Abstract
Background
Mutant TP53 interacts with other proteins to produce gain-of-function properties that contribute to cancer metastasis. However, the underlying mechanisms are still not fully understood.
Methods
Using immunoprecipitation and proximity ligation assays, we evaluated breast cancer anti-estrogen resistance 1 (BCAR1) as a novel binding partner of TP53R273H, a TP53 mutant frequently found in human cancers. The biological functions of their binding were examined by the transwell invasion assay. Clinical outcome of patients was analysed based on TP53 status and BCAR1 expression using public database.
Results
We discovered a novel interaction between TP53R273H and BCAR1. We found that BCAR1 translocates from the cytoplasm into the nucleus and binds to TP53R273H in a manner dependent on SRC family kinases (SFKs), which are known to enhance metastasis. The expression of full-length TP53R273H, but not the BCAR1 binding-deficient mutant TP53R273HΔ102–207, promoted cancer cell invasion. Furthermore, among the patients with mutant TP53, high BCAR1 expression was associated with a poorer prognosis.
Conclusions
The interaction between TP53R273H and BCAR1 plays an important role in enhancing cancer cell invasion. Thus, our study suggests a disruption of the TP53R273H–BCAR1 binding as a potential therapeutic approach for TP53R273H-harbouring cancer patients.

Similar content being viewed by others
Background
The transcription factor TP53 is one of the most important tumour suppressors. TP53 prevents tumour development and progression by regulating target genes involved in cell cycle arrest, senescence, apoptosis, autophagy, metabolic reprogramming, tumour microenvironment signalling, and the suppression of cancer cell migration and invasion.1,2,3,4,5,6,7 Given its central role in tumour suppression, it is not surprising that the TP53 pathway is frequently inactivated through mutations in TP53 itself or alterations in its regulators including CDKN2A (also known as p14ARF) and MDM2.8 In contrast to other tumour suppressors such as RB1, APC, NF1, NF2, and VHL, where deletion or nonsense mutations result in little or no protein expression of these tumour suppressors, most TP53 mutations are missense mutations that produce stable and long-lived proteins. A number of hotspot mutations including R273H are identified within the DNA-binding domain of TP53.
Mutation within the DNA-binding domain often leads to the loss of DNA-binding ability of TP53. Therefore, such mutant TP53 is unable to induce CDKN1A, BBC3/PUMA and BAX to trigger tumour-suppressive functions, and the mutations in TP53 have been known to be similar with deletion of TP53, often referred to as the loss-of-function of TP53.4,5 However, in vivo knock-in mice harbouring mutant forms of Trp53 (mouse orthologue corresponding to human TP53) developed more metastatic tumours as compared to Trp53-null mice.9,10 TP53 is mutated more in the advanced stage of human cancers, compared to the early stages, suggesting that mutant TP53 is involved in metastasis in human cancer as well.11,12 These findings shifted the paradigm of understanding of TP53 mutations. In addition to the loss-of-function properties, mutant TP53 have oncogenic gain-of-function properties often related to invasion or metastasis;13 for example, they can promote epithelial-to-mesenchymal transition (EMT),14,15 disrupt focal adhesion signalling pathways,16 augment filopodia formation,17 disrupt mammary tissue architecture,18 and promote integrin or growth factor receptor recycling.19,20 Several mechanisms are proposed through which mutant forms of TP53 exhibit their gain-of-function properties: (i) binding to proteins such as transcription factors or cofactors to repress or augment their activities, (ii) binding to other proteins to modulate their activities, and (iii) binding to certain DNA structural motifs.8,13,21,22,23,24 However, the gain-of-function properties of mutant TP53 are still not fully understood due to their cell-type and cell-context dependency regulated by multiple underlying mechanisms.13,25,26
To gain novel insights into these mechanisms, we looked for interacting partners of TP53R273H and identified BCAR1 as a novel mutant TP53R273H binding protein. We propose that the formation of the TP53R273H–BCAR1 complex is a novel mechanism of mutant TP53-mediated gain-of-functions for cancer cell invasion.
Methods
Cell lines, cell culture, and reagents
MDA-MB-468, A431, H1299, and HEK 293T cells were obtained from the American Type Culture Collection (ATCC, LGC Standards, Teddington, UK). U251MG cell line was purchased from CLS Cell Lines Service GmbH (Eppelheim, Germany). All cell lines were cultured in Dulbecco’s modified Eagle medium (DMEM, Thermo Fisher Scientific, Waltham, MA, USA) supplemented with 10% foetal bovine serum (FBS) (Thermo Fisher Scientific) and maintained in a 37 °C incubator with 5% CO2. SRC family kinase inhibitors (SFKs) CGP77675 and PP2 were purchased from Axon Medchem (Reston, VA, USA) and Sigma Aldrich (St. Louis, MO, USA), respectively. Leptomycin B was purchased from LC laboratories (Woburn, MA, USA). Matrigel was purchased from Corning (Corning, NY, USA). 4',6-diamidino-2-phenylindole (DAPI) was purchased from Nacalai Tesque (Nakagyo-ku, Kyoto, Japan).
Plasmids
To generate human BCAR1 expression constructs, full-length BCAR1 complementary DNA (cDNA) was amplified by RT-PCR using cDNA from MDA-MB-468 cells as a template. PCR products were cloned into pcDNA3.1(+)-FLAG3, pcDNA3.1(+)-MYC3, or pCMV-MYC3 vector backbones. The CMV-TP53R273H plasmid was generated by site-directed mutagenesis PCR using CMV-TP53WT as a template, which was kindly provided by Dr. Yanping Zhang (University of North Carolina at Chapel Hill, USA). Truncation mutants of TP53R273H (1–292, 102–393, and 293–393) were generated by PCR cloning, and deletion mutants of TP53R273H (Δ102–207 and Δ208–292) were generated by site-directed mutagenesis PCR using CMV-TP53R273H as a template. Cas9-EGFP was obtained from Addgene (Watertown, MA, USA), and the plasmid containing gRNA targeting TP53 was constructed as previously described.27 ScreenFect A (FUJIFILM Wako Pure Chemical Corporation, Osaka, Japan) or calcium phosphate transfection method was used for plasmid transfection according to the manufacturer’s instructions or as previously described,28 respectively. When FLAG- or MYC-tagged TP53R273H (or BCAR1) plasmid was transfected, we used the plasmid containing FLAG or MYC sequence as a negative control.
Indirect immunofluorescence analysis
H1299 cells were fixed with 4% paraformaldehyde in phosphate buffered saline (PBS), permeabilised with 0.2% Triton-X, and blocked with 5% bovine serum albumin (BSA) in PBS. Mouse anti-BCAR1 (1:1000, 21/p130[Cas], BD Biosciences, San Jose, CA, USA), rabbit anti-TP53 (1:1000, FL393, Santa Cruz Biotechnology, Dallas, TX, USA) and mouse anti-FLAG (1:1000, M2, Sigma Aldrich) were used as primary antibodies. Alexa Fluor 488- or Alexa Fluor 594-conjugated donkey anti-mouse IgG and Alexa Fluor 488- or Alexa Fluor 594-conjugated donkey anti-rabbit IgG (1:1000, Jackson ImmunoResearch, West Grove, PA, USA) were used as secondary antibodies. DAPI was used for staining the nuclei. Images of the nuclei, BCAR1, and TP53R273H were acquired using 405, 488, and 591 nm lasers on a confocal microscope (Carl Zeiss LSM 710, Oberkochen, Germany) equipped with oil-immersion objective lens (NA 1.40, ×63; Plan Apochromat, Carl Zeiss) and ZEN 2010 software (version 6.0.0.486; Carl Zeiss). Images were cropped using ImageJ software (version 1.45f; National Institutes of Health, Bethesda, MD, USA).
Co-immunoprecipitation
Cells were washed three times with PBS and lysed with ice-cold 0.1% NP40 lysis buffer (50 mM Tris-HCl [pH 7.5], 0.1% NP40, 150 mM NaCl, 50 mM NaF, 1 mM phenylmethylsulfonyl fluoride, 1 mM Na3VO4, and protease inhibitors). All subsequent steps were performed at 4 °C. Cell lysates were pre-cleared with 50 µL of Sepharose CL-4B beads (Sigma Aldrich) for 30 min, and 1 mg of total proteins were immunoprecipitated with 1 µg of the indicated antibodies (rabbit or goat anti-TP53 [FL393, Santa Cruz Biotechnology], mouse anti-FLAG [M2, Sigma Aldrich], mouse anti-MYC [9E10, Santa Cruz Biotechnology], or mouse anti-BCAR1 [21/p130[Cas], BD Biosciences]) for 16 h. After incubating the lysates with 10 µL of 50% Protein A/G agarose beads slurry (Thermo Fisher Scientific) for 2 h, the agarose beads were washed three times using ice-cold 0.1% NP40 lysis buffer before being subjected to SDS-polyacrylamide gel electrophoresis and protein analysis.
Proximity ligation assay and quantification of proximity ligation foci
Proximity ligation assay (PLA) was performed according to the manufacturer’s instructions (Sigma Aldrich). Briefly, H1299 or U251MG cells seeded onto µ-dish (ibidi, Grafelfing, Germany) were fixed with 4% paraformaldehyde in PBS. Cells were then permeabilised using 0.2% Triton-X and blocked with 5% BSA in PBS. For H1299 cells transfected with CMV-TP53R273H and/or pCMV-MYC3-BCAR1, mouse anti-TP53 (1:1000, DO-1, Santa Cruz Biotechnology) and rabbit anti-MYC (1:1000, Proteintech Group, Inc, Rosemont, IL, USA) were used as primary antibodies. For H1299 cells transfected with pcDNA3.1(+)-TP53R273H-FLAG3, TP53R273H (1–292)-FLAG3, and/or pcDNA3.1(+)-MYC3-BCAR1, mouse anti-FLAG (1:1000, M2, Sigma Aldrich) and rabbit anti-MYC (1:1000, Proteintech Group, Inc) were used as primary antibodies. For U251MG cells, rabbit anti-TP53 (1:1000, FL393, Santa Cruz Biotechnology) and mouse anti-BCAR1 (1:1000, 21/p130[Cas], BD Biosciences) were used as primary antibodies. Anti-mouse PLUS and anti-rabbit MINUS probes (1:500, Duolink, Sigma Aldrich) were used as secondary antibodies. Ligation and amplification reactions were performed according to the manufacturer’s instructions. To confirm the localisation of TP53R273H-FLAG3 and TP53R273H (1–292)-FLAG3, cells were also stained with Alexa Fluor 594-conjugated donkey anti-mouse IgG and CF680R-conjugated donkey anti-rabbit IgG (1:1000, Biotium, Fremont, CA, USA) as secondary antibodies after PLA assay. Cells were then washed, and the nuclei were stained with DAPI. Images of the nuclei, proximity ligated foci, TP53R273H, and BCAR1 were acquired using 405, 488, 591, and 688 nm lasers on a confocal microscope (Carl Zeiss LSM 710) equipped with oil-immersion objective lens (NA 1.40, ×63; Plan Apochromat, Carl Zeiss) and ZEN 2010 software (version 6.0.0.486; Carl Zeiss). Images were cropped using ImageJ software (version 1.45f; National Institutes of Health). The fluorescence intensity of proximity ligated foci was measured by ImageJ software using 8-bit images. Briefly, the tracing tool was used to outline individual cells, and the ‘measure’ function was used to obtain mean fluorescence intensity of each proximity ligated foci. Total fluorescence intensity in each cell was normalised by cell area and plotted as a dot in the graph. Statistical analysis of data was done by unpaired Student’s two-sided t test.
Three-dimensional spheroid invasion assay and quantification of the invasion area
U251MG cells (1 × 103) were seeded into ultra-low attachment 96-well round-bottom plates (Corning) containing 200 µL of DMEM supplemented with 10% FBS and cultured for 2 days to form spheroids. Following spheroid formation, 100 µL of DMEM was replaced with 100 µL of 100% Matrigel (Corning) and incubated at 37 °C for 1 h before overlaying with 100 µL of DMEM containing 10% FBS. This was set as day 0. Micrographs of the spheroids were taken at ×10 magnification using an inverted Phase Contrast microscope (IX73, Olympus, Tokyo, Japan) coupled with a DP71 camera (Olympus). The area of cells invading into Matrigel was measured by ImageJ software using 8-bit images of spheroids. Briefly, the ‘find edges’ function with a minimum and a maximum threshold value of 0 and 50, respectively, was applied to each spheroid image. The tracing tool was then used to outline the spheroids. Area of spheroids measured on day 0 was subtracted from the area of spheroids measured on day 2 to obtain the area of cells invading into Matrigel. The data are presented relative to the shControl cells.
Transwell invasion assay
Matrigel (Corning) diluted in serum-free DMEM was coated on the upper side of 8 μm pore-size transwell inserts (Corning) and allowed to dry. MDA-MB-468 (1 × 105), U251MG (1 × 105), or A431 (1 × 106) cells suspended in serum-free DMEM were then seeded onto the Matrigel-coated transwell inserts (Corning) and placed into 24-well plates containing DMEM supplemented with 20% FBS. Cells were allowed to invade across the transwell insert for 20 h (MDA-MB-468 and A431 cells) or 12 h (U251MG cells) before fixing and permeabilising with 4% paraformaldehyde and 0.1% Triton-X, respectively. Non-invading cells in the upper side of the transwell were removed by cotton-tipped swab before staining with 0.1% Crystal Violet solution (Sigma Aldrich). The excess stain was washed with water, and the inserts were allowed to dry. More than five micrographs per sample were taken at ×11.5 magnification using a dissection microscope (Olympus) coupled with a DP71 camera (Olympus). The area of invasion was measured by ImageJ software using 8-bit images of micrographs. Briefly, the ‘analyse particles’ function was applied to each transwell image. The data are presented as a percentage. Statistical analysis of data was done by unpaired Student’s two-sided t test.
Protein analysis
Cells were washed three times with PBS and lysed with 2% SDS lysis buffer (2% SDS, 50 mM Tris-HCl [pH 6.8], 10% glycerol). Protein concentration was determined using DC Protein Assay Reagents (BioRad Laboratories, Hercules, CA, USA). Equal amounts of total protein were separated by SDS-polyacrylamide gel electrophoresis and transferred to nitrocellulose membranes. Membranes were blocked with 5% non-fat milk in PBS containing 0.1% Tween-20 and blotted with the indicated antibodies (mouse anti-TP53 [1:1000, DO-1, Santa Cruz Biotechnology], rabbit anti-TP53 [1:1000, FL393, Santa Cruz Biotechnology], mouse anti-BCAR1 [1:1000, 21/p130[Cas], BD Biosciences], mouse anti-ACTIN [1:50,000, C4, Merck Millipore, Darmstadt, Germany], rabbit anti-phospho-p130 Cas/BCAR1 (Tyr165) [1:1000, Cell Signaling Technology, Danvers, MA, USA], mouse anti-FLAG [1:1000, M2, Sigma Aldrich], rabbit anti-FLAG [1:1000, Proteintech Group, Inc], or mouse anti-MYC [1:1000, 9E10, Santa Cruz Biotechnology]). Detection of blots was done with West Pico (Thermo Fisher Scientific) for secondary antibody conjugated with HRP (1:10,000, HRP-conjugated goat anti-mouse or anti-rabbit IgG, Jackson ImmunoResearch), or done with Odyssey Infrared Imaging System (LI-COR Biosciences, Lincoln, NE, USA) for fluorescent-labelled secondary antibody (1:10,000, DyLight 680- or 800-conjugated goat anti-mouse IgG or DyLight 680- or 800-conjugated goat anti-rabbit IgG, Thermo Fisher Scientific).
Affinity purification mass spectrometry, CRAPome, and Ingenuity Pathway Analyses
U251MG cells were transduced with empty vector control- or TP53R273H-FLAG-expressing adenovirus at large-scale. Cells were lysed with ice-cold 0.1% NP40 lysis buffer and subjected to co-immunoprecipitation using EZview Red anti-FLAG M2 Affinity Gel (M2, Sigma Aldrich). Proteins bound to the beads were eluted with 3xFLAG peptide (Sigma Aldrich), digested with trypsin, and then applied to a C18 column (Thermo Fisher Scientific). Bound proteins were washed with ultrapure water to remove any salt, eluted with acetonitrile, and concentrated using a SpeedVac vacuum concentrator (Thermo Fisher Scientific). Samples were analysed by LC-MS/MS. MS data were analysed using GPM software (http://gpmdb.thegpm.org) with default parameter settings. Proteins with the expectation values log(e)29 of −1.0 or lower were used for downstream analysis. A hundred and fifty-one proteins were identified after background subtraction. BioGRID (https://thebiogrid.org/) and PubMed were used to assess if the identified proteins are novel or reported. Identified proteins were queried on CRAPome database30 (http://www.crapome.org), and 62 proteins with CRAPome value < 15/411 were considered as specific TP53R273H binding candidates. These were annotated and analysed for molecular and cellular functions through the use of Ingenuity Pathway Analysis31 (QIAGEN Inc., https://www.qiagenbioinformatics.com/products/ingenuity-pathway-analysis).
Kaplan–Meier plots
We downloaded the clinical datasets of Molecular Taxonomy of Breast Cancer International Consortium (METABRIC) for breast cancer and The Cancer Genome Atlas (TCGA) PanCancer Atlas for lung adenocarcinoma from cBioportal database32,33 (https://www.cbioportal.org/). The data were sequenced on Illumina platform. We retrieved the RSEM normalised counts for BCAR1 gene along with TP53 mutation status (wild type or mutant), survival months, and status (alive or dead). After filtering off samples with missing information, METABRIC dataset has 1904 samples composed of TP53 mutated (n = 659) and TP53 wild type (n = 1245). TCGA PanCancer Atlas dataset has 501 samples composed of TP53 mutated (n = 260) and TP53 wild type (n = 241). Kaplan–Meier curves were plotted using SPSS (version 26) for the TP53 mutated group and TP53 wild-type group. Each group was further divided into two sub-groups based on whether it was above or below the median of BCAR1 expression. A log-rank test was used to compare the survival curves between BCAR1 high and low gene expression groups. P values < 0.05 were considered statistically significant.
Generation of TP53 –/– MDA-MB-468 clones
TP53 knockout MDA-MB-468 cells were generated as previously described.27 Briefly, MDA-MB-468 cells were transfected with either a gRNA targeting exon 4 of TP53 (GGCAGCTACGGTTTCCGGTCT) or empty vector together with Cas9-EGFP. Cells were sorted for GFP-positive cells using BD FACSArial II cell sorter (BD Biosciences) and plated sparsely onto tissue cultured plastic plates. Colonies of cells were picked using sterilised cloning cylinders and expanded. Protein analysis was performed to validate the absence of TP53.
Retroviral vectors and retroviral transduction
To generate retroviruses carrying shRNAs against human TP53 or BCAR1, TP53 targeting sequence #1 (GGTGAACCTTAGTACCTAA), TP53 targeting sequence #2 (GTAATCTACTGGGACGGAA), BCAR1 target sequence #1 (GGATGGAGGACTATGACTA), and BCAR1 target sequence #2 (GAGTTTGAGAAGACCCAGA) were cloned into a pSuper retro puro (Oligoengine) or pSuper retro hygro vector; the latter was generated by replacing the sequencing encoding puromycin with that encoding hygromycin. Retroviral transduction was performed as previously described.27 Briefly, retrovirus vector containing shRNA sequence together with amphotropic retroviral plasmids were co-transfected into HEK 293T cells to generate viral particles. Cells at 50% confluence were infected with retrovirus overnight in the presence of 8 µg/mL polybrene (Sigma Aldrich) and selected with 0.5 µg/mL puromycin or 300 µg/mL hygromycin for 4 days after transduction.
Small interfering RNA (siRNA) and transfection
Calcium phosphate transfection protocol was performed as described28 to introduce control siRNA (D-001810-10-20, ON-TARGETplus Non-targeting Pool, Dharmacon GE Healthcare Bio-Sciences, Lafayette, CO, USA) or siRNA targeting human TP53 or BCAR1 at a final concentration of 40 nM into H1299 or A431 cells.
TP53 targeting sequence forward: GUAAUCUACUGGGACGGAAdTdT
TP53 targeting sequence reverse: UUCCGUCCCAGUAGAUUACdTdT
BCAR1 targeting sequence #1 forward: GGAUGGAGGACUAUGACUAdTdT
BCAR1 targeting sequence #1 reverse: UAGUCAUAGUCCUCCAUCCdAdG
BCAR1 targeting sequence #2 forward: GAGUUUGAGAAGACCCAGAdTdT
BCAR1 targeting sequence #2 reverse: UCUGGGUCUUCUCAAACUCdCdT
Results
TP53R273H binds to BCAR1 in the nucleus
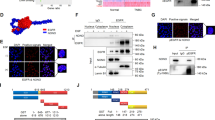
To uncover a novel mechanism by which mutant TP53 exhibits its gain-of-function properties, we looked for TP53R273H interacting proteins that contribute to cancer cell invasion. R273H mutation is one of the hotspot mutations in TP53 in human cancers.8,11,12 We first conducted a large-scale co-immunoprecipitation with a FLAG antibody following overexpression of TP53R273H-FLAG in U251MG cells (TP53R273H-harbouring human glioblastoma cell line34) (Supplementary Fig. S1). The proteins pulled down by the anti-FLAG antibody were subjected to mass spectrometry (Fig. 1a). After background subtraction, this approach identified 151 potential TP53R273H-associated proteins (Supplementary Table 1). Fifty-one out of 151 proteins were reported to bind to TP53. Known TP53R273H binding proteins such as MDM235 and NRDC36 were also in our list, validating the reliability of our approach in identifying novel TP53R273H interacting proteins. To further eliminate potential background contaminants from our mass spectrometry, we stringently selected proteins with a low CRAPome value of <15/411. We then stratified the remaining 62 proteins according to their molecular and cellular functions using the Ingenuity Pathway Analysis (IPA). The results show that many of the candidate interactors likely function in the cell cycle or cellular movement (Fig. 1b).
a A schematic workflow of affinity purification coupled to mass spectrometry. Immunoprecipitation with FLAG antibody was performed using U251MG cells (TP53R273H-harbouring human glioblastoma) overexpressing TP53R273H-FLAG or control vector. Proteins pulled down by FLAG antibody were subjected to mass spectrometry analysis. Proteins identified in cells expressing control vector were considered to bind non-specifically to the antibody (background). b Sixty-two proteins identified as candidate interactors were sorted based on their molecular and cellular functions using Ingenuity Pathway Analysis. The top five molecular and cellular functions are shown with the number of proteins under each category indicated. Bars represent the level of significance for the genes associated with the indicated molecular and cellular functions. The P value threshold (dotted line) is set at 0.05, indicating a maximum false discovery rate of 5%. c, d Co-immunoprecipitation of exogenous TP53R273H and FLAG-BCAR1. Lysates of H1299 cells transfected with the indicated plasmids were immunoprecipitated with anti-FLAG antibody (c) or with anti-TP53 antibody (d), and analysed by Western blot. A FLAG tag-containing vector was used as a negative control for FLAG-BCAR1 in (c). e–g Co-immunoprecipitation of endogenous TP53R273H and BCAR1. Lysates of U251MG (e) or MDA-MB-468 (MDA468) cells (f) expressing shControl (empty vector control) or shTP53(#2) were immunoprecipitated with the anti-TP53 antibody. Lysates of MDA468 cells (g) were immunoprecipitated with anti-BCAR1 antibody or control IgG. Immunoprecipitated proteins were analysed by Western blot. h Localisation of the exogenous TP53R273H–BCAR1 complexes. Confocal microscopy images of green fluorescence signals obtained from the proximity ligation assay are shown. H1299 cells were transfected with the indicated plasmids. Cells transfected with empty and/or MYC tag-containing plasmids served as negative controls. The nuclei were stained with DAPI. Scale bar represents 10 μm. i Quantifications of the proximity ligated foci. The images obtained in (h) were used for calculation. Bars represent the mean fluorescence intensity of proximity ligated foci in each cell normalised by cell area ± SD. Three or more cells were used for quantifications and indicated as dots. *p value < 0.05. Statistical analysis of data was done by unpaired Student’s two-sided t test. j Localisation of the endogenous TP53R273H–BCAR1 complexes in U251MG cells expressing shControl (empty vector control). Confocal microscopy images of green fluorescence signals obtained from proximity ligation assay are shown. U251MG cells expressing shTP53 or shBCAR1 served as negative controls. Fields enclosed by the yellow line show the magnified images of selected cells (arrowhead). Scale bar represents 10 μm. k Quantifications of the proximity ligated foci. The images obtained in (j) were used for calculation. Bars represent the mean fluorescence intensity of proximity ligated foci in each cell normalised by cell area ± SD. Eight or more cells were used for quantifications and indicated as dots. *P value < 0.05. Statistical analysis of data was done by unpaired Student’s two-sided t test.
Since we were looking for novel TP53R273H-interacting proteins that contribute to cancer cell invasion, we focused on the 17 proteins grouped under the ‘cellular movement’ category (Supplementary Table 2). Using IPA, we generated a functional network of the 17 proteins to connect with the individual cellular functions (Supplementary Fig. S2). The protein that had the highest numbers of the functional connections to the cellular movement was breast cancer anti-estrogen resistance 1 (BCAR1). BCAR1 functions as an adaptor protein downstream of several cell-surface receptors including growth factor receptors and integrins.37 Activation of these cell-surface receptors induces phosphorylation of 15 tyrosine motifs (YxxP) of BCAR1 by SRC family kinases (SFKs).38 BCAR1 promotes EMT, cell motility, and invasion37 and is overexpressed in many human cancers including breast,39,40 glioma,38 prostate,41 ovarian,42 and hepatocellular carcinoma.43 The increased expression of BCAR1 in these cancers is frequently associated with advanced stage, metastatic properties, and poor survival rates. Given the well-established importance of BCAR1 in cancer and invasion,37,44,45,46 we chose to focus the rest of our study on the TP53R273H−BCAR1 interaction.
To verify our mass spectrometry data, we ectopically expressed TP53R273H and FLAG-tagged BCAR1 in TP53-null H1299 cells and performed the co-immunoprecipitation assays. TP53R273H was co-immunoprecipitated with FLAG-BCAR1 (Fig. 1c). The binding was further confirmed by reciprocal co-immunoprecipitation (Fig. 1d). To demonstrate that endogenous TP53R273H and BCAR1 also interact under physiological conditions, we conducted co-immunoprecipitation assay using lysates of U251MG and MDA-MB-468 (TP53R273H-harbouring human breast cancer cell line24) cells expressing either empty vector control or shTP53. The amount of BCAR1 co-immunoprecipitated by TP53R273H was reduced in both cell lines expressing TP53 shRNA, corresponding to the reduced amount of TP53R273H pulled down (Fig. 1e, f), confirming their endogenous binding. The endogenous binding was also confirmed reciprocally by co-immunoprecipitating TP53R273H with BCAR1 (Fig. 1g).
To examine the localisation of the TP53R273H–BCAR1 complexes, we performed PLAs using H1299 cells ectopically expressing TP53R273H and MYC-tagged BCAR1. Confocal microscopy revealed that exogenous TP53R273H and MYC-BCAR1 were in close proximity mainly in the nucleus (Fig. 1h, i). Consistent with this, endogenous TP53R273H and BCAR1 were also in close proximity in the nucleus in U251MG cells expressing shControl (empty vector control) (Fig. 1j, k). Together, these results suggest that the binding between TP53R273H and BCAR1 occurs mainly in the nucleus.
Depletion of TP53R273H and BCAR1 reduces cancer cell invasion to a similar extent as single depletion of TP53R273H or BCAR1
It has been reported that TP53R273H19,20,36,47 as well as BCAR137,44,45,46 regulate cancer cell invasion. Consistent with these reports, the knockdown of TP53R273H reduced 3D spheroid invasion of U251MG cells (Fig. 2a–c). Knockdown of BCAR1 reduced 3D spheroid invasion to a similar extent as knockdown of TP53R273H (Fig. 2a–c). However, it is not known whether TP53R273H and BCAR1 work cooperatively or separately to promote invasion. If they work cooperatively, then the simultaneous depletion of both proteins should not further reduce 3D spheroid invasion as compared to individual depletion of each protein. Indeed, our results revealed that the invasiveness of U251MG cells was similar when TP53R273H and BCAR1 were depleted simultaneously or individually. We further evaluated cancer cell invasion using a transwell invasion assay. Transwell invasion assay can be done in a shorter time than 3D spheroid invasion assay, thereby minimising the possible influence of cell proliferation on the final readout. Consistent with 3D spheroid invasion assay (Fig. 2b, c), the invasiveness of U251MG cells measured by transwell assay was similar when TP53R273H and BCAR1 were depleted simultaneously or individually (Fig. 2d, e). We confirmed this observation in another cancer cell line, A431 cells (TP53R273H-harbouring human epidermoid carcinoma cell line24) (Supplementary Fig. S3). Taken together, these results suggest that TP53R273H and BCAR1 may contribute to cancer cell invasion in the same pathway.
a Lysates of U251MG cells transduced with a retrovirus expressing shControl (empty vector control), shTP53(#2), shBCAR1, or both shTP53(#2) and shBCAR1 were analysed by Western blot. b Phase-contrast micrographs of 3D spheroid invasion assay using U251MG cells expressing shControl (empty vector control), shTP53(#2), shBCAR1, or both shTP53(#2) and shBCAR1. Images were taken at the indicated time after spheroids were embedded into Matrigel. Representative images of each group are shown. Scale bar represents 100 μm. c Quantification of the area of cells invading into the Matrigel. The images obtained in (b) were used for calculation. Bars represent the mean area of cells invading into Matrigel relative to the control (the first bar, set as 100%) ± SD from three independent experiments (n = 3). d Bright-field micrographs of transwell invasion assay using U251MG cells expressing the indicated shRNA. Representative images of the transwells are shown. Scale bar represents 200 μm. e Quantification of the area of cells invading into the Matrigel. The images obtained in (d) were used for calculation. Bars represent the mean area of cells invading into Matrigel relative to the control (the first bar, set as 100%) ± SD from three independent experiments (n = 3).
A SRC family kinase inhibitor prevents BCAR1 nuclear import, TP53R273H–BCAR1 complex formation, and invasion
SFKs phosphorylate BCAR1 and promote cancer cell invasion.37,48 To evaluate whether SFKs are involved in the TP53R273H−BCAR1 interaction and cancer cell invasion, we first examined whether inhibition of SFKs prevents the invasiveness of U251MG and MDA-MB-468 cells. Consistent with previous findings,49 treatment with SFKs inhibitor CGP77675 abolished the invasion of U251MG and MDA-MB-468 cells (Fig. 3a, b).
a, b Bright-field micrographs of transwell invasion assay using U251MG (a) or MDA-MB-468 (MDA468) cells (b) in the absence or presence of CGP77675 (5 µM). Representative images of the transwells are shown. Scale bar represents 200 μm. Quantification of the invading area is shown on the right. Bars represent the mean invading area relative to the DMSO-treated cells (the first bar, set as 100%) ± SD from three independent experiments (n = 3). c Localisation of BCAR1 and TP53. H1299 cells transfected with Control (empty vector control) or TP53R273H were pre-treated with or without CGP77675 (5 µM) for 9 h, followed by incubation with DMSO, LMB (10 nM), and/or CGP77675 for another 16 h. Cells were then subjected to immunofluorescence staining. Nuclei (blue), endogenous BCAR1 (green) and TP53R273H (red). Scale bar represents 10 μm. d Co-immunoprecipitation of BCAR1 with endogenous TP53R273H. The cells were treated with DMSO, CGP77675 (5 µM), or PP2 (40 µM) for 16 h. Lysates of shControl (empty vector control)- or shTP53(#2)-expressing MDA-MB-468 (MDA468) cells were immunoprecipitated with anti-TP53 antibody and analysed by Western blot. e Co-immunoprecipitation of exogenous TP53R273H-FLAG with MYC-BCAR1 in the presence or absence of c-SRCY530F. Lysates of H1299 cells transfected with the indicated plasmids were immunoprecipitated with anti-MYC antibody and analysed by Western blot. An empty vector or MYC tag-containing plasmid was used as a negative control.
We next examined the effects of SFKs inhibition on the spatial localisation of TP53R273H and BCAR1 in cells by immunofluorescence staining. The specificity of the BCAR1 antibody was confirmed by the staining of H1299 cells expressing control siRNA or siRNA targeting BCAR1 (Supplementary Fig. S4). The majority of endogenous BCAR1 localised in the cytoplasm in TP53-null H1299 cells (Fig. 3c, panel b). Cytoplasmic localisation of BCAR1 was not affected by ectopic expression of TP53R273H (Fig. 3c, panel e), which primarily localised in the nucleus (Fig. 3c, panel f). This indicates that TP53R273H does not promote BCAR1 nuclear translocation. Consistent with this result, endogenous BCAR1 also localised in the cytoplasm regardless of the presence or absence of endogenous TP53R273H expression (Supplementary Fig. S5).
BCAR1 has been reported to shuttle between the nucleus and the cytoplasm.50,51,52 We speculated that BCAR1 nuclear export is stronger than nuclear import since we observed BCAR1 predominantly in the cytoplasm. Indeed, when the cells were treated with leptomycin B (LMB), an exportin inhibitor to inhibit protein export activity, BCAR1 was accumulated in the nucleus regardless of the TP53R273H expression (Fig. 3c, panels h, k). LMB treatment also slightly increased TP53R273H−BCAR1 binding (Supplementary Fig. S6). BCAR1 contains 15 tyrosine residues within its substrate domain, which are phosphorylated mainly by SFKs.37,38 One study has revealed that phosphorylation of BCAR1 is required for nuclear import,53 while another demonstrated that BCAR1 could be imported into the nucleus in the absence of phosphorylation.50 Since the role for phosphorylation in nuclear import of BCAR1 remains controversial, we examined whether SFKs-mediated BCAR1 phosphorylation is required for nuclear import of BCAR1 in our study. However, it was difficult to assess whether blocking BCAR1 phosphorylation with a SFKs inhibitor CGP77675 attenuates nuclear import since BCAR1 was already predominantly localised in the cytoplasm due to the strong nuclear export activity, regardless of treatment with CGP77675 (Fig. 3c, panels b, n). Therefore, we treated the cells with CGP77675 in the presence of LMB to make it easier to observe the effect of CGP77675 on BCAR1 nuclear transport. Nuclear accumulation of BCAR1 by LMB (Fig. 3c, panel h) was attenuated when cells were treated with both CGP77675 and LMB (Fig. 3c, panel t). This was also observed in cells expressing exogenous TP53R273H (Fig. 3c, panels k, w). These results suggest that SFKs-mediated phosphorylation of BCAR1 is required for nuclear translocation regardless of TP53R273H expression.
The amount of phosphorylated BCAR1 remained unchanged upon depletion of TP53R273H (Fig. 3d, lanes 1–2). These data led us to speculate that, regardless of TP53R273H expression, once BCAR1 gets phosphorylated, a subset of BCAR1 translocates into the nucleus where it interacts with TP53R273H. Hence, we expected that the interaction between TP53R273H and BCAR1 would be abolished if BCAR1 phosphorylation was inhibited by SFKs inhibitors, either CGP77675 or PP2.54 Indeed, the amount of endogenous BCAR1 that was co-immunoprecipitated with endogenous TP53R273H was markedly reduced in CGP77675- or PP2-treated cells as compared to DMSO-treated cells (Fig. 3d, lanes 1, 3–4). Furthermore, the expression of constitutively activated c-SRC (c-SRCY530F) enhanced the phosphorylation of BCAR1 remarkably and increased the amount of TP53R273H that was co-immunoprecipitated with BCAR1 (Fig. 3e, lanes 1–2).
Taken together, these data indicate that TP53R273H is not involved in SFKs-mediated phosphorylation of BCAR1 and translocation of phosphorylated BCAR1 into the nucleus, but rather suggest that TP53R273H forms a complex with the nuclear-imported BCAR1 that is phosphorylated by SFKs.
BCAR1 interacts with the DNA-binding domain region 102–207 of TP53R273H
To evaluate whether the interaction between TP53R273H and BCAR1 is crucial for promoting cancer cell invasion, we first generated FLAG-tagged full-length and deletion mutants of TP53R273H to identify which region is important for BCAR1 interaction (Fig. 4a). Our immunofluorescence staining revealed that two truncation mutants, TP53R273H (102–393) and TP53R273H (293–393), localised in the nucleus similar to TP53R273H full-length (Fig. 4b). Despite similar nuclear localisation patterns, full-length TP53R273H and TP53R273H (102–393) but not TP53R273H (293–393) was immunoprecipitated with endogenous BCAR1 (Fig. 4c), suggesting that BCAR1 binds to the region corresponding to amino acids 102–292 of TP53R273H. On the other hand, TP53R273H (1–292) did not bind efficiently to BCAR1 even though it contains the 102–292 region (Fig. 4c). We speculate that this is because TP53R273H (1–292) localised to the cytoplasm due to the lack of the nuclear localisation signal (NLS) of TP53R273H located in the 293–393 region (Fig. 4b). The reduction in the interaction between cytoplasmic TP53R273H (1–292) and BCAR1 further supports our earlier findings that the interaction takes place mainly in the nucleus (Fig. 1h, j). Consistent with these results, PLA revealed that the cells co-expressing TP53R273H (1–292) and MYC-BCAR1 have low fluorescence signals in the cytoplasm similar to the low signals found in the cytoplasm of some cells co-expressing TP53R273H full-length and MYC-BCAR1 (Fig. 1h and Supplementary Fig. S7). Taken together, these results suggest that BCAR1 binds to the 102–292 region of TP53R273H predominantly in the nucleus.
a Schematic of full-length and deletion mutants of TP53R273H-FLAG constructs and their binding status to BCAR1. Transactivation domain (TAD); DNA-binding domain (DBD); Nuclear localisation domain (NLS). b Localisation of full-length or deletion mutants of TP53R273H-FLAG. Fluorescence images of nuclei (blue) or TP53R273H-FLAG (red) are shown. H1299 cells were transfected with the indicated plasmids. The nuclei were stained with DAPI. Scale bar represents 20 μm. c Co-immunoprecipitation of endogenous BCAR1 with full-length or deletion mutants of TP53R273H-FLAG. Lysates of H1299 cells expressing the indicated constructs were immunoprecipitated with anti-FLAG antibody and analysed by Western blot. A FLAG tag-containing plasmid was used as a negative control. d Localisation of full-length or deletion mutants of TP53R273H-FLAG. Fluorescence images of nuclei (blue) or TP53R273H-FLAG (red) are shown. H1299 cells were transfected with the indicated plasmids. The nuclei were stained with DAPI. Scale bar represents 20 μm. e Co-immunoprecipitation of deletion mutants of TP53R273H-FLAG with exogenous MYC-BCAR1. Lysates of H1299 cells expressing the indicated constructs were immunoprecipitated with anti-MYC antibody and analysed by Western blot. A FLAG or MYC tag-containing plasmid was used as a negative control.
To further narrow down the binding region within 102–292 of TP53R273H, we generated TP53R273HΔ102–207 and TP53R273H Δ208–292 deletion mutants. Immunofluorescence staining confirmed that both TP53R273HΔ102–207 and TP53R273HΔ208–292 localised in the nucleus (Fig. 4d). We performed co-immunoprecipitation assays and found that MYC-BCAR1 bound to TP53R273H full-length and TP53R273HΔ208–292, but not TP53R273HΔ102–207 (Fig. 4e). These data suggest that BCAR1 binds to TP53R273H within the region corresponding to amino acids 102–207.
Disruption of the TP53R273H−BCAR1 interaction reduces invasion
To test whether the interaction between TP53R273H and BCAR1 is essential for cancer cell invasion, we evaluated the invasiveness of MDA-MB-468 TP53–/– cells expressing TP53R273H full-length or deletion mutants by transwell invasion assay. TP53R273H knockout MDA-MB-468 human breast cancer cells were generated using the CRISPR/Cas9 system (Fig. 5a), and exogenous TP53R273H full-length or deletion mutants were introduced by transient transfection (Fig. 5b). Consistent with the reported invasive phenotype of TP53R273H,19,20,36,47 TP53R273H full-length promoted transwell invasion compared to the empty vector control (Fig. 5c, d). As we expected, BCAR1 binding-deficient-TP53R273H (1–292) that does not localise in the nucleus did not promote transwell invasion as strongly as TP53R273H full-length (Fig. 5c, d). However, TP53R273H (102–393), which binds to BCAR1, also did not promote transwell invasion to a similar extent as TP53R273H full-length (Fig. 5c, d). This could be due to the lack of the transactivation domain of TP53R273H, which is essential for some of the mutant TP53 gain-of-function effects.18,55,56,57 Therefore, we decided to use the deletion mutants that retain the transactivation domain for further assessment (Fig. 5e). TP53R273HΔ208–292, which binds to BCAR1, promoted transwell invasion to a similar extent as TP53R273H full-length (Fig. 5f, g). By contrast, TP53R273HΔ102–207, which does not bind to BCAR1, did not promote transwell invasion (Fig. 5f, g). Together, these results suggest that the interaction between TP53R273H and BCAR1 in the nucleus (in the presence of an intact transactivation domain of TP53R273H) is responsible for promoting cancer cell invasion (Fig. 5h).
a Lysates of MDA-MB-468 (MDA468) CRISPR control or knockout cells were analysed by Western blot. b Lysates of MDA468 TP53–/– cells expressing the indicated constructs were analysed by Western blot. c Bright-field micrographs of transwell invasion assay using MDA468 TP53–/– cells expressing the indicated plasmids. Representative images of the transwells are shown. Scale bar represents 200 μm. d Quantification of the invading area. The images obtained in (c) were used for calculation. Bars represent the mean area of cells invading Matrigel relative to full-length (the second bar, set as 100%) ± SD from three independent experiments (n = 3). *P < 0.05. **P value < 0.01. Statistical analysis of data was done by unpaired Student’s two-sided t test. e Lysates of MDA468 TP53–/– cells expressing the indicated constructs were analysed by Western blot. f Bright-field micrographs of transwell invasion assay using MDA468 TP53–/– cells expressing the indicated plasmids. Representative images of the transwells are shown. Scale bar represents 200 μm. g Quantification of the invading area. The images obtained in (f) were used for calculation. Bars represent the mean area of cells invading Matrigel relative to full-length (the second bar, set as 100%) ± SD from three independent experiments (n = 3). **P value < 0.01. NS = not significant. Statistical analysis of data was done by unpaired Student’s two-sided t test. h Schematic model of the TP53R273H–BCAR1 complex-mediated cancer cell invasion. SRC family kinases (SFKs) promote BCAR1 phosphorylation and nuclear translocation. Full-length TP53R273H, but not the BCAR1 binding-deficient mutant (TP53R273HΔ102–207), enhances cancer cell invasion by binding to BCAR1 in the nucleus. P stands for phosphorylation.
Higher BCAR1 expression with TP53 mutations confers poorer overall survival in cancer patients
We next examined whether there is a positive correlation between TP53R273H and BCAR1 expression. The depletion of TP53R273H expression did not reduce the BCAR1 protein amounts in U251MG (Fig. 1e), MDA-MB-468 (Fig. 1f), and A431 cells (Supplementary Fig. S3). Conversely, overexpression of TP53R273H did not increase the endogenous BCAR1 protein amounts in H1299 cells (Fig. 4c). Furthermore, there was no clear positive correlation in the expression of TP53R273H and BCAR1 between TP53-null H1299 cancer cells and multiple TP53R273H-harbouring cancer cells (Supplementary Fig. S8). Likewise, the depletion of BCAR1 did not reduce TP53R273H expression in U251MG (Fig. 2a) and A431 cells (Supplementary Fig. S3). Together, these results indicate that there is no positive correlation between TP53R273H and BCAR1 expression.
As higher BCAR1 expression is associated with poorer prognosis,37,39,45,58 we next examined whether the TP53R273H–BCAR1 binding is affected by the amount of BCAR1 protein expression. We performed co-immunoprecipitation assays using the cells expressing the increasing amounts of BCAR1. We demonstrated that more TP53R273H was co-immunoprecipitated with the increasing amount of MYC-BCAR1 (Fig. 6a). This result led us to speculate that higher expression of BCAR1 in the presence of TP53R273H enhances the TP53R273H–BCAR1 protein binding, thereby adversely affecting the survival outcome of cancer patients. To extend our speculation to other hotspot mutants of TP53, we also showed that BCAR1 interacted with TP53R248W and TP53R175H to a similar extent as TP53R273H (Fig. 6b). This result indicates that BCAR1 can bind to other mutant TP53, and its binding is not restricted to TP53R273H specifically. We then retrieved the clinical data from cBioportal database for breast cancer (METABRIC dataset) and for lung adenocarcinoma (TCGA PanCancer Atlas dataset) and plotted the Kaplan−Meier curves for each TP53 status (wild type or mutant group) (Fig. 6c). All the TP53 mutations, including TP53R273H, were classified into the mutant group. We further divided each group into two sub-groups based on whether it was above or below the median of BCAR1 expression. The Kaplan−Meier curves showed that, among the patients with TP53 mutations, those with high BCAR1 expression had significantly shorter overall survival than those with low BCAR1 expression. In wild-type TP53 patients, on the other hand, there was no significant difference in overall survival between high and low BCAR1 expression. These data suggest that the TP53R273H and BCAR1 may work together to adversely affect overall survival in cancer patients.
a Co-Immunoprecipitation of exogenous TP53R273H-FLAG with MYC-BCAR1. H1299 cells were transfected with TP53R273H-FLAG together with the increasing amounts of MYC-BCAR1 plasmids. Lysates were immunoprecipitated with anti-MYC antibody and analysed by Western blot. An MYC tag-containing plasmid was used as a negative control. b Co-immunoprecipitation of exogenous FLAG-tagged various mutant TP53 with MYC-BCAR1. Lysates of H1299 cells transfected with the indicated plasmids were immunoprecipitated with anti-MYC antibody and analysed by Western blot. An MYC tag-containing plasmid was used as a negative control. c Kaplan−Meier plots using a dataset from METABRIC for breast cancer and a dataset from TCGA PanCancer Atlas for lung adenocarcinoma. The datasets were stratified according to the TP53 status (wild type or mutant) and BCAR1 expression levels; higher (high) or lower (low) than the median. Censored cases are depicted.
Discussion
Mutant TP53 interacts with multiple proteins to exert gain-of-function properties for tumour progression. We have discovered a novel interaction between TP53R273H and BCAR1 through the DNA-binding domain of TP53R273H and propose the importance of this interaction in promoting cancer cell invasion.
Many studies have shown that mutant forms of TP53 bind to several transcription factors, including SP1, ETS1, ETS2, VDR, SREBP-2, and NF-Y, altering expression of their target genes to produce gain-of-function properties.8,21,23 We showed that TP53R273H interacts with BCAR1 in the nucleus and that the TP53 transactivation domain and the nuclear localisation of both TP53R273H and BCAR1 are required to promote cancer cell invasion. These results suggest that the TP53R273H–BCAR1 complex may contribute to transcriptional regulation. However, BCAR1 is not a transcription factor, and it does not have any DNA-binding domain or transactivation domain. Therefore, BCAR1 may act as a bridge protein to recruit TP53R273H to certain transcription factors. For example, TOPBP1 is required for the interaction between TP53R273C and the transcription factor NF-Y to enhance NF-Y target gene expression.59 BCAR1 was reported to interact with zinc finger protein ZNF384, which is a transcription factor that regulates extracellular matrix remodelling.37,45,48,60,61 However, we could not detect ZNF384 in the proteins pulled down by TP53R273H in our mass spectrometry screen, although we isolated several other transcription factors including members of the zinc finger protein family. Further work is required to uncover the transcription factors involved in the TP53R273H−BCAR1 complex-mediated transcriptional regulation.
Most TP53 mutations are missense mutations that result in amino acid substitutions within the DNA-binding domain of TP53.8 These mutations are widely thought to result in loss of TP53 transcriptional function due to loss of its DNA-binding ability. Although some forms of TP53 with a mutated DNA-binding domain can still bind to TP53-responsive elements derived from CDKN1A, BAX and IGFBP3 promoters,62 TP53R273H used in our study does not directly bind to the responsive elements of wild-type TP53.63 However, we cannot completely eliminate the possibility that the TP53R273H−BCAR1 complexes directly bind to promoters since it has been shown that mutant p53 does bind to certain DNA structural motifs.8 Further studies are needed to address whether the TP53R273H−BCAR1 complex itself can access promoter regions of pro-invasive genes to enhance the invasiveness phenotype.
Mutant TP53 is reported to enhance cancer cell invasion by promoting RAB11FIP1/Rab coupling protein (RCP)-mediated integrin recycling via the endosomal pathway.19 It has not been examined whether BCAR1 is involved in this process. BCAR1 is an adaptor protein that is often recruited to integrins as part of the focal adhesion assembly, where BCAR1 can be phosphorylated by SFKs.37 A recent study has demonstrated that SFKs co-localise with endocytosed integrin and that treatment with SFKs inhibitors interrupts the integrin recycling process.64 Although we do not exclude the possibility that cytoplasmic BCAR1 and SFKs are involved in mutant TP53-dependent integrin recycling as an essential component for focal adhesion, we have found a previously unappreciated BCAR1 function in the nucleus.
In contrast to mutant TP53, wild-type TP53 suppresses SRC-induced invadopodia, actin-rich structures that promote cancer cell invasion, through the induction of tumour suppressor PTEN (phosphatase and tensin homologue).65 PTEN inactivates SRC kinase by dephosphorylating its tyrosine 418 residue.65 It is, therefore, possible that the mutation in TP53 results in the reduction of PTEN expression, leading to SRC activation, which promotes BCAR1 phosphorylation, nuclear import of BCAR1, and TP53R273H−BCAR1 complex formation, thereby increasing the invasive phenotype.
Recent evidence suggests that not all hotspot mutants of TP53, including those with mutations at residues R175, R248, and R273, have the same gain-of-function phenotypes, and each underlying mechanism may differ.13,66 Although we showed that TP53R175H, TP53R248W, and TP53R273H interact with BCAR1, we have not determined whether TP53R175H and TP53R248W bind to BCAR1 through the same 102–207 region of TP53R273H. Since TP53R175H is structurally different from TP53R273H and TP53R248W,67 TP53R175H might bind to BCAR1 differently. While the patients with mutant TP53 and high BCAR1 expression have shorter overall survival than the patients with low BCAR1 expression, we are unclear how much the mutant TP53−BCAR1 complex-mediated cancer cell invasion affects the reduction in overall survival. Besides the cancer cell invasion, different TP53 mutants may exert a variety of gain-of-function including drug resistance.24,68 Drug resistance is an important factor that influences the overall survival of patients.69 BCAR1 is also known to promote drug resistance.37,45 Therefore, the underlying mechanisms of how these interactions affect patient survival remain to be investigated.
High BCAR1 expression was not correlated with poorer overall survival among breast cancer and lung adenocarcinoma patients with wild-type TP53. This suggests, at least in part, high BCAR1 expression itself is insufficient to drive breast cancer progression without the cooperation of mutant TP53. However, BCAR1 also has other binding partners which contribute to tumour progression.37,70 These binding proteins can be differentially expressed in different cancers and may affect patient survival to various degrees. Therefore, depending on the types of cancers, high BCAR1 expression could adversely affect patient survival even without mutant TP53. That might be one of the reasons why we observed the different trends in survival curves between breast cancer and lung adenocarcinoma with wild-type TP53 and high/low BCAR1 expression, although there was no statistical significance in both cancer patients. There may be many other factors that affect the survival curves, such as various tumour-suppressive activities of wild-type TP53 in each tissue,1 which warrant further investigation.
Our results showed that TP53R273H did not promote nuclear translocation of BCAR1. Instead, the TP53R273H−BCAR1 complex formation is dependent on SFKs-mediated BCAR1 nuclear localisation. This may be a regulatory mechanism for cancer cells to avoid constitutive and excessive induction of pro-invasive genes by the TP53R273H−BCAR1 complex. The TP53R273H−BCAR1 complex may induce gene expression only when SFKs are activated by upstream signals triggered by either binding between the extracellular matrix and integrin receptors or binding between growth factors and cell-surface receptors such as receptor tyrosine kinases.37 The development of drugs that block BCAR1 phosphorylation and translocation into the nucleus, interfere with the TP53R273H−BCAR1 interaction, or degrade TP53R273H, could be effective strategies to inhibit the TP53R273H−BCAR1-mediated invasion in cancer. Supporting this idea, drugs designed to degrade mutant TP53, including 17-AAG (HSP90 inhibitor) and SAHA (histone deacetylase inhibitor), effectively reduce the invasiveness of grafted tumours of MDA-MB-468 cells carrying the TP53R273H mutation in mouse xenograft models.71 Taken together, the results presented here can potentially be used to explore novel therapeutic options for cancer patients carrying mutant TP53.
References
Bieging, K. T., Mello, S. S. & Attardi, L. D. Unravelling mechanisms of p53-mediated tumour suppression. Nat. Rev. Cancer 14, 359–370 (2014).
Brown, C. J., Lain, S., Verma, C. S., Fersht, A. R. & Lane, D. P. Awakening guardian angels: drugging the p53 pathway. Nat. Rev. Cancer 9, 862–873 (2009).
Guo, A. K., Hou, Y., Hirata, H., Yamauchi, S., Yip, A. K., Chiam, K. H. et al. Loss of p53 enhances NF‐κB‐dependent Lamellipodia formation. J. Cell. Physiol. 229, 696–704 (2014).
Kastenhuber, E. R. & Lowe, S. W. Putting p53 in context. Cell 170, 1062–1078 (2017).
Levine, A. J. p53: 800 million years of evolution and 40 years of discovery. Nat. Rev. Cancer 20, 471–480 (2020).
Vousden, K. H. & Prives, C. Blinded by the light: the growing complexity of p53. Cell 137, 413–431 (2009).
Yamauchi, S., Hou, Y. Y., Guo, A. K., Hirata, H., Nakajima, W., Yip, A. K. et al. p53-mediated activation of the mitochondrial protease HtrA2/Omi prevents cell invasion. J. Cell Biol. 204, 1191–1207 (2014).
Freed-Pastor, W. A. & Prives, C. Mutant p53: one name, many proteins. Genes Dev. 26, 1268–1286 (2012).
Lang, G. A., Iwakuma, T., Suh, Y. A., Liu, G., Rao, V. A., Parant, J. M. et al. Gain of function of a p53 hot spot mutation in a mouse model of Li-Fraumeni syndrome. Cell 119, 861–872 (2004).
Olive, K. P., Tuveson, D. A., Ruhe, Z. C., Yin, B., Willis, N. A., Bronson, R. T. et al. Mutant p53 gain of function in two mouse models of Li-Fraumeni syndrome. Cell 119, 847–860 (2004).
Rivlin, N., Brosh, R., Oren, M. & Rotter, V. Mutations in the p53 tumor suppressor gene: important milestones at the various steps of tumorigenesis. Genes Cancer 2, 466–474 (2011).
Yamamoto, S. & Iwakuma, T. Regulators of oncogenic mutant TP53 gain of function. Cancers (Basel) 11, 4 (2019).
Kim, M. P. & Lozano, G. Mutant p53 partners in crime. Cell Death Differ. 25, 161–168 (2018).
Kogan-Sakin, I., Tabach, Y., Buganim, Y., Molchadsky, A., Solomon, H., Madar, S. et al. Mutant p53(R175H) upregulates Twist1 expression and promotes epithelial-mesenchymal transition in immortalized prostate cells. Cell Death Differ. 18, 271–281 (2011).
Zhang, Y., Yan, W. & Chen, X. Mutant p53 disrupts MCF-10A cell polarity in three-dimensional culture via epithelial-to-mesenchymal transitions. J. Biol. Chem. 286, 16218–16228 (2011).
Araki, K., Ebata, T., Guo, A. K., Tobiume, K., Wolf, S. J. & Kawauchi, K. p53 regulates cytoskeleton remodeling to suppress tumor progression. Cell. Mol. Life Sci. 72, 4077–4094 (2015).
Arjonen, A., Kaukonen, R., Mattila, E., Rouhi, P., Hognas, G., Sihto, H. et al. Mutant p53-associated myosin-X upregulation promotes breast cancer invasion and metastasis. J. Clin. Invest. 124, 1069–1082 (2014).
Freed-Pastor, W. A., Mizuno, H., Zhao, X., Langerod, A., Moon, S. H., Rodriguez-Barrueco, R. et al. Mutant p53 disrupts mammary tissue architecture via the mevalonate pathway. Cell 148, 244–258 (2012).
Muller, P. A., Caswell, P. T., Doyle, B., Iwanicki, M. P., Tan, E. H., Karim, S. et al. Mutant p53 drives invasion by promoting integrin recycling. Cell 139, 1327–1341 (2009).
Muller, P. A., Trinidad, A. G., Timpson, P., Morton, J. P., Zanivan, S., van den Berghe, P. V. et al. Mutant p53 enhances MET trafficking and signalling to drive cell scattering and invasion. Oncogene 32, 1252–1265 (2013).
Aschauer, L. & Muller, P. A. Novel targets and interaction partners of mutant p53 gain-of-function. Biochem. Soc. Trans. 44, 460–466 (2016).
Kim, M. P., Zhang, Y. & Lozano, G. Mutant p53: multiple mechanisms define biologic activity in cancer. Front. Oncol. 5, 249 (2015).
Muller, P. A. & Vousden, K. H. p53 mutations in cancer. Nat. Cell Biol. 15, 2–8 (2013).
Muller, P. A. & Vousden, K. H. Mutant p53 in cancer: new functions and therapeutic opportunities. Cancer Cell 25, 304–317 (2014).
Lee, M. K., Teoh, W. W., Phang, B. H., Tong, W. M., Wang, Z. Q. & Sabapathy, K. Cell-type, dose, and mutation-type specificity dictate mutant p53 functions in vivo. Cancer Cell 22, 751–764 (2012).
Olszewski, M. B., Pruszko, M., Snaar-Jagalska, E., Zylicz, A. & Zylicz, M. Diverse and cancer type-specific roles of the p53 R248Q gain-of-function mutation in cancer migration and invasiveness. Int. J. Oncol. 54, 1168–1182 (2019).
Yeo, S. Y., Itahana, Y., Guo, A. K., Han, R., Iwamoto, K., Nguyen, H. T. et al. Transglutaminase 2 contributes to a TP53-induced autophagy program to prevent oncogenic transformation. Elife 5, e07101 (2016).
Jordan, M., Schallhorn, A. & Wurm, F. M. Transfecting mammalian cells: optimization of critical parameters affecting calcium-phosphate precipitate formation. Nucleic Acids Res. 24, 596–601 (1996).
Fenyö, D. & Beavis, R. C. A method for assessing the statistical significance of mass spectrometry-based protein identifications using general scoring schemes. Anal. Chem. 75, 768–774 (2003).
Mellacheruvu, D., Wright, Z., Couzens, A. L., Lambert, J.-P., St-Denis, N. A., Li, T. et al. The CRAPome: a contaminant repository for affinity purification–mass spectrometry data. Nat. Methods 10, 730 (2013).
Krämer, A., Green, J., Pollard, J. Jr & Tugendreich, S. Causal analysis approaches in ingenuity pathway analysis. Bioinformatics 30, 523–530 (2014).
Cerami, E., Gao, J., Dogrusoz, U., Gross, B. E., Sumer, S. O., Aksoy, B. A. et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2, 401–404 (2012).
Gao, J., Aksoy, B. A., Dogrusoz, U., Dresdner, G., Gross, B., Sumer, S. O. et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 6, pl1–pl1 (2013).
Leroy, B., Girard, L., Hollestelle, A., Minna, J. D., Gazdar, A. F. & Soussi, T. Analysis of TP 53 mutation status in human cancer cell lines: a reassessment. Hum. Mutat. 35, 756–765 (2014).
Lukashchuk, N. & Vousden, K. H. Ubiquitination and degradation of mutant p53. Mol. Cell. Biol. 27, 8284–8295 (2007).
Coffill, C. R., Muller, P. A., Oh, H. K., Neo, S. P., Hogue, K. A., Cheok, C. F. et al. Mutant p53 interactome identifies nardilysin as a p53R273H-specific binding partner that promotes invasion. EMBO Rep. 13, 638–644 (2012).
Cabodi, S., del Pilar Camacho-Leal, M., Di Stefano, P. & Defilippi, P. Integrin signalling adaptors: not only figurants in the cancer story. Nat. Rev. Cancer 10, 858–870 (2010).
Barrett, A., Pellet-Many, C., Zachary, I. C., Evans, I. M. & Frankel, P. p130Cas: a key signalling node in health and disease. Cell. Signal. 25, 766–777 (2013).
Dorssers, L. C., Grebenchtchikov, N., Brinkman, A., Look, M. P., van Broekhoven, S. P., de Jong, D. et al. The prognostic value of BCAR1 in patients with primary breast cancer. Clin. Cancer Res. 10, 6194–6202 (2004).
Tornillo, G., Bisaro, B., Camacho-Leal Mdel, P., Galie, M., Provero, P., Di Stefano, P. et al. p130Cas promotes invasiveness of three-dimensional ErbB2-transformed mammary acinar structures by enhanced activation of mTOR/p70S6K and Rac1. Eur. J. Cell Biol. 90, 237–248 (2011).
Dai, Y., Qi, L., Zhang, X., Li, Y., Chen, M. & Zu, X. CrkI and p130(Cas) complex regulates the migration and invasion of prostate cancer cells. Cell Biochem. Funct. 29, 625–629 (2011).
Nick, A. M., Stone, R. L., Armaiz-Pena, G., Ozpolat, B., Tekedereli, I., Graybill, W. S. et al. Silencing of p130cas in ovarian carcinoma: a novel mechanism for tumor cell death. J. Natl Cancer Inst. 103, 1596–1612 (2011).
Guo, C., Liu, Q. G., Yang, W., Zhang, Z. L. & Yao, Y. M. Relation among p130Cas, E-cadherin and beta-catenin expression, clinicopathologic significance and prognosis in human hepatocellular carcinoma. Hepatobiliary Pancreat. Dis. Int. 7, 490–496 (2008).
Pincini, A., Tornillo, G., Orso, F., Sciortino, M., Bisaro, B., Leal Mdel, P. et al. Identification of p130Cas/ErbB2-dependent invasive signatures in transformed mammary epithelial cells. Cell Cycle 12, 2409–2422 (2013).
Tornillo, G., Defilippi, P. & Cabodi, S. Cas proteins: dodgy scaffolding in breast cancer. Breast Cancer Res. 16, 443 (2014).
Wang, Y. & McNiven, M. A. Invasive matrix degradation at focal adhesions occurs via protease recruitment by a FAK-p130Cas complex. J. Cell Biol. 196, 375–385 (2012).
Ahn, J.-H., Kim, T. J., Lee, J. H. & Choi, J.-H. Mutant p53 stimulates cell invasion through an interaction with Rad21 in human ovarian cancer cells. Sci. Rep. 7, 1–11 (2017).
Cunningham-Edmondson, A. C. & Hanks, S. K. p130Cas substrate domain signaling promotes migration, invasion, and survival of estrogen receptor-negative breast cancer cells. Breast Cancer 1, 39–52 (2009).
Recchia, I., Rucci, N., Festuccia, C., Bologna, M., MacKay, A. R., Migliaccio, S. et al. Pyrrolopyrimidine c-Src inhibitors reduce growth, adhesion, motility and invasion of prostate cancer cells in vitro. Eur. J. Cancer 39, 1927–1935 (2003).
Charoonpatrapong-Panyayong, K., Shah, R., Yang, J., Alvarez, M., Pavalko, F. M., Gerard-O’Riley, R. et al. Nmp4/CIZ contributes to fluid shear stress induced MMP-13 gene induction in osteoblasts. J. Cell. Biochem. 102, 1202–1213 (2007).
Chen, C. H., Ho, Y. C., Ho, H. H., Chang, I. C., Kirsch, K. H., Chuang, Y. J. et al. Cysteine-rich protein 2 alters p130Cas localization and inhibits vascular smooth muscle cell migration. Cardiovasc. Res. 100, 461–471 (2013).
Kim, W., Seok Kang, Y., Soo Kim, J., Shin, N. Y., Hanks, S. K. & Song, W. K. The integrin-coupled signaling adaptor p130Cas suppresses Smad3 function in transforming growth factor-beta signaling. Mol. Biol. Cell 19, 2135–2146 (2008).
Sakai, R., Iwamatsu, A., Hirano, N., Ogawa, S., Tanaka, T., Mano, H. et al. A novel signaling molecule, p130, forms stable complexes in vivo with v-Crk and v-Src in a tyrosine phosphorylation-dependent manner. EMBO J. 13, 3748–3756 (1994).
Nam, J. S., Ino, Y., Sakamoto, M. & Hirohashi, S. Src family kinase inhibitor PP2 restores the E-cadherin/catenin cell adhesion system in human cancer cells and reduces cancer metastasis. Clin. Cancer Res. 8, 2430–2436 (2002).
Matas, D., Sigal, A., Stambolsky, P., Milyavsky, M., Weisz, L., Schwartz, D. et al. Integrity of the N-terminal transcription domain of p53 is required for mutant p53 interference with drug-induced apoptosis. EMBO J. 20, 4163–4172 (2001).
Scian, M. J., Stagliano, K. E., Anderson, M. A., Hassan, S., Bowman, M., Miles, M. F. et al. Tumor-derived p53 mutants induce NF-kappaB2 gene expression. Mol. Cell. Biol. 25, 10097–10110 (2005).
Kalo, E., Buganim, Y., Shapira, K. E., Besserglick, H., Goldfinger, N., Weisz, L. et al. Mutant p53 attenuates the SMAD-dependent transforming growth factor beta1 (TGF-beta1) signaling pathway by repressing the expression of TGF-beta receptor type II. Mol. Cell. Biol. 27, 8228–8242 (2007).
Deng, B., Sun, Z., Jason, W. & Yang, P. Increased BCAR1 predicts poor outcomes of non-small cell lung cancer in multiple-center patients. Ann. Surg. Oncol. 20, 701–708 (2013).
Liu, K., Ling, S. & Lin, W. C. TopBP1 mediates mutant p53 gain of function through NF-Y and p63/p73. Mol. Cell. Biol. 31, 4464–4481 (2011).
Janssen, H. & Marynen, P. Interaction partners for human ZNF384/CIZ/NMP4-zyxin as a mediator for p130CAS signaling? Exp. Cell Res. 312, 1194–1204 (2006).
Nakamoto, T., Yamagata, T., Sakai, R., Ogawa, S., Honda, H., Ueno, H. et al. CIZ, a zinc finger protein that interacts with p130(cas) and activates the expression of matrix metalloproteinases. Mol. Cell. Biol. 20, 1649–1658 (2000).
Ludwig, R. L., Bates, S. & Vousden, K. H. Differential activation of target cellular promoters by p53 mutants with impaired apoptotic function. Mol. Cell. Biol. 16, 4952–4960 (1996).
Zhu, J., Sammons, M. A., Donahue, G., Dou, Z., Vedadi, M., Getlik, M. et al. Gain-of-function p53 mutants co-opt chromatin pathways to drive cancer growth. Nature 525, 206–211 (2015).
Nader, G. P., Ezratty, E. J. & Gundersen, G. G. FAK, talin and PIPKIgamma regulate endocytosed integrin activation to polarize focal adhesion assembly. Nat. Cell Biol. 18, 491–503 (2016).
Mukhopadhyay, U. K., Mooney, P., Jia, L., Eves, R., Raptis, L. & Mak, A. S. Doubles game: Src-Stat3 versus p53-PTEN in cellular migration and invasion. Mol. Cell. Biol. 30, 4980–4995 (2010).
Sabapathy, K. & Lane, D. P. Therapeutic targeting of p53: all mutants are equal, but some mutants are more equal than others. Nat. Rev. Clin. Oncol. 15, 13–30 (2018).
Joerger, A. C. & Fersht, A. R. Structural biology of the tumor suppressor p53. Annu. Rev. Biochem. 77, 557–582 (2008).
Mantovani, F., Collavin, L. & Del Sal, G. Mutant p53 as a guardian of the cancer cell. Cell Death Differ. 26, 199–212 (2019).
Vasan, N., Baselga, J. & Hyman, D. M. A view on drug resistance in cancer. Nature 575, 299–309 (2019).
Leal, Md. P. C., Sciortino, M., Tornillo, G., Colombo, S., Defilippi, P. & Cabodi, S. p130Cas/BCAR1 scaffold protein in tissue homeostasis and pathogenesis. Gene 562, 1–7 (2015).
Alexandrova, E. M., Yallowitz, A. R., Li, D., Xu, S., Schulz, R., Proia, D. A. et al. Improving survival by exploiting tumour dependence on stabilized mutant p53 for treatment. Nature 523, 352–356 (2015).
Acknowledgements
We thank Dr. Yanping Zhang and Dr. Keiko Kawauchi for the CMV-TP53WT and CMV-c-SRCY530F plasmids, respectively. We thank Dr. Keiko Kawauchi and Ms. Kimberley Kang for helpful discussions regarding the manuscript. We thank Dr. Katrina Woolcock, Life Science Editors, and Dr. Eddy Saputra Leman for editorial assistance. We thank Ms. Nguyen Thi Mai Nhi and Mr. Tan Ter Yong for assisting with the project.
Author information
Authors and Affiliations
Contributions
A.K.G., Y.I., and H.Y.C. performed experiments. A.K.G., Y.I., and K.I. designed the experiments and analysed the results. V.P.S. and S.G. analysed the datasets from cBioportal and generated the Kaplan−Meier plots. A.K.G., Y.I., V.P.S., and K.I. wrote the manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
All cell lines were purchased from ATCC or CLS Cell Lines Service GmbH. Clinical data were obtained from publicly available cBioportal database (https://www.cbioportal.org/).
Data availability
The data and materials reported in this article are available by the corresponding author upon reasonable request. Clinical datasets are available from public database described in the “Methods” section.
Competing interests
The authors declare no competing interests.
Funding information
This work was supported by the Duke-NUS Medical School core grant (to K.I.), the Singapore Ministry of Health’s National Medical Research Council grant (NMRC/OFIRG/0006/2016 to K.I.), the Singapore Ministry of Education AcRF Tier 2 fund (MOE 2017-T2-1-081 to K.I.), and the Khoo Postdoctoral Fellowship Award (Duke-NUS-KPFA/2016/0009 to A.K.G.).
Additional information
Note This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution 4.0 International (CC BY 4.0).
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Guo, A.K., Itahana, Y., Seshachalam, V.P. et al. Mutant TP53 interacts with BCAR1 to contribute to cancer cell invasion. Br J Cancer 124, 299–312 (2021). https://doi.org/10.1038/s41416-020-01124-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41416-020-01124-9