Programme funding source had no role in study design, data collection, data analysis, data interpretation or writing of the report. Genotyping of the OncoArray was funded by the NIH Grant U19 CA148065, and Cancer UK Grant C1287/A16563 and the PERSPECTIVE project supported by the Government of Canada through Genome Canada and the Canadian Institutes of Health Research (grant GPH-129344) and, the Ministère de l’Économie, Science et Innovation du Québec through Genome Québec and the PSR-SIIRI-701 grant, and the Quebec Breast Cancer Foundation. Funding for the iCOGS infrastructure came from the European Community’s Seventh Framework Programme under grant agreement no. 223175 (HEALTH-F2-2009-223175) (COGS), Cancer Research UK (C1287/A10118, C1287/A10710, C12292/A11174, C1281/A12014, C5047/A8384, C5047/A15007, C5047/A10692 and C8197/A16565), the National Institutes of Health (CA128978) and Post-Cancer GWAS initiative (1U19 CA148537, 1U19 CA148065 and 1U19 CA148112—the GAME-ON initiative), the Department of Defence (W81XWH-10-1-0341), the Canadian Institutes of Health Research (CIHR) for the CIHR Team in Familial Risks of Breast Cancer and Komen Foundation for the Cure, the Breast Cancer Research Foundation and the Ovarian Cancer Research Fund. The DRIVE Consortium was funded by U19 CA148065. The Australian Breast Cancer Family Study (ABCFS) was supported by grant UM1 CA164920 from the National Cancer Institute (USA). The content of this paper does not necessarily reflect the views or policies of the National Cancer Institute or any of the collaborating centres in the Breast Cancer Family Registry (BCFR) nor does mention of trade names, commercial products, or organisations imply endorsement by the USA Government or the BCFR. The ABCFS was also supported by the National Health and Medical Research Council of Australia, the New South Wales Cancer Council, the Victorian Health Promotion Foundation (Australia) and the Victorian Breast Cancer Research Consortium. J.L.H. is a National Health and Medical Research Council (NHMRC) Senior Principal Research Fellow. M.C.S. is a NHMRC Senior Research Fellow. The ABCS study was supported by the Dutch Cancer Society [grants NKI 2007-3839; 2009 4363]. The Australian Breast Cancer Tissue Bank (ABCTB) was supported by the National Health and Medical Research Council of Australia, The Cancer Institute NSW and the National Breast Cancer Foundation. The AHS study is supported by the intramural research program of the National Institutes of Health, the National Cancer Institute (grant number Z01-CP010119), and the National Institute of Environmental Health Sciences (grant number Z01-ES049030). The work of the BBCC was partly funded by ELAN-Fond of the University Hospital of Erlangen. The BBCS is funded by Cancer Research UK and Breast Cancer Now and acknowledges NHS funding to the NIHR Biomedical Research Centre, and the National Cancer Research Network (NCRN). The BCEES was funded by the National Health and Medical Research Council, Australia and the Cancer Council Western Australia and acknowledges funding from the National Breast Cancer Foundation (JS). For BIGGS, ES is supported by NIHR Comprehensive Biomedical Research Centre, Guy’s & St. Thomas’ NHS Foundation Trust in partnership with King’s College London, United Kingdom. IT is supported by the Oxford Biomedical Research Centre. The BREast Oncology GAlician Network (BREOGAN) is funded by Acción Estratégica de Salud del Instituto de Salud Carlos III FIS PI12/02125/Cofinanciado FEDER; Acción Estratégica de Salud del Instituto de Salud Carlos III FIS Intrasalud (PI13/01136); Programa Grupos Emergentes, Cancer Genetics Unit, Instituto de Investigacion Biomedica Galicia Sur. Xerencia de Xestion Integrada de Vigo-SERGAS, Instituto de Salud Carlos III, Spain; Grant 10CSA012E, Consellería de Industria Programa Sectorial de Investigación Aplicada, PEME I + D e I + D Suma del Plan Gallego de Investigación, Desarrollo e Innovación Tecnológica de la Consellería de Industria de la Xunta de Galicia, Spain; Grant EC11-192. Fomento de la Investigación Clínica Independiente, Ministerio de Sanidad, Servicios Sociales e Igualdad, Spain; and Grant FEDER-Innterconecta. Ministerio de Economia y Competitividad, Xunta de Galicia, Spain. The BSUCH study was supported by the Dietmar-Hopp Foundation, the Helmholtz Society and the German Cancer Research Center (DKFZ). CBCS is funded by the Canadian Cancer Society (grant # 313404) and the Canadian Institutes of Health Research. CCGP is supported by funding from the University of Crete. The CECILE study was supported by Fondation de France, Institut National du Cancer (INCa), Ligue Nationale contre le Cancer, Agence Nationale de Sécurité Sanitaire, de l’Alimentation, de l’Environnement et du Travail (ANSES), Agence Nationale de la Recherche (ANR). The CGPS was supported by the Chief Physician Johan Boserup and Lise Boserup Fund, the Danish Medical Research Council and Herlev and Gentofte Hospital. The CNIO-BCS was supported by the Instituto de Salud Carlos III, the Red Temática de Investigación Cooperativa en Cáncer and grants from the Asociación Española Contra el Cáncer and the Fondo de Investigación Sanitario (PI11/00923 and PI12/00070). The American Cancer Society funds the creation, maintenance, and updating of the CPS-II cohort. The California Teachers Study and the research reported in this publication were supported by the National Cancer Institute of the National Institutes of Health under award number U01-CA199277; P30-CA033572; P30-CA023100; UM1-CA164917; and R01-CA077398. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute or the National Institutes of Health. The collection of cancer incidence data used in the California Teachers Study was supported by the California Department of Public Health pursuant to California Health and Safety Code Section 103885; Centers for Disease Control and Prevention’s National Program of Cancer Registries, under cooperative agreement 5NU58DP006344; the National Cancer Institute’s Surveillance, Epidemiology and End Results Program under contract HHSN261201800032I awarded to the University of California, San Francisco, contract HHSN261201800015I awarded to the University of Southern California, and contract HHSN261201800009I awarded to the Public Health Institute. The opinions, findings, and conclusions expressed herein are those of the author(s) and do not necessarily reflect the official views of the State of California, Department of Public Health, the National Cancer Institute, the National Institutes of Health, the Centers for Disease Control and Prevention or their Contractors and Subcontractors, or the Regents of the University of California, or any of its programs. The coordination of EPIC is financially supported by the European Commission (DG-SANCO) and the International Agency for Research on Cancer. The national cohorts are supported by: Ligue Contre le Cancer, Institut Gustave Roussy, Mutuelle Générale de l’Education Nationale, Institut National de la Santé et de la Recherche Médicale (INSERM) (France); German Cancer Aid, German Cancer Research Center (DKFZ), Federal Ministry of Education and Research (BMBF) (Germany); the Hellenic Health Foundation, the Stavros Niarchos Foundation (Greece); Associazione Italiana per la Ricerca sul Cancro-AIRC-Italy and National Research Council (Italy); Dutch Ministry of Public Health, Welfare and Sports (VWS), Netherlands Cancer Registry (NKR), LK Research Funds, Dutch Prevention Funds, Dutch ZON (Zorg Onderzoek Nederland), World Cancer Research Fund (WCRF), Statistics Netherlands (The Netherlands); Health Research Fund (FIS), PI13/00061 to Granada, PI13/01162 to EPIC-Murcia, Regional Governments of Andalucía, Asturias, Basque Country, Murcia and Navarra, ISCIII RETIC (RD06/0020) (Spain); Cancer Research UK (14136 to EPIC-Norfolk; C570/A16491 and C8221/A19170 to EPIC-Oxford), Medical Research Council (1000143 to EPIC-Norfolk, MR/M012190/1 to EPIC-Oxford) (United Kingdom). The ESTHER study was supported by a grant from the Baden Württemberg Ministry of Science, Research and Arts. Additional cases were recruited in the context of the VERDI study, which was supported by a grant from the German Cancer Aid (Deutsche Krebshilfe). FHRISK is funded from NIHR grant PGfAR 0707-10031. The GC-HBOC (German Consortium of Hereditary Breast and Ovarian Cancer) is supported by the German Cancer Aid (grant no 110837, coordinator: Rita K. Schmutzler, Cologne). This work was also funded by the European Regional Development Fund and Free State of Saxony, Germany (LIFE—Leipzig Research Centre for Civilization Diseases, project numbers 713-241202, 713-241202, 14505/2470, 14575/2470). The GENICA was funded by the Federal Ministry of Education and Research (BMBF) Germany grants 01KW9975/5, 01KW9976/8, 01KW9977/0 and 01KW0114, the Robert Bosch Foundation, Stuttgart, Deutsches Krebsforschungszentrum (DKFZ), Heidelberg, the Institute for Prevention and Occupational Medicine of the German Social Accident Insurance, Institute of the Ruhr University Bochum (IPA), Bochum, as well as the Department of Internal Medicine, Evangelische Kliniken Bonn gGmbH, Johanniter Krankenhaus, Bonn, Germany; gefördert durch die Deutsche Forschungsgemeinschaft (DFG) im Rahmen der Exzellenzstrategie des Bundes und der Länder—EXC 2180—390900677; German Cancer Consortium (DKTK) and German Cancer Research Center (DKFZ). The GESBC was supported by the Deutsche Krebshilfe e. V. [70492] and the German Cancer Research Center (DKFZ). The HABCS study was supported by the Claudia von Schilling Foundation for Breast Cancer Research, by the Lower Saxonian Cancer Society, and by the Rudolf Bartling Foundation. The HEBCS was financially supported by the Helsinki University Hospital Research Fund, the Finnish Cancer Society, and the Sigrid Juselius Foundation. The HMBCS was supported by a grant from the Friends of Hannover Medical School and by the Rudolf Bartling Foundation. The HUBCS was supported by a grant from the German Federal Ministry of Research and Education (RUS08/017), B.M. was supported by grant 17-44-020498, 17-29-06014 of the Russian Foundation for Basic Research, D.P. was supported by grant 18-29-09129 of the Russian Foundation for Basic Research, E.K. was supported by the program for support the bioresource collections №007-030164/2, and the study was performed as part of the assignment of the Ministry of Science and Higher Education of the Russian Federation (№АААА-А16-116020350032-1). Financial support for KARBAC was provided through the regional agreement on medical training and clinical research (ALF) between Stockholm County Council and Karolinska Institutet, the Swedish Cancer Society, The Gustav V Jubilee foundation and Bert von Kantzows foundation. The KARMA study was supported by Märit and Hans Rausings Initiative Against Breast Cancer. The KBCP was financially supported by the special Government Funding (EVO) of Kuopio University Hospital grants, Cancer Fund of North Savo, the Finnish Cancer Organizations, and by the strategic funding of the University of Eastern Finland. kConFab is supported by a grant from the National Breast Cancer Foundation, and previously by the National Health and Medical Research Council (NHMRC), the Queensland Cancer Fund, the Cancer Councils of New South Wales, Victoria, Tasmania and South Australia, and the Cancer Foundation of Western Australia. Financial support for the AOCS was provided by the United States Army Medical Research and Materiel Command [DAMD17-01-1-0729], Cancer Council Victoria, Queensland Cancer Fund, Cancer Council New South Wales, Cancer Council South Australia, The Cancer Foundation of Western Australia, Cancer Council Tasmania and the National Health and Medical Research Council of Australia (NHMRC; 400413, 400281, 199600). G.C.T. and P.W. are supported by the NHMRC. R.B. was a Cancer Institute NSW Clinical Research Fellow. L.M.B.C. is supported by the ‘Stichting tegen Kanker’. D.L. is supported by the FWO. The MABCS study is funded by the Research Centre for Genetic Engineering and Biotechnology “Georgi D. Efremov”, MASA. The MARIE study was supported by the Deutsche Krebshilfe e.V. [70-2892-BR I, 106332, 108253, 108419, 110826, 110828], the Hamburg Cancer Society, the German Cancer Research Center (DKFZ) and the Federal Ministry of Education and Research (BMBF) Germany [01KH0402]. MBCSG is supported by grants from the Italian Association for Cancer Research (AIRC) and by funds from the Italian citizens who allocated the 5/1000 share of their tax payment in support of the Fondazione IRCCS Istituto Nazionale Tumori, according to Italian laws (INT-Institutional strategic projects “5x1000”). The MCBCS was supported by the NIH grants CA192393, CA116167, CA176785 an NIH Specialized Program of Research Excellence (SPORE) in Breast Cancer (CA116201), and the Breast Cancer Research Foundation and a generous gift from the David F. and Margaret T. Grohne Family Foundation. The Melbourne Collaborative Cohort Study (MCCS) cohort recruitment was funded by VicHealth and Cancer Council Victoria. The MCCS was further augmented by Australian National Health and Medical Research Council grants 209057, 396414 and 1074383 and by infrastructure provided by Cancer Council Victoria. Cases and their vital status were ascertained through the Victorian Cancer Registry and the Australian Institute of Health and Welfare, including the National Death Index and the Australian Cancer Database. The MEC was supported by NIH grants CA63464, CA54281, CA098758, CA132839 and CA164973. The MISS study is supported by funding from ERC-2011-294576 Advanced grant, Swedish Cancer Society, Swedish Research Council, Local hospital funds, Berta Kamprad Foundation, Gunnar Nilsson. The MMHS study was supported by NIH grants CA97396, CA128931, CA116201, CA140286 and CA177150. The work of MTLGEBCS was supported by the Quebec Breast Cancer Foundation, the Canadian Institutes of Health Research for the “CIHR Team in Familial Risks of Breast Cancer” program—grant # CRN-87521 and the Ministry of Economic Development, Innovation and Export Trade—grant # PSR-SIIRI-701. The NBCS has received funding from the K.G. Jebsen Centre for Breast Cancer Research; the Research Council of Norway grant 193387/V50 (to A.-L. Børresen-Dale and V.N. Kristensen) and grant 193387/H10 (to A.-L. Børresen-Dale and V.N. Kristensen), South Eastern Norway Health Authority (grant 39346 to A.-L. Børresen-Dale) and the Norwegian Cancer Society (to A.-L. Børresen-Dale and V.N. Kristensen). The NBHS was supported by NIH grant R01 CA100374. Biological sample preparation was conducted the Survey and Biospecimen Shared Resource, which is supported by P30 CA68485. The Northern California Breast Cancer Family Registry (NC-BCFR) and Ontario Familial Breast Cancer Registry (OFBCR) were supported by grants U01 CA164920 and U01CA167551 from the USA National Cancer Institute of the National Institutes of Health. The content of this paper does not necessarily reflect the views or policies of the National Cancer Institute or any of the collaborating centres in the Breast Cancer Family Registry (BCFR) or the Colon Cancer Family Registry (CCFR) nor does mention of trade names, commercial products, or organisations imply endorsement by the USA Government or the BCFR or CCFR. The Carolina Breast Cancer Study (NCBCS) was funded by Komen Foundation, the National Cancer Institute (P50 CA058223, U54 CA156733, U01 CA179715), and the North Carolina University Cancer Research Fund. The NHS was supported by NIH grants P01 CA87969, UM1 CA186107, and U19 CA148065. The NHS2 was supported by NIH grants UM1 CA176726 and U19 CA148065. The OBCS was supported by research grants from the Finnish Cancer Foundation, the Academy of Finland (grant number 250083, 122715 and Center of Excellence grant number 251314), the Finnish Cancer Foundation, the Sigrid Juselius Foundation, the University of Oulu, the University of Oulu Support Foundation and the special Governmental EVO funds for Oulu University Hospital-based research activities. The ORIGO study was supported by the Dutch Cancer Society (RUL 1997-1505) and the Biobanking and Biomolecular Resources Research Infrastructure (BBMRI-NL CP16). The PBCS was funded by Intramural Research Funds of the National Cancer Institute, Department of Health and Human Services, USA. Genotyping for PLCO was supported by the Intramural Research Program of the National Institutes of Health, NCI, Division of Cancer Epidemiology and Genetics. The PLCO is supported by the Intramural Research Program of the Division of Cancer Epidemiology and Genetics and supported by contracts from the Division of Cancer Prevention, National Cancer Institute, National Institutes of Health. PROCAS is funded from NIHR grant PGfAR 0707-10031. The RBCS was funded by the Dutch Cancer Society (DDHK 2004-3124, DDHK 2009–4318). The SASBAC study was supported by funding from the Agency for Science, Technology and Research of Singapore (A*STAR), the US National Institute of Health (NIH) and the Susan G. Komen Breast Cancer Foundation. The SBCS was supported by Sheffield Experimental Cancer Medicine Centre and Breast Cancer Now Tissue Bank. SEARCH is funded by Cancer Research UK (C490/A10124, C490/A16561) and supported by the UK National Institute for Health Research Biomedical Research Centre at the University of Cambridge. The University of Cambridge has received salary support for PDPP from the NHS in the East of England through the Clinical Academic Reserve. The Sister Study (SISTER) is supported by the Intramural Research Program of the NIH, National Institute of Environmental Health Sciences (Z01-ES044005 and Z01-ES049033). The SMC is funded by the Swedish Cancer Foundation and the Swedish Research Council (VR 2017-00644) grant for the Swedish Infrastructure for Medical Population-based Life-course Environmental Research (SIMPLER). The SZBCS was supported by Grant PBZ_KBN_122/P05/2004 and the program of the Minister of Science and Higher Education under the name “Regional Initiative of Excellence” in 2019–2022 project number 002/RID/2018/19 amount of financing 12,000,000 PLN. The TNBCC was supported by: a Specialized Program of Research Excellence (SPORE) in Breast Cancer (CA116201), a grant from the Breast Cancer Research Foundation, a generous gift from the David F. and Margaret T. Grohne Family Foundation. The UCIBCS component of this research was supported by the NIH [CA58860, CA92044] and the Lon V Smith Foundation [LVS39420]. The UKBGS is funded by Breast Cancer Now and the Institute of Cancer Research (ICR), London. ICR acknowledges NHS funding to the NIHR Biomedical Research Centre. The USRT Study was funded by Intramural Research Funds of the National Cancer Institute, Department of Health and Human Services, USA.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Members of the NBCS Collaborators, AOCS Group, ABCTB Investigators and kConFab Investigators are listed above Acknowledgements.
A full list of authors and their affiliations appear at the end of the paper
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Johnson, N., Maguire, S., Morra, A. et al. CYP3A7*1C allele: linking premenopausal oestrone and progesterone levels with risk of hormone receptor-positive breast cancers. Br J Cancer 124, 842–854 (2021). https://doi.org/10.1038/s41416-020-01185-w
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41416-020-01185-w
This article is cited by
-
Common variability in oestrogen-related genes and pancreatic ductal adenocarcinoma risk in women
Scientific Reports (2022)