Abstract
Background
Oestrogen Receptor 1 (ESR1) mutations are frequently acquired in oestrogen receptor (ER)-positive metastatic breast cancer (MBC) patients who were treated with aromatase inhibitors (AI) in the metastatic setting. Acquired ESR1 mutations are associated with poor prognosis and there is a lack of effective therapies that selectively target these cancers.
Methods
We performed a proteomic kinome analysis in ESR1 Y537S mutant cells to identify hyperactivated kinases in ESR1 mutant cells. We validated Recepteur d’Origine Nantais (RON) and PI3K hyperactivity through phospho-immunoblot analysis, organoid growth assays, and in an in vivo patient-derived xenograft (PDX) metastatic model.
Results
We demonstrated that RON was hyperactivated in ESR1 mutant models, and in acquired palbociclib-resistant (PalbR) models. RON and insulin-like growth factor 1 receptor (IGF-1R) interacted as shown through pharmacological and genetic inhibition and were regulated by the mutant ER as demonstrated by reduced phospho-protein expression with endocrine therapies (ET). We show that ET in combination with a RON inhibitor (RONi) decreased ex vivo organoid growth of ESR1 mutant models, and as a monotherapy in PalbR models, demonstrating its therapeutic efficacy. Significantly, ET in combination with the RONi reduced metastasis of an ESR1 Y537S mutant PDX model.
Conclusions
Our results demonstrate that RON/PI3K pathway inhibition may be an effective treatment strategy in ESR1 mutant and PalbR MBC patients. Clinically our data predict that ET resistance mechanisms can also contribute to CDK4/6 inhibitor resistance.

Similar content being viewed by others
Background
Over 600,000 women die each year from metastatic breast cancer (MBC) worldwide.1 Despite effective endocrine and targeted therapies to treat oestrogen receptor (ER)-positive breast cancer (BC), 20% of women develop recurrence and/or metastasis.2 The most common mechanism of endocrine therapy (ET) resistance is the acquisition of Oestrogen Receptor 1 (ESR1) mutations, which are characterised as having hormone hypersensitive or ligand-independent transcriptional activity.3,4,5,6 The current clinical approach for patients with ESR1 mutations is to treat with a selective oestrogen receptor degrader (SERD) in combination with a cyclin-dependent kinase (CDK) 4/6 inhibitor.7 However, there are no effective therapies that can selectively target ESR1 mutant cancers, and patients with cancers expressing ESR1 mutations have a worse progression-free (PFS) and overall survival (OS) compared to cancers expressing only wild-type (WT) ESR1.3,8,9,10,11,12 ctDNA analysis in the SoFEA and PALOMA-3 clinical trials found that ESR1 mutations were present in 39% and 25% of patients, respectively. Patients in these trials were treated with an aromatase inhibitor (AI) or fulvestrant (Ful) containing regimens, demonstrating the ESR1 mutant clinical resistance to both of these ETs. Therefore, there is an unmet need to identify actionable targets in ESR1 mutant cancers, which are required to better control metastatic disease.
The addition of CDK4/6 inhibitors to ET has improved PFS and OS in ER-positive BC.13,14,15,16,17,18,19 However, an analysis of the PALOMA-3 clinical trial showed that there was a selection for the ESR1 Y537S mutation at end of treatment in patients who were treated with Ful alone or in combination with the CDK4/6 inhibitor palbociclib.20 These data demonstrated that the ESR1 Y537S mutation was intrinsically resistant to this combination in ESR1 mutant-positive MBC patients. Therefore, we interrogated the kinome of ESR1 mutant and palbociclib resistant (PalbR) models with the goal of discovering novel therapeutic strategies for this patient population.
We and others have previously shown that the insulin-like growth factor-1 receptor (IGF-1R) signalling pathway was upregulated in ESR1 mutant BCs and was associated with ET resistance.21,22 Inhibition of IGF-1R restored ET sensitivity, and this finding is an encouraging outcome for the clinical co-targeting of ER and IGF-1R. However, IGF-1R inhibitors have had limited clinical utility in BC, potentially due to the reliance on insulin receptor to propagate insulin growth factor 2 (IGF-2) signalling, as well as IGF-1R inhibitor induced hyperglycemia.23,24 In this study we aimed to identify additional kinases that may cooperate with IGF-1R, and found that recepteur d’origine nantais (RON) kinase activation is an escape driver pathway with therapeutic vulnerability in ESR1 mutant MBC.
RON is a growth factor receptor tyrosine kinase (RTK) that is expressed on resident macrophages and epithelial cells. RON can be activated by its ligand, macrophage stimulating protein (MSP),25 or by ligand-independent dimerisation in RON-overexpressed cancers.26 The oncogenic potential of RON signalling has been demonstrated in several epithelial cancer types, where RON propagates signalling pathways involved in cell motility, invasion, epithelial-mesenchymal transition, and metastasis.27,28,29 In BC, RON primarily signals through the PI3K and MAPK pathways.28,30 Previous studies have shown that overexpression of RON is sufficient to induce metastasis in ER-positive BC cell lines, and pharmacological inhibition with a small molecule inhibitor selective for RON delayed progression in RON-overexpressing ER-positive BC patient-derived xenograft (PDX) models.31,32 Additionally, knockout of the RON tyrosine kinase domain resulted in reduced lung metastasis in an ER-negative mouse tumour model.33 RON overexpression has been shown to confer tamoxifen (Tam) resistance in ER-positive BC cell lines;30 however, the mechanisms by which the mutant ER can regulate RON activity has not been reported. Although previous studies have shown that RON activation could cross talk with ER signalling,30 our data demonstrated that the mutant ER could hyperactivate RON signalling to promote therapeutic resistance, and that RON inhibition inhibited metastasis of an ESR1 Y537S mutant PDX model. Herein, we use a proteomic approach to identify hyperactivated kinase pathways in ESR1 mutant models. Our data demonstrated that ESR1 mutations induce RTK/PI3K hyperactivation and identified that RON and IGF-1R cooperate to promote ET resistance and metastasis. We also demonstrated that ESR1 mutant and PalbR models exhibited a shared transcriptional reprogramming that suggests acquired ESR1 mutations may predispose cancers to palbociclib resistance.
Methods
KinoBead proteomics
KinoBead proteomics (KiP) analysis was developed using modified techniques.34 Briefly, MCF-7 ESR1 WT and ESR1 Y537S cells were cultured in minimal essential media (MEM) supplemented with 10% foetal bovine serum (FBS), 1X non-essential amino acids (NEAA), and 1X penicillin-streptomycin for 72 h before harvesting for KiP analysis. Fold changes are represented as mass spectrometry (MS) intensity based absolute quantification (IBAQ) values for each kinase.
siRNA knockdown
siRNA transfections were performed as previously described.21 Cells were seeded in 6-well plates and transfected with indicated siRNA (Silencer Select Negative Control No.1 4390843, MST1R s8996, s8997, s8998, PIK3R1 s10535, PIK3R3 s16151, PIK3R4 s105, GAB2 s19052 from Life Technologies, Carlsbad, CA and ESR1 J-003401-11-0005, J-003401-12-0005 from Dharmacon, Lafayette, CO) and Lipofectamine RNAiMAX reagent (Life Technologies, Carlsbad, CA) using the manufacturer’s suggested protocol. Two days post transfection, cells were harvested for downstream analysis.
MTT growth assay
MTT growth assays were performed as previously described.35 Treated plates were analysed at day 7. The absorbance was measured at 570 nm with background subtraction at 655 nm.
Immunoblot analysis
Cells were prepared and harvested for immunoblot analysis as previously described.35 Membranes were incubated with primary antibodies overnight: RON (cat# 2654S), PR (cat# 8757S), c-Jun (cat# 9165S), CCND1 (cat# 2978S), IGF-1R (cat# 3027S), phospho-IGF-1R/Insulin Receptor (cat# 3021S), p-AKT S473 (cat# 4060S), AKT (cat# 9272S), p-p44/42 (cat# 4370S), p44/42 (cat# 4695S), p85 (cat# 4257S), p55 (cat# 11889S) were purchased from Cell Signaling Technologies, Danvers, MA. GAPDH (cat# sc-25778) was purchased from Santa Cruz Biotechnology, Dallas, TX. p-RON was purchased from R&D Systems, Minneapolis, MN. Oestrogen Receptor α (cat# MA5-13304) was purchased from Life Technologies, Carlsbad, CA. Membranes were washed and incubated with HRP-linked secondary antibody (cat# NXA931 and NA934V) purchased from MilliporeSigma, Burlington, MA before developing.
Organoid growth assay
Ex vivo organoid growth assays were developed using a modified protocol.36 Primary tumours were harvested from athymic nude (xenograft models) or SCID/Beige (PDX models) mice when tumour reached a volume of 800 mm3. Metastatic tumours were harvested at necropsy. Tumours were minced and digested in Advanced DMEM/F12 media (Corning, Tewksbury, MA) with 5% FBS, 50 µg/mL gentamicin, 5 µg/mL insulin, 1 mg/mL collagenase A, and 300 µg/mL trypsin by shaking for 30 min. Organoids were collected by differential centrifugation and plated in 24-well plates at 50 organoids per well. Organoids were supplemented with Advanced DMEM/F12 media containing 10% FBS, 1X penicillin-streptomycin, 1X Insulin-Transferrin-Selenium (ITS), 4 µg/mL basic fibroblast growth factor (bFGF) and 50 µg/mL epidermal growth factor (EGF). Two days after plating, the media was changed, and inhibitors were added. 4-Hydroxytamoxifen (Tam), BMS-777607/ASLAN002 (RONi), and taselisib (PI3Ki) were purchased from Selleck Chemicals, Houston, TX. Media and inhibitors were changed twice a week for 2 weeks. Organoids were imaged and analysed using GelCount software (Oxford Optronix LTD., Abingdon, UK). Data is represented as the number of organoids greater than 100 µm. Each treatment had a minimum of three replicates for analysis.
In vivo tumour growth and metastasis experiments
In all, 5 × 106 MCF-7 ESR1 WT, Y537S, or LTED cells were injected into the mammary fat pad of 8-week old oophorectomised athymic nude mice. For PDX tumour transplants, tumour pieces were transplanted into 8-week old SCID/Beige mice. Tumour size was measured twice per week using callipers. When tumours reached 350 mm3, animals were randomised to treatment as indicated. Experimenters were unblinded to group allocation. β-oestradiol was purchased from MilliporeSigma, Burlington, MA. BMS-777607/ASLAN002 (RONi, cat # S1561) was purchased from Selleck Chemicals, Houston, TX and palbociclib (cat # HY-50767) was purchased from MedChemExpress, Monmouth Junction, NJ. Tamoxifen citrate (cat #T9262) was purchased from Sigma-Aldrich. Mice were treated with 50 mg/kg RONi by oral gavage three times per week. Mice were treated with 60 mg/kg/day tamoxifen citrate in corn oil by subcutaneous injection. Mice were treated with 125 mg/kg palbociclib five days per week. When tumour sizes reached 800 mm3, survival surgery was performed to remove the primary tumour, and animals remained on treatment. For survival surgery procedures, mice were treated with subcutaneous injection of 1 mg/kg buprenorphine one hour prior to surgery and maintained on oxygen and isoflurane at a flow rate of 2.5% during the surgical procedure. Mice had 5 days of post-surgical monitoring and were treated with 2.5 mg/kg meloxicam for 3 days post surgery. Four months after transplantation, animals were euthanised by isoflurane inhalation followed by cervical dislocation and necropsy. Necropsy was performed to collect metastatic tumours and major organs (lungs, liver, and brain). Tumours and organs were FFPE and prepared for H&E and IHC analysis. A portion of tumour was flash frozen to be used for immunoblot analysis. Sample size was calculated to detect differences of 50% between any two groups with 80% power. Time to tumour halving and time to tumour doubling were estimated using Kaplan-Meier analysis and the Generalized Wilcoxon test adjusted for multiple comparisons. Metastatic frequency was analysed using Fisher’s Exact test. Analysis was performed in GraphPad Prism 8.
qRT-PCR
Cells were plated in 6-well plates and treated as indicated for two days before harvesting. RNA was extracted using RNeasy Mini Kit (cat# 74104, Qiagen, Germantown, MD) following manufacturer’s protocol. Reverse transcription of RNA was performed using iScript RT Supermix (cat# 1708842; BioRad, Hercules, CA). qPCR was performed on the BioRad CFX96. Analysis was performed using CFX Maestro software. GAPDH was used for normalisation.
ER transactivation assays
ER transactivation assays were performed as previously described.35 At 24 h post transfection, cells were treated as indicated. After 24 h, cells were harvested and luciferase signal was measured using the Luciferase Assay System (Promega, Madison, WI). β-galactosidase expression was used for transfection efficiency normalisation.
Statistical analysis
For in vitro MTT assays, data is represented by mean of triplicates and standard deviation. One- or two-way ANOVA was performed as indicated. For qRT-PCR experiments, data is represented by mean of triplicates and standard error of the mean. Two-way ANOVA was performed for statistical analysis. For transactivation assays and ER-ChIP, data is represented by mean of triplicates and standard deviation. Student’s t-test was used for statistical analysis. In ex vivo organoid experiments, data is represented by mean number of organoids of triplicates and standard deviation. Student’s t-test or one-way ANOVA was used for statistical analysis, as indicated. For all one-way ANOVA analyses, the Tukey multiple comparisons test was performed. For all two-way ANOVA analyses, the Dunnett’s multiple comparisons test was performed. For in vivo studies, the Mantel-Cox test was used for primary tumour survival analysis and the Fisher’s Exact test was used for metastatic frequency analysis. Statistical analyses were performed in GraphPad Prism 8.0.
Additional Methods including cell culture, generation of CRISPR/Cas9 models, ddPCR, microarray analysis, ChIP-seq and ChIP-PCR are in Supplementary Materials.
Results
ESR1 mutant models exhibited oestrogen-independent growth and enhanced metastatic propensity
ESR1 mutations are principally acquired in patients who were treated with an AI in the metastatic setting.3 Thus, we developed ER-positive BC cell lines that express ESR1 mutations utilising CRISPR/Cas9 to knock-in homozygous ESR1 Y537S mutations in MCF-7 and T47D cell lines. We also generated a long-term oestrogen-deprived (LTED) model by culturing MCF-7 cells in hormone-deprived media for 6 months. MCF-7 ESR1 mutant and LTED cells were relatively growth resistant to Tam and Ful, in vitro (Fig. 1a). T47D ESR1 Y537S cells were relatively resistant to low concentrations of Tam and Ful, but were ET sensitive at supraphysiological concentrations (Fig. 1b). These results are consistent with previous studies that demonstrated ESR1 mutations are relatively resistant to Tam and Ful and required higher concentrations of ET to suppress ESR1 mutant cell growth.6 MCF-7 and T47D ESR1 Y537S cells, and the MCF-7 LTED cells exhibited an oestrogen-independent upregulation of ER-regulated proteins cultured in hormone deprived (5% charcoal-stripped FBS) media. The MCF-7 ESR1 Y537S model constitutively overexpressed progesterone receptor (PR), Cyclin D1 (CCND1), and c-Jun proteins, and the MCF-7 LTED model overexpressed PR and CCND1 (Fig. 1c). T47D ESR1 Y537S cells constitutively overexpressed PR (Fig. 1d). PR mRNA expression was also upregulated in MCF-7 ESR1 Y537S and LTED cells, but not in T47D ESR1 Y537S cells given that T47D cells have PR amplification (Fig. S1). We observed differences in total ER protein levels between ESR1 WT and ESR1 mutant cells. We postulated that differences in cellular localisation and insufficient cellular nuclear extraction with RIPA buffer contributed to this observation. To explore this, we performed subcellular fractionation of MCF-7 and T47D cells that were cultured in hormone-deprived conditions (Figs. 1c, d and S2). ER was localised in both the cytoplasm and nucleus of ESR1 WT cells; however, ER was almost exclusively localised in the nucleus of MCF-7 and T47D ESR1 Y537S and MCF-7 LTED cells. We conclude that mutant ER protein was localised to the nucleus as a consequence of its constitutive activation.
a MTT growth assay of Tam and Ful-treated MCF-7 and b T47D cells. Percent survival is normalised to no treatment controls. Graphs represent mean + standard deviation (N = 3 replicates) one-way ANOVA was used for statistical analysis. c MCF-7 cells were cultured in charcoal-stripped serum supplemented media, underwent whole cell lysis, and analysed by immunoblot for ER-regulated proteins (left). Values below PR, CCND1 and c-Jun represent normalised densitometry of each protein compared to GAPDH. PR was not detected in WT cells; thus, relative expression was normalised to Y537S cells. Cells were cultured in charcoal-stripped supplemented media and underwent cellular fractionation (right). Nuclear fraction is shown. d T47D cells were cultured in charcoal-stripped serum supplemented media and analysed by immunoblot for ER-regulated proteins (left). Values below PR, CCND1 and c-Jun represent normalised densitometry of each protein compared to GAPDH. PR was not detected in WT cells. Cells were cultured in charcoal-stripped supplemented media and underwent cellular fractionation (right). e Venn diagram of upregulated MSigDB Hallmark gene sets in MCF-7 ESR1 Y537S and LTED cells compared to MCF-7 ESR1 WT, and T47D ESR1 Y537S compared to T47D ESR1 WT. The numbers shown are of Hallmarks with a false discovery rate < 0.25. f In vivo individual tumour growth curves of MCF-7 ESR1 WT, ESR1 Y537S and LTED tumours. g In vivo primary tumour growth of MCF-7 WT, ESR1 Y537S and LTED tumours shown by time to tumour halving after E2 withdrawal. The Mantel-Cox test was used for statistical analysis (WT N = 5, Y537S N = 8, LTED N = 9). h Metastatic frequency in MCF-7 ESR1 WT-, ESR1 Y537S- and LTED-transplanted mice (WT N = 5, Y537S N = 8, LTED N = 9). Metastatic frequency is represented by the total number of mice per group that exhibited a macrometastasis. Fisher’s Exact test was used for statistical analysis. P < 0.05 was considered statistically significant in all tests. (*p < 0.05, **p < 0.01, ***p < 0.001).
We found that the LTED model spontaneously acquired polyclonal ESR1 mutations, corroborating previous reports of BC cell lines acquiring ESR1 mutations under LTED conditions.37 We quantified allele frequencies of ESR1 WT, Y537C, Y537N, Y537S, and D538G mutations using digital droplet (dd)PCR in MCF-7 ESR1 WT and LTED cells, and detected the Y537N and Y537C mutations in the LTED cells at a frequency of 31% and 21%, respectively (Fig. S3a), but did not detect these ESR1 mutations in the WT cells (Fig. S3b). The acquisition of polyclonal ESR1 mutations in a single cell line suggests that certain ESR1 mutant subpopulations may influence each other and co-evolve, as opposed to a single dominant ESR1 mutant clone outcompeting other ESR1 mutant-expressing cells. To define an ESR1 mutant transcriptional signature, we performed microarray analyses of MCF-7 and T47D ESR1 WT and mutant cell lines, and performed Gene Set Enrichment Analyses to identify upregulated Molecular Signature Database (MSigDB) cancer Hallmarks in ESR1 mutant cells compared to WT cells (Fig. 1e and Tables S1–3). The three ESR1 mutant models had a shared upregulation of oestrogen response, interferon response, mTORC signalling, and genes downregulated by KRAS activation. Additional upregulated pathways in ESR1 mutant cells include genes involved in EMT, DNA repair, and mitogenic signalling. These data demonstrate that different ESR1 mutations express a shared oestrogen-independent transcriptome that contributes to ET resistance.
We next performed in vivo experiments to determine the oestrogen-independent growth and metastatic frequency of ESR1 mutant-expressing tumours. We used MCF-7 ESR1 WT, ESR1 Y537S, and LTED cells to inject orthotopically into oophorectomised immune-deficient mice that were treated with oestradiol (E2) until primary tumours reached 350-700 mm3, and then E2 was removed to simulate AI treatment. We found MCF-7 ESR1 WT tumours regressed after E2 withdrawal, whereas Y537S mutant tumours continued to grow (Fig. 1f, g). The MCF-7 LTED primary tumours exhibited static tumour growth upon oestrogen withdrawal but did not regress. Primary tumours were resected after reaching 800 mm3, and at four months post-transplantation mice were euthanised. We determined metastatic frequency by assessing the presence of tumours in the omentum and major organs. While none of the mice transplanted with MCF-7 ESR1 WT cells exhibited metastases, the frequency of metastasis in mice transplanted with Y537S mutant cells was 63% (p = 0.0310), and the frequency in mice transplanted with the LTED cells was 89% (p = 00014). (Fig. 1h). The latter findings suggest that cells co-expressing endogenous WT and mutant ER undergo a transcriptional reprogramming that promotes a metastatic phenotype. Collectively through the generation of engineered and spontaneously acquired polyclonal ESR1 mutant models, these data demonstrate that ESR1 mutations promote an intrinsic ET-resistant and metastatic phenotype.
ESR1 mutations induced global kinome reprogramming
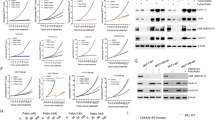
To identify activated kinases that might contribute to ESR1 mutant-driven ET resistance and metastasis, we performed a KiP screen. MCF-7 ESR1 WT and ESR1 Y537S cell lysates were incubated with kinase inhibitor-bound Sepharose beads that preferentially capture kinases in the active conformation.34,38 We analysed the kinome pulldowns by mass spectrometry and performed relative quantitative analysis between the MCF-7 ESR1 WT and ESR1 Y537S cells. There were 169 kinases identified in both ESR1 WT and Y537S cells, 13 kinases uniquely identified in WT cells, and 18 kinases unique to Y537S (Figs. 2a and S4 and Table S4). We identified 87 hyperactivated kinases (69 hyperactivated by at least 1.5-fold and 18 Y537S-unique) in the MCF-7 ESR1 Y537S cells compared to MCF-7 ESR1 WT cells. To determine which kinase pathways were activated in the MCF-7 ESR1 Y537S model, we performed KEGG pathway analysis of the 87 kinases (Fig. 2b). The MAPK signalling pathway was the most significantly upregulated, and additional upregulated pathways downstream of RTK activation include mTOR, PI3K-Akt, and Ras signalling. Given that several of the ESR1 Y537S most activated kinases were RTKs that classically signal through the PI3K and MAPK pathways (Fig. 2c), we focused on validating the expression of selected RTKs in ESR1 mutant cells. RON and IGF-1R were among the top hyperactivated kinases in the MCF-7 ESR1 Y537S cells (14- and 2.5-fold, respectively). We analysed the hormone-independent expression of RON and IGF-1R in ESR1 WT and ESR1 mutant cells by culturing cells in media supplemented with charcoal-stripped serum and performing immunoblot analysis. There were increased RON levels in MCF-7 ESR1 Y537S, MCF-7 LTED, and in T47D ESR1 Y537S cells (Fig. 2d, e). To determine if RON was targetable in the ESR1 mutant cells, we treated MCF-7 cells with an inhibitor selective for RON and the closely related MET receptor (BMS-777607/ASLAN002; RONi), and observed that this inhibitor decreased both RON and IGF-1R phosphorylation levels (Fig. 2f). Given this observation, we postulated that RON and IGF-1R may interact to activate the PI3K signalling pathway, which has previously been shown in pancreatic carcinoma models.39 Since the IGF-1R pathway is directly regulated by ER genomic binding,40 we also performed siRNA knockdowns of ER and RON. As expected, ER knockdown reduced IGF-1R levels (Fig. 2g). ER knockdown also significantly suppressed p-RON levels, demonstrating the mutant ER regulation of RON activity. We also observed reduced IGF-1R levels with knockdown of RON, which supported our findings that RON and IGF-1R may cooperate to promote PI3K pathway activation. We have previously shown that the upregulation of the IGF-1R signalling pathway is one mechanism of ET resistance in ESR1 mutant models, and that IGF-1R inhibition restored ET sensitivity.21 However, IGF-1R inhibitors have not yet been proven to be clinically useful in BC,23 thus we reasoned that inhibition of the RON signalling pathway may be an alternative approach to reduce both signalling pathways and restore ET sensitivity. Taken together, these data show that ESR1 mutant models have selectively reprogrammed their kinome, and that targeting of RON may be an optimal therapeutic approach to inhibiting both of these pathways.
a Flowchart demonstrating the total number of kinases captured in KiP experiment and the selection of kinases used to perform KEGG pathway analysis. Hyperactivated was defined as Y537S/WT ≥ 1.5-fold. Unique was defined as having corresponding peptide detection by MS in only MCF-7 ESR1 WT or MCF-7 ESR1 Y537S cells. b KEGG pathway analysis of the hyperactivated kinases in ESR1 Y537S cells. Analysis was performed using the DAVID Bioinformatics Functional Annotation Tool. c Y537S/WT quantitative ratio of kinases in KiP analysis categorised by RTKs, PI3K, and MAPK pathways. Red bars represent Y537S-unique activated kinases. d MCF-7 cells were cultured in charcoal-stripped serum-supplemented media and analysed by immunoblot for phosphorylated and total RON. Numbers below each protein represent normalised densitometry of each protein compared to GAPDH. e T47D cells were cultured in charcoal-stripped serum-supplemented media and analysed by immunoblot for phosphorylated and total RON. Numbers below each protein represent normalised densitometry of each protein compared to GAPDH. f MCF-7 cells were cultured in full serum-supplemented media and treated with BMS-777607/ASLAN002 (RONi) for 90 min and analysed by immunoblot for phosphorylated and total RON and IGF-1R. Numbers below each protein represent normalised densitometry of each protein compared to GAPDH. g MCF-7 cells were cultured in full serum-supplemented media and transfected with indicated siRNA. Immunoblot analysis for phosphorylated and total IGF-1R was performed two days post transfection. Numbers below each protein represent normalised densitometry of each protein compared to GAPDH.
ET reduced RON/PI3K pathway activation
ESR1 mutations are relatively resistant to ET including AIs, Tam, and Ful, where higher doses of drug are required to reduce mutant ER activity.4 The plasmaMATCH clinical trial evaluated the efficacy of extended dose Ful in patients with pre-existing ESR1 mutations and found this arm did not meet the pre-specified criteria for efficacy in this population despite having elevated Ful exposure compared to exposures achieved in standard of care dosing.41 Despite this, MBC patients are often treated with Ful in combination with a CDK4/6 inhibitor given the clinical data that the combination of Ful and a CDK4/6 inhibitor extended PFS compared to Ful monotherapy.10,18,19 Therefore, to test the hypothesis that components of the reprogrammed kinome contributes to ET resistance, we treated MCF-7 ESR1 WT and MCF-7 ESR1 Y537S cells with Tam and analysed the kinome using KiP analyses. Tam treatment reduced 49/88 of the Y537S hyperactivated kinases by at least 50% (Table S5), including RTKs such as EPHA2, EPHB2, EPHB3, EPHB4, FGFR4, IGF-1R, INSR, RON and RET (Fig. 3a). Other kinases involved in the PI3K and MAPK signalling pathway including MAP2K1, INSR, CDK4, RPS6KA5 and RPS6KA6 were also reduced by Tam, suggesting that the mutant ER selectively regulated the activation of these pathways. Tam significantly reduced RON activity by 74%, however, Tam-treated MCF-7 ESR1 Y537S had 3.5-fold higher RON levels compared to untreated MCF-7 ESR1 WT cells by KiP analysis. We then confirmed these findings using phospho-immunoblot analysis of RON (Fig. 3b). p-RON was elevated in ESR1 Y537S and LTED cells compared to ESR1 WT cells (6.3-fold and 2.7-fold, respectively). Both Tam and Ful reduced p-RON levels in ESR1 mutant models, but Tam treatment incompletely reduced p-RON levels; Tam-treated Y537S cells had a 2.4-fold increase and MCF-7 LTED cells had a 1.2-fold increase in p-RON levels compared to untreated WT cells. We additionally found that ET reduced p-RON levels in T47D with RON/MSP overexpression, indicating that RON activation can be inhibited by blocking ER, even with induction of exogenous RON activity (Fig. S5). Total RON protein and RNA were not consistently reduced with ET (Fig. 3b, c), and there were no mutant ER genomic binding sites within 100 kb of the RON transcription start site as analysed by ChIP-seq (data not shown), suggesting that RON activation may be through indirect regulation of RON levels by the mutant ER.
a Fold change of kinases in KiP analysis in MCF-7 Y537S cells after treatment with Tam. X-values represent fold change of Tam-treated Y537S cells compared to untreated Y537S cells. b MCF-7 cells were cultured in 10% FBS supplemented media and treated with 1 µM Tam and 1 µM Ful for 48 h. Immunoblot analysis of phosphorylated and total RON was performed. Numbers below each protein represent normalised densitometry of each protein compared to GAPDH. c MCF-7 cells were cultured in 10% FBS supplemented media and treated with Tam and Ful for 48 h. qRT-PCR analysis of RON was performed. Graphs represent mean + standard error of the mean (N = 3 replicates). Two-way ANOVA was used for statistical analysis. d MCF-7 cells were cultured in 10% FBS supplemented media and treated with RONi for ninety minutes. Immunoblot analysis was performed for phosphorylated and total AKT and p44/42 MAPK. e MCF-7 cells were cultured in charcoal-stripped serum supplemented media and treated with RONi for 24 hours and ER transactivation assay was performed. Graphs represent mean + standard deviation (N = 3 replicates). Student’s t-test was used for statistical analysis. f MCF-7 cells were cultured in 10% FBS supplemented media and treated with 1 µM Tam and 1 µM Ful for 48 h. Immunoblot of phosphorylated and total AKT and p44/42 MAPK was performed. g T47D cells were cultured in 10% FBS supplemented media and treated with Tam and Ful for 48 h. Immunoblot of phosphorylated and total AKT and p44/42 MAPK was performed. P < 0.05 was considered statistically significant in all tests. (*p < 0.05, **p < 0.01, ***p < 0.001).
Given our data that RON inhibition or knockdown reduced IGF-1R activity, we hypothesised that RON inhibition may be a unique strategy to decrease both receptor signalling pathways for therapeutic benefit. To determine whether there was an indication for combining ET with RONi, we first analysed the downstream signalling effects of RON inhibition in cells with mutant ER. Treatment with the RONi reduced downstream p-AKT and resulted in a concomitant increase in p-p44/42 MAPK (Fig. 3d), presumably due to a relief of negative feedback repression of receptor signalling proteins.42 Furthermore, RONi treatment increased ER transactivation in MCF-7 ESR1 WT and MCF-7 ESR1 Y537S cells as demonstrated using ERE-luciferase reporter assays (Fig. 3e), suggesting that RON inhibition could alter crosstalk between ER and RTK signalling pathways, leading to ligand-independent activation of ER. We hypothesised that in the context of RON inhibition, the mutant ER may exhibit an adaptive response to RTK inhibition by further enhancing ER activity. To determine the effects of ET on PI3K pathway activation, we treated MCF-7 and T47D cells with Tam and Ful and found that ET reduced p-AKT and p-p44/42 MAPK (Fig. 3f, g). Given the incomplete reduction of RON activity with ET and the implications of MAPK reactivation, these data suggest that a combination of ET and RON inhibition may provide adequate vertical inhibition of the RON signalling pathway required to suppress reactivation of additional bypass pathways.
The Y537S mutant ER regulated key mediators of the RON/PI3K pathway
Our data demonstrated that acquisition of ESR1 mutations results in a dynamic transcriptional and kinome reprogramming of breast tumour cells. RON was hyperactivated in ESR1 mutant cells, however, the ability of ET to reduce RON signalling was not due to reduction in total RON RNA or protein. Whether ESR1 mutants directly regulate signalling proteins downstream of RTKs is an imperative question to determine sensitivity of ET in combination with RTK inhibitors, as it has been shown that downstream signalling proteins are activated by positive feedback loops and crosstalk with other RTKs.43 To determine possible mechanisms by which the mutant ER activates PI3K pathway signalling, we performed an integrated analysis of kinome, transcriptome, and Y537S mutant ER ChIP-seq datasets. We interrogated these datasets for kinases that were both hyperactivated and transcriptionally upregulated in MCF-7 ESR1 Y537S cells and exhibited ER genomic binding sites within 100kB of the selected gene body. We identified GRB2 Associated Binding Protein 2 (GAB2), an adaptor protein involved in RTK/PI3K signalling,44 that was elevated at the RNA level in the ESR1 Y537S cells. Y537S ER ChIP-seq analysis revealed an ER binding site within intron 2 of GAB2 (Fig. 4a). We determined using ChIP-qPCR that there was a 30% increase in mutant receptor binding at this site (Fig. 4b). We then performed qRT-PCR on Tam- and Ful-treated MCF-7 cells to determine if ET could affect mRNA expression of GAB2, and found its RNA was elevated by 5.1-fold and was reduced by 35% with Ful treatment (Fig. 4c). These data suggest that ER binds to a transcriptional regulatory region of GAB2, and that the enhanced binding by the mutant receptor could lead to an upregulation of its expression.
a ER ChIP-Seq track for GAB2 gene body in MCF-7 ESR1 Y537S cells. b ER ChIP-qPCR in MCF-7 ESR1 WT and MCF-7 ESR1 Y537S cells at the binding site highlighted in a. Graphs represent mean + standard deviation (N = 3 replicates). Student’s t-test was performed for statistical analysis. c MCF-7 cells were cultured in 10% FBS supplemented media and treated with 1 µM Tam and 1 µM Ful for 48 h. qRT-PCR analysis of GAB2 was performed. Graphs represent mean + standard error of the mean (N = 3 replicates). Two-way ANOVA was performed for statistical analysis. d MCF-7 cells were cultured in 10% FBS supplemented media and transfected with scrambled (Scr) siRNA and GAB2 siRNA. Immunoblot analysis of phosphorylated and total AKT and p44/42 MAPK was performed 48 h post transfection. e T47D cells were cultured in 10% FBS supplemented media and transfected with GAB2 siRNA. Immunoblot analysis of phosphorylated and total AKT and p44/42 MAPK was performed 48 h post transfection. P < 0.05 was considered statistically significant for all tests. (*p < 0.05, **p < 0.01, ***p < 0.001).
Our data showed that the mutant ER upregulated GAB2 expression and that ET reduced p-AKT, which is downstream of GAB2 signalling. To determine if GAB2 was a mediator of PI3K pathway activation in ESR1 mutant cells, we performed a GAB2 siRNA experiment, and analysed p-AKT and p-p44/42 MAPK using immunoblot. p-AKT was reduced with GAB2 knockdown in MCF-7 and T47D ESR1 WT and ESR1 mutant cells (Fig. 4d, e). Consistent with our other findings, p-p44/42 MAPK levels were increased with PI3K pathway inhibition, indicating relief of negative feedback suppression resulting in activation of the MAPK pathway. These data demonstrate that the mutant receptor transcriptionally upregulates the GAB2/PI3K axis to further promote PI3K pathway activation, and that ET in combination with a RONi may provide robust inhibition of this pathway.
ESR1 mutant intrinsic resistance predisposed cells to palbociclib resistance
The preferred first-line therapy for women with ER-positive MBC is an AI in combination with a CDK4/6 inhibitor.7 However, patients eventually develop resistance to this combination and require alternative endocrine and biologic combination therapies to further control metastatic disease progression.15,45 Therefore, there is a critical need to understand the mechanisms of resistance to CDK4/6 inhibitors in this setting. To investigate the role of CDK4/6 inhibitor resistance in the context of ESR1 mutations, we developed MCF-7 and T47D ESR1 WT and ESR1 Y537S PalbR cell lines by treating the cells with palbociclib for 6 months. By 6 months, MCF-7 PalbR models were relatively resistant to the three clinically approved CDK4/6 inhibitors palbociclib, ribociclib and abemaciclib as shown in MTT growth assays (Figs. 5a and S6a), suggesting there are common resistance mechanisms between the CDK4/6 inhibitors. Although T47D PalbR cells were resistant to palbociclib, resistance to ribociclib and abemaciclib differed from the MCF-7 model (Figs. 5b and S1). The T47D ESR1 WT PalbR cells remained sensitive to ribociclib, and the T47D ESR1 Y537S PalbR cells remained sensitive to abemaciclib. This supports other data showing cancers with acquired palbociclib resistance can retain sensitivity to other CDK4/6 inhibitors.46 We next performed transcriptomic analyses of the MCF-7 ESR1 WT and PalbR models to evaluate whether transcriptional reprogramming was also associated with the development of CDK4/6 inhibitor resistance. Interestingly, there was a significant overlap of upregulated genes between MCF-7 ESR1 WT PalbR and MCF-7 ESR1 Y537S (Fig. 5c, p < 1.62 × 10−196 based on hypergeometric distribution), demonstrating there are shared mechanisms between ET resistance driven by acquired ESR1 mutations and those leading to palbociclib resistance. We performed GSEA on the shared upregulated genes and identified pathways including interferon and oestrogen responses, among others that are known to be involved in CDK4/6 inhibitor and ET resistance, suggesting that expression of a subset of genes involved in ER signalling are maintained in the PalbR setting. ddPCR analysis of the PalbR models did not detect ESR1 mutations (Fig. S7) confirming that acquired ESR1 mutations in this model was not contributing to the shared transcriptional phenotype between the two models. Transcriptional profiling revealed RTK upregulation in MCF-7 PalbR models, including EPHA2, EPHA4, FGFR4, IGF-1R, RON, and RET (Fig. 5d). The upregulation of these RTKs suggests that an upregulation of RTK signalling is a shared resistance mechanism between ESR1 mutations and acquired palbociclib resistance. Given our data that RON was intrinsically activated in ESR1 mutant models, we sought to expand our findings in the PalbR models to determine if RON may be effectively targeted in the PalbR setting, using a strategy including a RON inhibitor. RON RNA expression was increased in MCF-7 ESR1 Y537S PalbR cells by 1.8-fold compared to ESR1 WT cells. Furthermore, p-RON levels were increased in the MCF-7 ESR1 Y537S cells by 2.1-fold, in ESR1 Y537S PalbR cells by 2.6-fold, and in the T47D WT PalbR cells by 2.7-fold as analysed by immunoblot (Fig. 5e, f). We conclude that acquired ESR1 mutations predispose cells to palbociclib resistance, and that ET resistance via RON hyperactivation may be a biomarker for palbociclib resistance.
a MTT growth assay of MCF-7 and MCF-7 PalbR models after treatment with palbociclib. Mean + standard deviations are shown (N = 3 replicates). Two-way ANOVA was used for statistical analysis. b MTT growth assay of T47D and T47D PalbR models after treatment with palbociclib. Mean with standard deviations are shown (N = 3 replicates). One-way ANOVA was used for statistical analysis. c Venn diagram of upregulated genes in MCF-7 ESR1 Y537S and MCF-7 WT PalbR cells compared to MCF-7 WT. GSEA Hallmarks are of the shared upregulated genes. d Microarray profiling of RTKs in MCF-7 WT PalbR cells and ESR1 Y537S cells compared to MCF-7 WT. X-values represent microarray expression fold change ratios of average values per condition (N = 3 replicates/condition). ANOVA was performed for statistical analysis. e MCF-7 cells were cultured in 10% FBS supplemented media and analysed by immunoblot for phosphorylated and total RON. Numbers below each protein represent normalised densitometry of each protein compared to GAPDH. f T47D cells were cultured in 10% FBS supplemented media and analysed by immunoblot for phosphorylated and total RON. Numbers below each protein represent normalised densitometry of each protein compared to GAPDH. g Organoid growth assays with RONi treatment in MCF-7 WT PalbR primary tumour, h MCF-7 ESR1 Y537S PalbR primary tumour, and i) MCF-7 ESR1 Y537S PalbR metastatic tumour. Bar graphs in g–i represent mean number of organoids + standard deviation (N = 3 replicates). Student’s t-test was used for statistical analysis. P < 0.05 was considered statistically significant for all tests. (*p < 0.05, **p < 0.01, ***p < 0.001).
To determine if PalbR models were responsive to RON inhibition, we transplanted MCF-7 ESR1 WT PalbR and MCF-7 ESR1 Y537S PalbR cells into mice and treated with palbociclib for up to three months. The MCF-7 ESR1 WT PalbR tumours were indeed resistant to palbociclib; in vivo median survival was 11.0 vs 5.0 weeks, (p = 0.0276; Fig. S8). We then developed organoids from primary tumours from both models, and a metastatic tumour from the MCF-7 ESR1 Y537S PalbR model to test their responsiveness to the RONi. Organoid growth was significantly reduced with RONi treatment compared to untreated control in all three PalbR organoid models tested (Fig. 5g–i). The RONi reduced organoid growth in MCF-7 ESR1 WT PalbR primary tumour organoids by 61% (p < 0.0001), the MCF-7 ESR1 Y537S primary tumour organoids by 25% (p = 0.0008), and the MCF-7 ESR1 Y537S metastatic tumour organoids by 42% (p < 0.0001). Collectively, these data demonstrate that ESR1 mutant and PalbR models express a shared ET-resistant transcriptional phenotype, and that targeting common upregulated pathways such as the RON pathway may overcome the therapeutic resistance in both of these settings.
Inhibition of the RON signalling pathway restored ET sensitivity
Our data supported that the combination of ET with the RONi could be an effective strategy to suppress the RON signalling pathway and the MAPK signalling pathway. Therefore, we next determined if inhibition of the RON signalling pathway could reduce the growth and metastasis of ESR1 mutant models. We first developed organoids derived from a cohort of PDX and xenografted primary and metastatic tumours that expressed either ESR1 WT or ESR1 mutations (BCM-15034: ESR1 WT; WHIM20: ESR1 Y537S, E2 independent;47 HCI-005: ESR1 L536P, E2 dependent;48 HCI-013: ESR1 Y537S, E2 dependent49). By 4 months, the mice injected with MCF-7 ESR1 Y537S cells developed distant metastases in vivo. Primary tumours from ESR1 WT and mutant models, and metastatic tumours from the MCF-7 ESR1 Y537S model were harvested and utilised for ex vivo organoid development. To test the effects of RON and downstream PI3K inhibition on ESR1 mutant tumour growth, we treated organoid cultures ex vivo with ET in combination with the RONi or PI3K inhibitors (taselisib; PI3Ki) (Fig. 6a, b). HCI-005 organoid growth was significantly inhibited with Tam monotherapy and was further suppressed with the RON and PI3K inhibitors. Tam alone had no effect on HCI-013 organoid growth, but enhanced the growth of BCM-15034, MCF-7 ESR1 Y537S primary and metastatic, and MCF-7 LTED organoids. This data suggests that these models were all resistant to Tam and that Tam was acting as an agonist in some of the models. However, the addition of RON or PI3K inhibitors suppressed organoid growth in the majority of ESR1 mutant models tested. These data show that RON activation is a potential therapeutic vulnerability in ESR1 mutant-expressing tumours.
a Representative images of organoids after a 2-week treatment with indicated inhibitors. b Heat map quantitation of treated organoids. Cell values represent quantitative ratio of the number of organoids in each treatment to the indicated comparison; Tam is relative to –E2 control; Tam + RONi and Tam + PI3Ki is relative to Tam (N = 3 replicates/treatment). One-way ANOVA and Tukey test was performed for multiple comparisons. Asterisks indicate indicated comparison was statistically significant (*p < 0.05). c Individual primary tumour growth curves of WHIM20 tumours in mice treated with -E2, RONi, Tam, or Tam + RONi. d In vivo primary tumour growth of WHIM20 PDX model with indicated treatments, represented as time to tumour doubling. The Mantel-Cox test was used for statistical analysis (-E2 N = 9, RONi N = 10, Tam N = 9, Tam + RONi N = 7) Primary tumours that did not reach predefined tumour doubling criteria were excluded from analysis. e Metastatic frequency of WHIM20 tumours (-E2 N = 11, RONi N = 11, Tam N = 7, Tam + RONi N = 7). Metastatic frequency is represented by the total number of mice per group that exhibited a macrometastasis. Animals that died prior to the defined endpoint were excluded from analysis. Fisher’s Exact test was used for statistical analysis. f Immunoblot of the RON signalling pathway in WHIM20 primary tumours with indicated treatment. Each lane represents an individual primary tumour from the indicated treatment group. Five tumours per group were analysed by immunoblot. P < 0.05 was considered statistically significant for all tests. (*p < 0.05, **p < 0.01, ***p < 0.001).
We next determined if RON inhibition could reduce primary and metastatic tumour growth in vivo. We performed orthotopic transplants of the WHIM20 ESR1 Y537S PDX model into mice and randomised them to -E2, Tam, RONi, or Tam + RONi treatments. Tam and Tam + RONi treatments significantly slowed primary tumour growth compared to -E2 (time to tumour doubling was 5.5 weeks, 6.5 weeks vs 2.0 weeks, respectively; Fig. 6c, d). In this study, mice were treated with 60 mg/kg Tam which results in higher exposure than what is seen clinically and may contribute to the activity seen in WHIM20 tumours.50 Mice underwent survival surgery to remove the primary tumours and continued treatments. At four months post-transplant, mice were euthanised, and major organs were harvested to determine distant metastatic frequencies. The metastatic frequency in the -E2 group was 90%, and treatment with Tam alone or Tam in combination with a RONi significantly reduced metastatic frequency to 29% (p = 0.0128) and 14% (p = 0.0025), respectively (Fig. 6e). To determine if the RON signalling pathway was indeed inhibited with these treatments, we performed immunoblot analysis of the treated primary tumours (Fig. 6f). As expected, Tam monotherapy reduced the expression of IGF-1R. Although the RONi monotherapy had no significant effects on the phosphorylated protein levels in the RON signalling pathway, the combination of Tam and a RONi significantly reduced pathway activation, as demonstrated by reduced p-RON, p-AKT, and p-p44/42 MAPK levels. We postulate that the addition of Tam may augment the pharmacodynamic effect of the RONi, as we have shown that the mutant receptor up-regulated downstream effectors of the RON pathway. The longer time to tumour doubling and lower metastatic frequency observed in the combination arm compared to Tam alone were not significant; however, this may be due to the limited sample size in each arm. The enhanced inhibition of the RON and IGF-1R signalling pathway in the combination arm is a promising effect that may contribute to the enhanced therapeutic benefit in this study. Collectively, our ex vivo organoid growth assays and in vivo spontaneous metastasis models demonstrated that Tam in combination with RONi may be a unique therapeutic approach to inhibit ESR1 mutant-expressing tumour growth and metastasis.
Discussion
MBC continues to be therapeutically challenging despite available endocrine and targeted therapies. Acquired resistance to these therapies occurs in a significant number of patients and therefore additional biologic targeted agents are required to control metastatic disease.2,20,51 Adding to this complexity is the acquisition of ESR1 mutations that are primarily found in patients treated with AIs in the metastatic setting.3 ESR1 mutations were first discovered in MBC tumours,5 and are now considered to be the most common mechanism of ET resistance in ER-positive MBC. Analysis of metastatic tumours and ctDNA have demonstrated the clinical resistance of ESR1 mutations to ET in several clinical trials.10,52 In the SoFEA clinical trial in which patients were treated with exemestane or Ful, 39% of patients exhibited ESR1 mutations. Patients in the exemestane group who had an ESR1 mutation had a worse PFS compared to those whose tumours expressed WT ER. In the PALOMA-3 trial in which patients were treated with Ful or Ful in combination with palbociclib, ESR1 mutations were detected in 25% of patients. In a meta-analysis of ESR1 mutations detected in clinical trials, the presence of an ESR1 mutation was prognostic for worse PFS and OS. In a subgroup analysis of individual ESR1 mutations, the D538G, but not the Y537S mutation, was associated with worse PFS.12 Despite this, more recent studies demonstrated the selective resistance of the Y537S mutation with Ful therapy, suggesting clinical resistance of this mutation.20 In the MONALEESA-2 study, patients who progressed 12 months after completing adjuvant AI therapy and who had de novo metastatic disease received either anastrozole or anastrozole with ribociclib.53 ESR1 mutations were present in only 4% of patients in this relatively ET sensitive cohort and therefore it was not possible to correlate with clinical outcome. Unfortunately, there are no clinical agents that can effectively target the mutant ER directly, so we therefore aimed at identifying targets that could restore ET sensitivity and reduce metastatic burden in MBC models. We found that RON and IGF-1R were hyperactivated kinases in ESR1 mutant models, and up until now the molecular mechanisms of this hyperactivation in ESR1 mutant BCs have not been explored. In this study, we have demonstrated that RON activation is a shared resistance mechanism in ESR1 mutant and PalbR models, and that inhibition of the RON signalling pathway was an effective therapy in both up-front ET-resistant and in the PalbR setting.
Similar to patients, polyclonal ESR1 mutations were acquired in MCF-7 cells when cultured long-term in hormone-deprived media, simulating clinical AI treatment. A previous study showed that monoclonal ESR1 mutations were acquired and enriched in LTED conditions in multiple ER-positive BC cell lines.37 Interestingly, our study demonstrated polyclonal acquisition of ESR1 Y537N and Y537C mutations in our ER-positive BC cell line, supporting clinical data that shows a positive selection of both monoclonal and polyclonal ESR1 mutant-expressing cells in AI-resistant MBC patients.3,54 In line with the clinical scenario of MBCs acquiring polyclonal ESR1 mutations, a recent study found that the levels of co-expressing WT and mutant ERs influence the cellular response to SERDs and selective oestrogen receptor modulators (SERMs); cells that overexpress higher levels of the mutant relative to WT ER were less sensitive to growth inhibition with standard of care ET.55 Although pharmacological differences in cells that co-express WT and mutant ER were demonstrated in that study, our cell line with spontaneously acquired polyclonal ESR1 mutations exhibited a significantly shared ET-resistant transcriptional signature and metastatic proclivity with our engineered homozygous ESR1 Y537S model, suggesting that these are dominant phenotypes that are shared among different ESR1 mutations.
We observed that RON was hyperactivated in MCF-7 and T47D ESR1 mutant models, suggesting RON and other RTK activation may contribute to ET resistance observed in ESR1 mutant tumours. Additionally, we found that the mutant ER upregulated RTK/PI3K signalling by at least three means: the mutant ER transcriptionally upregulated RTKs such as IGF-1R to directly promote activation of the pathway, transcriptionally upregulated RTKs such as IGF-1R cooperated with RON to promote PI3K pathway activation, and the mutant ER directly upregulated the transcription of PI3K effector proteins such as GAB2, to promote activation. We and others have previously shown that the IGF-1R signalling pathway is upregulated in ESR1 mutant BCs,21,22 and that its inhibition restores ET sensitivity. In this study, we developed a unique hypothesis to target this pathway by identifying kinases that may interact with IGF-1R, such as RON. Using this approach, therapeutic inhibition of RON resulted in inhibition of both the RON and IGF-1R pathways. We hypothesise that this approach may help overcome limitations of IGF-1R inhibitors used in the clinic such as on-target off-tumour induced hyperglycaemia since there is no appreciable RON expression in pancreatic epithelial cells,56 there were no hyperglycaemic adverse events reported in an BMS-777607/ASLAN002 Phase 1 clinical trial.57 In addition, we found that ET in combination with a RONi may prevent compensatory activation of the MAPK signalling pathway. GAB2 is a scaffolding protein that facilitates RTK pathway activation through interaction with PI3K subunits, and its overexpression in BC has been associated with PI3K and MAPK hyperactivity.58,59 This finding has important clinical implications for the use RTK inhibitors; upregulation of downstream signalling molecules may overcome therapeutic effects of RTK inhibitors and may require additional pathway suppression. This study showed that ET significantly reduced expression or activation of downstream signalling molecules, which may augment the effects of RON inhibition. These data demonstrate that the mutant ER has an inter-related network of kinase upregulation that collectively promote a metastatic phenotype.
Based on our data, targeting the RON/PI3K signalling pathway may be a unique and efficacious strategy for the selective treatment of ESR1 mutant-positive and/or palbociclib-resistant MBC. Our ex vivo ESR1 mutant organoid modelling and in vivo metastasis experiments demonstrated profound inhibition of ESR1 mutant organoid growth, and a significant reduction of metastasis. In a Phase 1 clinical trial (NCT01721148) evaluating the safety of the RON/MET inhibitor BMS-777607/ASLAN002 in solid cancers, BMS-777607/ASLAN002 was well-tolerated and five patients exhibited stable disease.57 There are currently no Phase 2 trials evaluating the therapeutic efficacy of BMS-777607/ASLAN002, however, given our data we hypothesise that ESR1-mutant expressing, RON-activated BCs may demonstrate sensitivity to vertical inhibition of the RON pathway using ET and a RONi. Our proteomic and immunoblot experiments demonstrated that ESR1 mutant-induced RON/PI3K activation was incompletely reduced with ET alone and that additional RON pathway blockade was required to achieve maximum therapeutic benefit. Although we did not demonstrate the therapeutic efficacy of triplet combinations of ET with RON and PI3K inhibition, it has been shown that triplet blockade using ET, the pan-HER family inhibitor neratinib, and the mTOR inhibitor everolimus had enhanced therapeutic benefit compared to either inhibitor used alone in HER2-positive BC cell lines,60 and combinatorial therapies have become standard of care for the clinical management of MBC.61
Clinical management of ESR1 mutant MBC patients includes treatment with Ful in combination with a CDK4/6 inhibitor.7 However, the PALOMA-3 clinical study showed that patients who were treated with Ful in combination with palbociclib showed increased levels of the ESR1 Y537S mutation at the end of treatment.20 The clinical resistance of the ESR1 mutations to ET warrants investigation of new SERDs and biologic therapies that can more potently target the mutant ER. Ongoing clinical trials are investigating the use of novel SERDs such as AZD9833 (SERENA-2; NCT04214288), SAR439859 (AMEERA-3; NCT04059484) and GDC-9545 (NCT03332797) in ESR1 mutant expressing BCs. Importantly, additional ongoing clinical trials are prospectively investigating the efficacy of ET combined with CDK4/6 inhibitors in ESR1 mutant selected patients (NCT04432454, NCT02738866, NCT04256941). Studies such as these and other studies that stratify patients based on ESR1 mutation status are crucial to determine the therapeutic efficacy in these populations.
Our preclinical study demonstrates for the first time that mechanistically the intrinsic ET-resistant phenotype of ESR1 mutations may predispose BCs to palbociclib resistance due to an overlap in resistance bypass pathway activation. Microarray analyses of ESR1 mutant and WT PalbR models showed a significant overlap of transcriptionally upregulated genes between them. Specifically, the interferon α response and interferon γ response were the most significantly upregulated MSigDB Hallmark pathways in these models. In the NeoPalAna neoadjuvant clinical trial in which patients were treated with anastrozole in combination with palbociclib, the interferon γ response Hallmark was also significantly upregulated in primary tumours after 12 weeks of palbociclib treatment.62 Preclinical validation in an MMTV-rtTa/tetO-HER2 mouse BC model also demonstrated that the interferon α response and interferon γ response Hallmarks were upregulated in abemaciclib-treated mice. These studies and our findings support that upregulation of interferon signalling is a common response and adaptive resistance mechanism to CDK4/6 inhibition. There is no clear therapeutic approach selectively targeting interferon signalling for the application to the CDK4/6 inhibitor-resistant population, and further investigation is needed to determine if this would be an effective approach in these patients. Furthermore, our MCF-7 ESR1 WT PalbR and ESR1 Y537S gene signatures exhibited an upregulation of early and late response to oestrogen responses, showing that ESR1 WT models retained oestrogen signalling after acquiring resistance to palbociclib. Previous work found that ER-positive PalbR cell lines lost ER signalling and were intrinsically resistant to ET.63 However, the MCF-7 PalbR cell line characterised in our study expressed a subset of genes that were shared with the ESR1 Y537S cells and contributed to upregulated oestrogen response Hallmarks. Taken together, our data show that the intrinsic ET-resistant phenotype of ESR1 mutations may predispose to the development of palbociclib resistance.
We have additionally observed that ET in combination with the mutant PI3K-α potent, β-sparing inhibitor taselisib was effective at reducing the growth of ESR1 mutant BC organoids. This is a promising result that may support the use of pan-PI3K inhibitors in ESR1 mutant BCs. However, the clinical use of taselisib is advanced and MBC is no longer being pursued given the Phase 3 clinical data that demonstrated only a modest improvement of PFS and significant toxicity.64 PIK3CA-mutant MBCs progressing on CDK4/6 inhibitor therapy are successfully treated with Ful in combination with the PI3K α-specific inhibitor alpelisib.7 Importantly, the Phase 2 BYLieve clinical trial demonstrated that ER-positive advanced BC patients who were pre-treated with ET in combination with a CDK4/6 inhibitor had a meaningful therapeutic benefit with the combination of ET and alpelisib, suggesting that alpelisib maintains its efficacy in some patients with palbociclib-resistant tumours.65 However, a separate study demonstrated that PTEN loss and ESR1 mutations were common alterations that may promote resistance to an AI and alpelisib.66 In our study, we observed an upregulation of RTK signalling including activation of RON in PalbR models. Therapeutic blockade of RON decreased ex vivo organoid growth in PalbR models suggesting its potential therapeutic efficacy in tumours with acquired palbociclib resistance. Further investigation of the use of Ful with PI3K inhibitors such as alpelisib in the context of palbociclib-resistant, ESR1 mutant-expressing tumours is thus warranted.
In conclusion, our data demonstrate that there is a shared transcriptional signature between ESR1 mutant and PalbR models that includes upregulation of RTK signalling that may promotes therapeutic resistance to both ET and CDK4/6 inhibition. Our data suggest that the intrinsic ESR1 mutant ET resistance mechanisms can predict CDK4/6i resistance. Our experimental modelling demonstrated that the RON/IGF-1R/PI3K signalling pathway was an escape mechanism in both models. Clinical analysis of Ful and PI3K inhibitors are needed to demonstrate if this may be an effective therapeutic approach in ESR1 mutant and PalbR populations. Further investigation of the clinical use of a RONi in combination with ET should also be considered in ESR1 mutant and PalbR MBCs.
References
(WHO) IAfRoCIaWHO. GLOBOCAN 2018: Breast 2019. http://gco.iarc.fr/today/data/factsheets/cancers/20-Breast-fact-sheet.pdf. (2018)
Early Breast Cancer Trialists’ Collaborative G. Effects of chemotherapy and hormonal therapy for early breast cancer on recurrence and 15-year survival: an overview of the randomised trials. Lancet. 365, 1687–1717 (2005).
Schiavon, G., Hrebien, S., Garcia-Murillas, I., Cutts, R. J., Pearson, A., Tarazona, N. et al. Analysis of ESR1 mutation in circulating tumor DNA demonstrates evolution during therapy for metastatic breast cancer. Sci. Transl. Med. 7, 313ra182 (2015).
Toy, W., Shen, Y., Won, H., Green, B., Sakr, R. A., Will, M. et al. ESR1 ligand-binding domain mutations in hormone-resistant breast cancer. Nat. Genet. 45, 1439–1445 (2013).
Zhang, Q. X., Borg, A., Wolf, D. M., Oesterreich, S. & Fuqua, S. A. An estrogen receptor mutant with strong hormone-independent activity from a metastatic breast cancer. Cancer Res. 57, 1244–1249 (1997).
Toy, W., Weir, H., Razavi, P., Lawson, M., Goeppert, A. U., Mazzola, A. M. et al. Activating ESR1 mutations differentially affect the efficacy of ER antagonists. Cancer Discov. 7, 277–287 (2017).
Network NCC. Breast Cancer (Version 4.2020). https://www.nccn.org/professionals/physician_gls/pdf/breast_blocks.pdf. (2020)
Chandarlapaty, S., Chen, D., He, W., Sung, P., Samoila, A., You, D. et al. Prevalence of ESR1 mutations in cell-free DNA and outcomes in metastatic breast cancer: a secondary analysis of the BOLERO-2 clinical trial. JAMA Oncol. 2, 1310–1315 (2016).
Clatot, F., Perdrix, A., Augusto, L., Beaussire, L., Delacour, J., Calbrix, C. et al. Kinetics, prognostic and predictive values of ESR1 circulating mutations in metastatic breast cancer patients progressing on aromatase inhibitor. Oncotarget 7, 74448–74459 (2016).
Fribbens, C., O’Leary, B., Kilburn, L., Hrebien, S., Garcia-Murillas, I., Beaney, M. et al. Plasma ESR1 mutations and the treatment of estrogen receptor-positive advanced breast cancer. J. Clin. Oncol. 34, 2961–2968 (2016).
Spoerke, J. M., Gendreau, S., Walter, K., Qiu, J., Wilson, T. R., Savage, H. et al. Heterogeneity and clinical significance of ESR1 mutations in ER-positive metastatic breast cancer patients receiving fulvestrant. Nat. Commun. 7, 11579 (2016).
Zhang, K., Hong, R., Xu, F., Xia, W., Kaping, L., Qin, G. et al. Clinical value of circulating ESR1 mutations for patients with metastatic breast cancer: a meta-analysis. Cancer Manag. Res. 10, 2573–2580 (2018).
Slamon, D. J., Neven, P., Chia, S., Fasching, P. A., De Laurentiis, M., Im, S. A. et al. Overall survival with ribociclib plus fulvestrant in advanced breast cancer. N. Engl. J. Med. 382, 514–524 (2020).
Sledge, G. W., Jr. Toi, M., Neven, P., Sohn, J., Inoue, K., Pivot, X., et al. The effect of abemaciclib plus fulvestrant on overall survival in hormone receptor-positive, ERBB2-negative breast cancer that progressed on endocrine therapy-MONARCH 2: a randomized clinical trial. JAMA Oncol. 6, 116–124 (2019).
Finn, R. S., Martin, M., Rugo, H. S., Jones, S., Im, S. A., Gelmon, K. et al. Palbociclib and letrozole in advanced breast cancer. N. Engl. J. Med. 375, 1925–36. (2016).
Turner, N. C., Slamon, D. J., Ro, J., Bondarenko, I., Im, S. A., Masuda, N. et al. Overall survival with palbociclib and fulvestrant in advanced breast cancer. N. Engl. J. Med. 379, 1926–1936 (2018).
Turner, N. C., Ro, J., Andre, F., Loi, S., Verma, S., Iwata, H. et al. Palbociclib in hormone-receptor-positive advanced breast cancer. N. Engl. J. Med. 373, 209–219 (2015).
Sledge, G. W. Jr., Toi, M., Neven, P., Sohn, J., Inoue, K., Pivot, X. et al. MONARCH 2: abemaciclib in combination with fulvestrant in women with HR+/HER2- advanced breast cancer who had progressed while receiving endocrine therapy. J. Clin. Oncol. 35, 2875–2884 (2017).
Slamon, D. J., Neven, P., Chia, S., Fasching, P. A., De Laurentiis, M., Im, S. A. et al. Phase III randomized study of ribociclib and fulvestrant in hormone receptor-positive, human epidermal growth factor receptor 2-negative advanced breast cancer: MONALEESA-3. J. Clin. Oncol. 36, 2465–2472 (2018).
O’Leary, B., Cutts, R. J., Liu, Y., Hrebien, S., Huang, X., Fenwick, K. et al. The genetic landscape and clonal evolution of breast cancer resistance to palbociclib plus fulvestrant in the PALOMA-3 Trial. Cancer Discov. 8, 1390–1403 (2018).
Gelsomino, L., Gu, G., Rechoum, Y., Beyer, A. R., Pejerrey, S. M., Tsimelzon, A. et al. ESR1 mutations affect anti-proliferative responses to tamoxifen through enhanced cross-talk with IGF signaling. Breast Cancer Res. Treat. 157, 253–265 (2016).
Li, Z., Levine, K. M., Bahreini, A., Wang, P., Chu, D., Park, B. H. et al. Upregulation of IRS1 enhances IGF1 response in Y537S and D538G ESR1 mutant breast cancer cells. Endocrinology 159, 285–296 (2018).
Robertson, J. F., Ferrero, J. M., Bourgeois, H., Kennecke, H., de Boer, R. H., Jacot, W. et al. Ganitumab with either exemestane or fulvestrant for postmenopausal women with advanced, hormone-receptor-positive breast cancer: a randomised, controlled, double-blind, phase 2 trial. Lancet Oncol. 14, 228–235 (2013).
Gualberto, A. & Pollak, M. Emerging role of insulin-like growth factor receptor inhibitors in oncology: early clinical trial results and future directions. Oncogene 28, 3009–3021 (2009).
Wang, M. H., Julian, F. M., Breathnach, R., Godowski, P. J., Takehara, T., Yoshikawa, W. et al. Macrophage stimulating protein (MSP) binds to its receptor via the MSP beta chain. J. Biol. Chem. 272, 16999–17004 (1997).
Chao, K. L., Tsai, I. W., Chen, C. & Herzberg, O. Crystal structure of the Sema-PSI extracellular domain of human RON receptor tyrosine kinase. PLoS ONE 7, e41912 (2012).
Wang, D., Shen, Q., Chen, Y. Q. & Wang, M. H. Collaborative activities of macrophage-stimulating protein and transforming growth factor-beta1 in induction of epithelial to mesenchymal transition: roles of the RON receptor tyrosine kinase. Oncogene 23, 1668–1680 (2004).
Faham, N., Zhao, L. & Welm, A. L. mTORC1 is a key mediator of RON-dependent breast cancer metastasis with therapeutic potential. npj Breast Cancer 4, 36 (2018).
Zinser, G. M., Leonis, M. A., Toney, K., Pathrose, P., Thobe, M., Kader, S. A. et al. Mammary-specific Ron receptor overexpression induces highly metastatic mammary tumors associated with beta-catenin activation. Cancer Res. 66, 11967–11974 (2006).
McClaine, R. J., Marshall, A. M., Wagh, P. K. & Waltz, S. E. Ron receptor tyrosine kinase activation confers resistance to tamoxifen in breast cancer cell lines. Neoplasia 12, 650–658 (2010).
Liu, X., Zhao, L., Derose, Y. S., Lin, Y. C., Bieniasz, M., Eyob, H. et al. Short-form ron promotes spontaneous breast cancer metastasis through interaction with phosphoinositide 3-kinase. Genes Cancer 2, 753–762 (2011).
Bieniasz, M., Radhakrishnan, P. & Faham, N., De La, J-P. & Welm, A. L. Preclinical efficacy of ron kinase inhibitors alone and in combination with PI3K inhibitors for treatment of sfron-expressing breast cancer patient-derived xenografts. Clin. Cancer Res. 21, 5588–5600 (2015).
Peace, B. E., Toney-Earley, K., Collins, M. H. & Waltz, S. E. Ron receptor signaling augments mammary tumor formation and metastasis in a murine model of breast cancer. Cancer Res. 65, 1285–1293 (2005).
Daub, H., Olsen, J. V., Bairlein, M., Gnad, F., Oppermann, F. S., Korner, R. et al. Kinase-selective enrichment enables quantitative phosphoproteomics of the kinome across the cell cycle. Mol. Cell 31, 438–448 (2008).
Rechoum, Y., Rovito, D., Iacopetta, D., Barone, I., Ando, S., Weigel, N. L. et al. AR collaborates with ERalpha in aromatase inhibitor-resistant breast cancer. Breast Cancer Res. Treat. 147, 473–485 (2014).
Sachs, N., de Ligt, J., Kopper, O., Gogola, E., Bounova, G., Weeber, F. et al. A living biobank of breast cancer organoids captures disease heterogeneity. Cell 172, 373–386 e10 (2018).
Martin, L. A., Ribas, R., Simigdala, N., Schuster, E., Pancholi, S., Tenev, T. et al. Discovery of naturally occurring ESR1 mutations in breast cancer cell lines modelling endocrine resistance. Nat. Commun. 8, 1865 (2017).
Duncan, J. S., Whittle, M. C., Nakamura, K., Abell, A. N., Midland, A. A., Zawistowski, J. S. et al. Dynamic reprogramming of the kinome in response to targeted MEK inhibition in triple-negative breast cancer. Cell 149, 307–321 (2012).
Jaquish, D. V., Yu, P. T., Shields, D. J., French, R. P., Maruyama, K. P., Niessen, S. et al. IGF1-R signals through the RON receptor to mediate pancreatic cancer cell migration. Carcinogenesis 32, 1151–1156 (2011).
Lee, A. V., Jackson, J. G., Gooch, J. L., Hilsenbeck, S. G., Coronado-Heinsohn, E., Osborne, C. K. et al. Enhancement of insulin-like growth factor signaling in human breast cancer: estrogen regulation of insulin receptor substrate-1 expression in vitro and in vivo. Mol. Endocrinol. 13, 787–796 (1999).
Turner, N., Kingston, B., Kilburn, L., Kernaghan, S., Wardley, A. M., Macpherson, I. et al. Abstract GS3-06: results from the plasmaMATCH trial: a multiple parallel cohort, multi-centre clinical trial of circulating tumour DNA testing to direct targeted therapies in patients with advanced breast cancer (CRUK/15/010). Cancer Res. 80, GS3–06-GS3 (2020).
Chandarlapaty, S., Sawai, A., Scaltriti, M., Rodrik-Outmezguine, V., Grbovic-Huezo, O., Serra, V. et al. AKT inhibition relieves feedback suppression of receptor tyrosine kinase expression and activity. Cancer Cell 19, 58–71 (2011).
Rexer, B. N., Chanthaphaychith, S., Dahlman, K. & Arteaga, C. L. Direct inhibition of PI3K in combination with dual HER2 inhibitors is required for optimal antitumor activity in HER2+ breast cancer cells. Breast Cancer Res. 16, R9 (2014).
Horst, B., Gruvberger-Saal, S. K., Hopkins, B. D., Bordone, L., Yang, Y., Chernoff, K. A. et al. Gab2-mediated signaling promotes melanoma metastasis. Am. J. Pathol. 174, 1524–1533 (2009).
McCartney, A., Migliaccio, I., Bonechi, M., Biagioni, C., Romagnoli, D., De Luca, F. et al. Mechanisms of resistance to CDK4/6 inhibitors: potential implications and biomarkers for clinical practice. Front Oncol. 9, 666 (2019).
Wander, S. A., Zangardi, M., Niemierko, A., Kambadakone, A., Kim, L. S., Xi, J. et al. A multicenter analysis of abemaciclib after progression on palbociclib in patients (pts) with hormone receptor-positive (HR+)/HER2− metastatic breast cancer (MBC). J. Clin. Oncol. 37, 1057 (2019).
Li, S., Shen, D., Shao, J., Crowder, R., Liu, W., Prat, A. et al. Endocrine-therapy-resistant ESR1 variants revealed by genomic characterization of breast-cancer-derived xenografts. Cell Rep. 4, 1116–1130 (2013).
DeRose, Y. S., Wang, G., Lin, Y. C., Bernard, P. S., Buys, S. S., Ebbert, M. T. et al. Tumor grafts derived from women with breast cancer authentically reflect tumor pathology, growth, metastasis and disease outcomes. Nat. Med. 17, 1514–1520 (2011).
Sikora, M. J., Cooper, K. L., Bahreini, A., Luthra, S., Wang, G., Chandran, U. R. et al. Invasive lobular carcinoma cell lines are characterized by unique estrogen-mediated gene expression patterns and altered tamoxifen response. Cancer Res. 74, 1463–1474 (2014).
Joseph, J. D., Darimont, B., Zhou, W., Arrazate, A., Young, A., Ingalla, E., et al. The selective estrogen receptor downregulator GDC-0810 is efficacious in diverse models of ER+ breast cancer. Elife. 5, e15828 (2016).
Baselga, J., Campone, M., Piccart, M., Burris, H. A. 3rd, Rugo, H. S., Sahmoud, T. et al. Everolimus in postmenopausal hormone-receptor-positive advanced breast cancer. N. Engl. J. Med. 366, 520–529 (2012).
Dustin, D., Gu, G. & Fuqua, S. A. W. ESR1 mutations in breast cancer. Cancer 125, 3714–3728 (2019).
Hortobagyi, G. N., Stemmer, S. M., Burris, H. A., Yap, Y. S., Sonke, G. S., Paluch-Shimon, S. et al. Updated results from MONALEESA-2, a phase III trial of first-line ribociclib plus letrozole versus placebo plus letrozole in hormone receptor-positive, HER2-negative advanced breast cancer. Ann. Oncol. 29, 1541–1547 (2018).
Kuang, Y., Siddiqui, B., Hu, J., Pun, M., Cornwell, M., Buchwalter, G. et al. Unraveling the clinicopathological features driving the emergence of ESR1 mutations in metastatic breast cancer. npj Breast Cancer 4, 22 (2018).
Andreano, K. J., Baker, J. G., Park, S., Safi, R., Artham, S., Oesterreich, S. et al. The dysregulated pharmacology of clinically relevant ESR1 mutants is normalized by ligand-activated WT receptor. Mol. Cancer Ther. 19, 1395–1405 (2020).
Thomas, R. M., Toney, K., Fenoglio-Preiser, C., Revelo-Penafiel, M. P., Hingorani, S. R., Tuveson, D. A. et al. The RON receptor tyrosine kinase mediates oncogenic phenotypes in pancreatic cancer cells and is increasingly expressed during pancreatic cancer progression. Cancer Res. 67, 6075–6082 (2007).
Roohullah, A., Cooper, A., Lomax, A. J., Aung, J., Barge, A., Chow, L. et al. A phase I trial to determine safety and pharmacokinetics of ASLAN002, an oral MET superfamily kinase inhibitor, in patients with advanced or metastatic solid cancers. Invest. N. Drugs 36, 886–894 (2018).
Brummer, T., Schramek, D., Hayes, V. M., Bennett, H. L., Caldon, C. E., Musgrove, E. A. et al. Increased proliferation and altered growth factor dependence of human mammary epithelial cells overexpressing the Gab2 docking protein. J. Biol. Chem. 281, 626–637 (2006).
Bentires-Alj, M., Gil, S. G., Chan, R., Wang, Z. C., Wang, Y., Imanaka, N. et al. A role for the scaffolding adapter GAB2 in breast cancer. Nat. Med. 12, 114–121 (2006).
Ribas, R., Pancholi, S., Rani, A., Schuster, E., Guest, S. K., Nikitorowicz-Buniak, J. et al. Targeting tumour re-wiring by triple blockade of mTORC1, epidermal growth factor, and oestrogen receptor signalling pathways in endocrine-resistant breast cancer. Breast Cancer Res. 20, 44 (2018).
Sammons, S., Shastry, M., Dent, S., Anders, C. & Hamilton, E. Practical treatment strategies and future directions after progression while receiving CDK4/6 inhibition and endocrine therapy in advanced HR(+)/HER2(−) breast cancer. Clin. Breast Cancer 20, 1–11 (2020).
Goel, S., DeCristo, M. J., Watt, A. C., BrinJones, H., Sceneay, J., Li, B. B. et al. CDK4/6 inhibition triggers anti-tumour immunity. Nature 548, 471–475 (2017).
Kettner, N. M., Vijayaraghavan, S., Durak, M. G., Bui, T., Kohansal, M., Ha, M. J. et al. Combined inhibition of STAT3 and DNA repair in palbociclib-resistant ER-positive breast cancer. Clin. Cancer Res. 25, 3996–4013 (2019).
Baselga, J., Dent, S. F., Cortés, J., Im, Y.-H., Diéras, V., Harbeck, N. et al. Phase III study of taselisib (GDC-0032) + fulvestrant (FULV) v FULV in patients (pts) with estrogen receptor (ER)-positive, PIK3CA-mutant (MUT), locally advanced or metastatic breast cancer (MBC): primary analysis from SANDPIPER. J. Clin. Oncol. 36, LBA1006–LBA1006 (2018).
Rugo, H. S., Lerebours, F., Ciruelos, E., Drullinsky, P., Borrego, M. R., Neven, P. et al. Alpelisib (ALP) + fulvestrant (FUL) in patients (pts) with PIK3CA-mutated (mut) hormone receptor-positive (HR+), human epidermal growth factor receptor 2-negative (HER2–) advanced breast cancer (ABC) previously treated with cyclin-dependent kinase 4/6 inhibitor (CDKi) + aromatase inhibitor (AI): BYLieve study results. J. Clin. Oncol. 38, 1006 (2020).
Razavi, P., Dickler, M. N., Shah, P. D., Toy, W., Brown, D. N., Won, H. H. et al. Alterations in PTEN and ESR1 promote clinical resistance to alpelisib plus aromatase inhibitors. Nat. Cancer 1, 382–393 (2020).
Acknowledgements
We would like to thank Arnoldo Corona-Rodriguez for his technical expertise in animal handling and surgical procedures.
Author information
Authors and Affiliations
Contributions
Concept and design: D.D., G.G., S.A.W.F. Development of methodology: D.D., G.G., D.W.C., D.L., C.C. Acquisition of data: D.D., G.G., A.R.B., S.K.H., D.G.E., H.L., T.L.G., D.C., B.-J.K., S.L.G., C.C., J.-P.D.L.O. Analysis and interpretation of data: D.D., G.G., A.R.B., S.H., T.L.G., D.W.C., S.L., A.L.W., M.J.E., S.A.W.F. Writing and review of paper: D.D., S.A.W.F. Administrative, technical or material support: A.R.B., S.L., A.L.W., M.J.E. Study supervision: S.A.W.F., D.D.
Corresponding author
Ethics declarations
Ethical approval and consent to participate
Animal experiments were performed in accordance with the protocol AN-1875 approved by the BCM Institutional Animal Care and Use Committee.
Data availability
All data and additional analysis generated in this study are available from the corresponding author on reasonable request. All microarray data are uploaded to GEO upon manuscript publication.
Competing interests
ALW previously received research support from Aslan Pharmaceuticals. SAWF is a Subject Editor, member of the Editorial Board of BJC and Guest Editor of the Special Issue on Metastasis.
Funding information
This study is supported by BCRF 19-055, NIH R01 CA207270, NIH R01 CA072038, CPRIT MIRA RP180712 (Kelly Hunt, PI), to S.A.W.F.; NIH R01 CA166422 to A.L.W. D.D. received training support from NIH T32 2389301302. SKH received training support from NIH T32 T32GM008231-30. TLG received training support from the Translational Breast Cancer Research Training Program NIH T32 CA203690-02. J-PDLO received support from the Susan G. Komen Postdoctoral Fellowship PDF12230127. M.J.E. was also supported by Cancer Prevention & Research Institutes of Texas Scholar (CPRIT) established investigator recruitment award CPRIT RR140033. M.J.E. is a Susan G. Komen Scholar and McNair Medical Institute Scholar. This project was supported by the Cytometry and Cell Sorting Core at Baylor College of Medicine with funding from the CPRIT Core Facility Support Award (CPRIT-RP180672), the NIH (CA125123 and RR024574) and the assistance of Joel M. Sederstrom. This project was supported in part by the C-BASS Core at Baylor College of Medicine with funding from the NIH (P30 CA125123) and the expert assistance of Dr. Jun Xu.
Additional information
Note This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution 4.0 International (CC BY 4.0).
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Dustin, D., Gu, G., Beyer, A.R. et al. RON signalling promotes therapeutic resistance in ESR1 mutant breast cancer. Br J Cancer 124, 191–206 (2021). https://doi.org/10.1038/s41416-020-01174-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41416-020-01174-z
This article is cited by
-
Proteomic profiling reveals that ESR1 mutations enhance cyclin-dependent kinase signaling
Scientific Reports (2024)
-
An immune-humanized patient-derived xenograft model of estrogen-independent, hormone receptor positive metastatic breast cancer
Breast Cancer Research (2021)
-
Metastasis: complexity thwarts precision targeting
British Journal of Cancer (2021)