Abstract
Background
The androgen receptor (AR) is discussed as a prognostic and/or predictive marker in breast cancer patients.
Methods
AR mRNA expression was analysed by RT-qPCR in breast cancer patients treated in the neoadjuvant TECHNO (n = 118, HER2-positive) and PREPARE trial (n = 321, HER2-positive and -negative). In addition, mRNA expression of the AR transcript variants 1 (AR1) and 2 (AR2) was measured.
Results
Regarding subtypes, high AR mRNA levels were frequent in HER2-positive (61.3%, 92/150) and luminal tumours (60.0%, 96/160) but almost absent in triple-negative tumours (4.3%, 3/69) (p < 0.0001). Overall, high AR mRNA levels were found to be associated with lower pathological complete remission (pCR) rates (OR 0.77 per unit, 95% CI 0.67–0.88, p = 0.0002) but also with better prognosis in terms of longer disease-free survival (DFS) (HR 0.57, 95% CI 0.39–0.85, p = 0.0054) and overall survival (OS) (HR 0.43, 95% CI, 0.26–0.71, p = 0.0011). In the PREPARE trial, a survival difference for patients with high and low AR1 mRNA levels could only be seen in the standard chemotherapy arm but not in the dose-dense treatment arm (OS: HR 0.41; 95% CI 0.22–0.74 vs. HR 1.05; 95% CI 0.52–2.13; p = 0.0459).
Conclusions
We provide evidence that AR mRNA predicts response to chemotherapy in breast cancer patients.
Similar content being viewed by others
Background
Androgen receptor (AR) is expressed in normal breast epithelial cells and in ~70–90% of invasive breast carcinomas.1 Alternative splicing of the AR gene results in multiple transcript variants encoding different isoforms. The AR consists of a full-length 110 K AR protein (AR1) and an 87 K N-terminally truncated AR isoform (AR2). The distribution of both AR isoforms varies in different human tissues2 and their role in breast cancer has not been studied in detail so far.
AR is frequently co-expressed with the oestrogen receptor (ER) and progesterone receptor,3 but is less frequently expressed in HER2-positive tumours.4,5 The emerging role of AR in breast cancer patients is due to results supporting the prognostic value of AR in both ER-positive and ER-negative tumours.6,7,8,9,10 As a relevant percentage of triple-negative breast cancers also express AR, it has been identified as a potential new therapeutic target in this subset of patients with limited therapeutic options.4 In contrast to smaller studies in which no effect of AR expression in HER2-positive breast cancer was observed, a current meta-analysis reported that higher AR mRNA expression levels were found to be associated with improved overall survival in both uni- and multivariate analyses also for women with HER2-enriched breast cancer.11
Although most publications discuss AR as a prognostic marker in breast cancer patients,6,9,12,13 some reports also found an association between AR and therapy response. First, AR was assumed to be a predictive marker for response to endocrine treatment in breast cancer patients.14,15 Then, AR was evaluated in ER-positive tumours and a prognostic role of AR could be basically seen in chemo-endocrine treated patients.7 Additionally, Park et al. described no effect of high AR expression levels on chemotherapy benefit in ER-positive patients, but concluded that patients with low AR expression could be ideal candidates for chemotherapy treatment.16 Our group has subsequently shown that although AR predicts response to adjuvant chemotherapy rather than to endocrine treatment, the worst response rates were observed in patients with low AR expression.17 According to data from the neoadjuvant Gepartrio trial, Loibl et al. also reported that low AR expression determined by immunohistochemistry was associated with shorter disease-free and overall survival in 673 patients receiving chemotherapy with TAC.18
Despite this published data, the biological role of the AR expression in breast cancer is still not clear. Up to now, no consequences for therapy decisions based on AR expression in breast cancer therapy could be drawn. Moreover, the potential role of the two isoforms in the context of current therapeutic strategies has not been defined.
In our previous study, we could show that low AR mRNA was a predictor for shorter survival in breast cancer patients receiving adjuvant chemotherapy.17 The aim of this study was to validate these findings in a defined therapeutic context and to investigate the androgen receptor (AR) and in addition its two isoforms (AR1 and AR2) in breast cancer patients receiving neoadjuvant chemotherapy in two trials: the TECHNO trial (HER2-positive patients) and the PREPARE trial (HER2-positive and -negative patients).
Methods
TECHNO trial
The TECHNO trial was a multicentre, prospective, open-label, phase II clinical trial investigating neoadjuvant chemotherapy plus trastuzumab in HER2-positive breast cancer patients.19 Patients (n = 217) received four 3-week cycles of epirubicin 90 mg/m2 and cyclophosphamide 600 mg/m2 followed by the combination of four 3-week cycles of paclitaxel 175 mg/m2 with trastuzumab 6 mg/kg every 3 weeks (8 mg/kg as loading dose) (EC → Pac + trastuzumab) followed by surgery. Trastuzumab was continued as single agent postoperatively to complete 1 year of treatment. The primary endpoint was pathological complete remission (pCR), defined as absence of invasive breast cancer in the breast and axillary lymph nodes in all surgically excised specimens. Eligible patients had confirmed HER2-overexpressing primary breast cancer defined as a 3+ staining intensity by immunohistochemistry (IHC) using the DAKO HerceptTest (DAKO, Glostrup, Denmark) or a 2 + staining intensity centrally confirmed for HER2 gene amplification by fluorescent in situ hybridisation (FISH; PathVision Abbott; Abbott Park, IL). Tumours were either ≥2 cm based on clinical or ultrasound assessment or were diagnosed clinically as inflammatory breast cancer.
PREPARE trial
The PREPARE trial was a multicentre, prospective, open-label, phase II clinical trial investigating a standard chemotherapy treatment consisting of neoadjuvant four 3 week cycles of epirubicin 90 mg/m(2) plus cyclophosphamide 600 mg/m(2) followed by four 3 week cycles of paclitaxel 175 mg/m(2) (EC → T), (n = 370), versus a dose-dense treatment arm with epirubicin 150 mg/m(2) followed by paclitaxel 225 mg/m(2) with pegfilgrastim support every two weeks followed by CMF (cyclophosphamide 500 mg/m(2), methotrexate 40 mg/m2, fluorouracil 600 mg/m(2)) on days 1 and 8 every 2 weeks (E(dd) → T(dd) → CMF), every 28 days (n = 363). Patients were randomly allocated to either simultaneous darbepoetin alfa (DA) (n = 356) or none (n = 377). Primary endpoint was pCR.20
Assessment of AR mRNA Expression by RT-qPCR
Tumour specimens were assessed by RT-qPCR as previously described.2 In short, for RNA extraction from formalin-fixed, paraffin-embedded tissue, a single 10-μm curl was processed according to a commercially available bead-based extraction method (Xtract kit; STRATIFYER Molecular Pathology GmbH, Cologne, Germany). RNA was eluted with 100 μl of elution buffer. DNA was digested, and RNA eluates were then stored at −80 °C until use. Primers for the AR transcript variant 1 (AR1; RefSeq NM_000044.3) and transcript variant 2 (AR2; RefSeq NM_001011645.2) were designed. The mRNA expression levels of the genes of interest (GOI), AR, AR1, AR2 and ESR1as well as the reference gene (REF) CALM2 were determined by RT-qPCR, which involves the reverse transcription of RNA and subsequent amplification of cDNA executed successively in a one-step reaction. Each patient sample or control was analysed in triplicate in a Siemens Versant PCR System (Siemens, DE) according to the following protocol: 5 min at 50 °C and 20 s at 95 °C followed by 40 cycles of 15 s at 95 °C and 60 s at 60 °C. Forty amplification cycles were applied, and the cycle threshold (CT) values of the four GOIs and the REF gene for each sample were estimated as the median of the triplicate measurements. These were then normalised against the median expression levels of the REF gene using the 40-ΔCT method to ensure that the gene expression obtained by the test corresponds to the normalised log2 mRNA expression levels:
Molecular subtype by RT-qPCR
In order to define molecular subtypes of the tumours, an additional RNA-based analysis with Mammatyper® was started. The MammaTyper® is a molecular in vitro diagnostic tool for the assessment of the expression levels of the four cancer biomarkers (HER2, ERα, PR and Ki-67) that are required for clinical management of breast cancer patients in clinical practice. We measured mRNA transcripts of the corresponding genes (ERBB2, ESR1, PGR and MKI67) with MammaTyper® in RNA samples retrieved from FFPE tissue as described before.2,21
Study cohort
Of the initial cohort of patients in both trials (Intention-to-treat (ITT)-population n = 950), 477 tumour samples could be analysed (50.2%). Statistical analyses were performed on those 439 patients for whom at least one gene expression (AR, AR1, AR2 or ESR1) was available after quality control. In order to define molecular subtypes of the tumours, an additional independent RNA-based analysis with Mammatyper® was performed as described above. Table 1 contains a more detailed overview. The MammaTyper classifies samples into six molecular subtypes, some of which had only very few patients in our analysis set. For meaningful statistical analysis, we combined some of the classes (luminal A and B to luminal, luminal HER2-enriched to HER2-positive).
There was no relevant imbalance between the whole study cohort and the subgroup of patients in our analysis regarding clinical and histopathological parameters (data not shown). 118 patients of the cohort (26.9%) participated in the TECHNO trial and 321 patients (73.1%) in the PREPARE trial. Median age of the patients was 48 years (range 25–67). Regarding tumour subtypes, 42.2% of patients (n = 160) had luminal, 39.6% (n = 150) HER2-positive and 18.2% (n = 69) triple-negative tumours. 47.4% of patients (n = 198) had node-positive disease, 15.7% of patients (n = 69) achieved a pCR. Detailed patient characteristics of the study cohort are shown in Table 2.
Statistical analysis
AR, AR1, AR2 and ESR1 were analysed as continuous variables and in addition were dichotomised using the median as cut-off as described before.22
Pathological complete remission (pCR) was defined as no evidence of invasive or non-invasive cells in tissue of breast and lymph nodes. Disease-free survival (DFS) was computed from the date of study inclusion to the date of first metastasis or recurrence. Overall survival (OS) was computed to the date of death.
Dichotomised biomarkers were compared to binary variables by Fishers exact tests, to multi-level categorial variables by Chi-square tests and to continuous variables by Wilcoxon tests. For the clinical endpoint pCR as dependent variable logistic regression models were constructed, endpoints DFS and OS were analysed by Cox regression models. Univariate and multivariate regression models adjusting for subtype (luminal vs HER2-positive vs triple-negative), tumour size (T1–2 vs T3-4), nodal status (N0 vs N +), grading (G1-2 vs G3) and age (continuous) were built. Survival curves were compared with the logrank test.
All tests were performed at a significance level of p = 0.05 (two-sided).
The analysis was performed according to the REporting recommendations for tumour MARKer prognostic studies (REMARK) criteria on reporting of biomarkers.23
Results
Given that the mRNA expression patterns of AR1 and AR2 were correlated in the nonparametric spearman correlation (rs: 0.6493, P < .0001) we report results exclusively for AR1. Furthermore, AR2 had no additional prognostic information.
Androgen receptor expression and clinical variables
A positive association between AR1 and oestrogen receptor status (immunohistochemistry) (p < 0.0001) and progesterone receptor status (p < 0.0001) (Table 3) was observed. We found an inverse association between AR1 and grading (p < 0.0001) and pathological complete remission (pCR) (p = 0.0670). Regarding subtypes, high AR1 expression was more frequent in HER2-positive (62.0%, 93/150) and luminal tumours (65.0%, 104/160) whereas high AR1 expression was almost absent in triple-negative tumours (4.3%, 3/69) (p < 0.0001). No association between AR1 mRNA levels and nodal status, tumour size or age could be found (Table 3).
Androgen receptor expression and pCR
In the entire cohort, higher AR1 expression levels showed a significant association with lower pCR rates (OR per unit 0.77, 95% CI 0.67–0.88, p = 0.0002) in univariate analysis while not significant in a multivariate model (Table 4). This association with pCR could not be seen in the TECHNO trial but in the PREPARE trial in which high AR1 expression levels were associated with lower pCR rates (OR 0.66, 95% CI 0.55–0.78, p < .0001). In accordance, a non-significant trend to lower pCR rates in patients with high AR1 expression was observed in the triple-negative subgroup (OR 0.74, 95% CI 0.49–1.12, p = 0.1546) compared with the HER2-positive subgroup (OR 1.40, 95% CI 0.95–2.06, p = 0.0920) (Table 4). High ESR1 expression was associated with lower pCR rates (OR 0.73, 95% CI 0.65–0.82, p < 0.0001) in univariate analysis, even in the subgroup of patients with luminal tumours (OR 0.60, 95% CI 0.40–0.91, p = 0.0149), but not in multivariate analysis (data not shown).
Androgen receptor expression and survival
To address the impact of AR mRNA expression levels on prognosis, we compared the risk of relapse among all patients, and within different breast cancer subtypes. Overall, high AR1 mRNA levels compared to low AR1 mRNA levels were found to be associated with better prognosis in terms of DFS (HR 0.76, 95% CI 0.56–1.04, p = 0.0860) and OS (HR 0.60, 95% CI 0.40–0.89, p = 0.0103) in univariate analysis and in multivariate analysis (DFS: HR 0.62, 95% CI 0.41–0.92, p = 0.0173; OS: HR 0.50, 95% CI, 0.30–0.83, p = 0.0080) (Tables 5 and 6). Within different subgroups, no significant association between AR1 mRNA and survival could be seen.
For other markers, only in luminal patients and only in univariate analysis, high ESR1 expression was associated with better DFS (HR 0.76, 95% CI 0.63–0.92, p = 0.0043; data not shown).
Androgen receptor expression and treatment
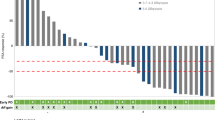
Next, we hypothesised that the worse prognosis of patients with low AR1 mRNA expression could be modified by treatment. In the PREPARE trial, a dose-dense treatment arm was compared with a standard treatment arm. Regarding overall survival, patients with high AR1 mRNA levels receiving the standard treatment had better survival compared to patients with low AR1 mRNA levels (HR 0.41 (95% CI 0.22–0.74) while for patients with dose-dense treatment the survival did not differ depending on the AR1 level (HR 1.05 (95% CI 0.52–2.13), interaction p = 0.0459, Fig. 1).
Discussion
Our data show that AR expression is a prognostic marker in chemotherapy-treated breast cancer patients. In our cohort, measuring the isoform AR1 could be of interest for the prediction of therapy response to dose-dense treatment.
In patients that did not receive any systemic treatment, we had already shown that the AR had no additional prognostic information.17 In contrast, in chemotherapy-treated patients, low AR mRNA expression was associated with shorter event-free survival.
Here, we could show that the group of patients with low AR1 mRNA levels also had worse survival rates compared with patients with high AR1 mRNA levels. But the worse prognosis might be overcome by dose-dense chemotherapy treatment in those patients with low AR1 mRNA levels. In line with data from bladder cancer, we could show that AR1 mRNA expression is the isoform, which gives further prognostic information in breast cancer patients.22
A potential drawback of our study is its retrospective nature and assessment of a biomarker that was not prospectively defined. We had AR data in 44.0% of patients (n = 418/950) of the whole study cohort. In addition, AR could be determined only on RNA level since additional paraffin-embedded tissue was not available from most patients. Therefore, a correlation between immunohistochemical analysis and mRNA levels could not be examined. Regarding AR positivity, immunohistochemical analysis reveals AR positivity in 65–80% of breast cancer patients.9 However, for AR, no uniform staining and scoring system is established and quantification is difficult.
In addition, in prostate cancer, no correlation between staining intensity and mRNA expression of the AR protein could be observed.24 Therefore, quantitative measurement by mRNA as applied in our study could be a valid alternative. Androgen receptor-targeted treatments for breast cancer are in development and have shown promising preliminary results.25,26,27,28 One direction in preclinical and clinical research is the use of AR antagonists in triple-negative breast cancer25,26,28,29,30 but up to now, no reliable biomarker has been identified to predict response. It is of interest that in our study very few patients with triple-negative breast cancer had high AR1 mRNA levels whereas in immunohistochemical studies AR-positivity rates of 12–32% in triple-negative tumours were described. This discrepancy might be one possible explanation why AR evaluation by immunohistochemistry does not predict response to AR-directed therapies.
In addition to the prognostic role of AR in breast cancer patients, there is growing evidence that AR predicts response to chemotherapy treatment rather than to endocrine treatment with the worst response rates in patients with low AR expression. According to data from the neoadjuvant Gepartrio trial, Loibl et al. reported that low AR expression determined by immunohistochemistry was associated with shorter disease-free and overall survival in 673 patients receiving chemotherapy with TAC.18 In this trial, AR-positive tumours had lower pCR rates (12.8 vs. 25.4%) and AR expression added independent predictive information for pCR.18 Within the non-pCR subgroup, AR positivity selected a group with a significant better survival but not within the pCR group.18 Interestingly, also in our cohort, patients with low AR1 mRNA levels had a higher probability to achieve a pCR than those with high AR1 mRNA levels. Despite this fact, patients with low AR1 mRNA levels had worse survival rates. However, we have to admit that the pCR rate of 15.7% in our study cohort is lower compared to pCR rates reached in recent neoadjuvant trials. This difference might be explained in part by the high number of patients with luminal tumours in our cohort who have lower pCR rates. In addition, high AR1 mRNA levels were rare in patients with triple-negative breast cancer who, on the other hand, have higher pCR rates.
To our knowledge, we are the first to document that the worse prognosis of patients with low AR1 mRNA levels might be compensated to a certain degree by dose-dense chemotherapy treatment. Survival differences could be mainly seen regarding overall survival rates whereas no significant interaction could be seen for AR1 mRNA levels, chemotherapy treatment and disease-free survival.
There is increasing evidence that dose-dense chemotherapy is beneficial in breast cancer patients.31,32 Currently node-positivity often guides the decision towards dose-dense treatment. We could show that AR1 mRNA may add information in the decision for or against dose-dense chemotherapy treatment, which would have to be validated in further studies.
In conclusion, we provide evidence that there seems to be an interaction between AR expression and chemotherapy-responsiveness in breast cancer patients. Determination of the isoform AR 1 might deliver additional information for the prediction of therapy response to dose-dense chemotherapy treatment.
References
Gonzalez, L. O., Corte, M. D., Vazquez, J., Junquera, S., Sanchez, R., Alvarez, A. C. et al. Androgen receptor expresion in breast cancer: relationship with clinicopathological characteristics of the tumors, prognosis, and expression of metalloproteases and their inhibitors. BMC Cancer 8, 149 (2008).
Breyer, J., Wirtz, R. M., Laible, M., Schlombs, K., Erben, P., Kriegmair, M. C. et al. ESR1, ERBB2, and Ki67 mRNA expression predicts stage and grade of non-muscle-invasive bladder carcinoma (NMIBC). Virchows Arch. 469, 547–552 (2016).
Kuenen-Boumeester, V., Van der Kwast, T. H., Claassen, C. C., Look, M. P., Liem, G. S., Klijn, J. G. et al. The clinical significance of androgen receptors in breast cancer and their relation to histological and cell biological parameters. Eur. J. Cancer 32A, 1560–1565 (1996).
Ogawa, Y., Hai, E., Matsumoto, K., Ikeda, K., Tokunaga, S., Nagahara, H. et al. Androgen receptor expression in breast cancer: relationship with clinicopathological factors and biomarkers. Int. J. Clin. Oncol. 13, 431–435 (2008).
Collins, L. C., Cole, K. S., Marotti, J. D., Hu, R., Schnitt, S. J. & Tamimi, R. M. Androgen receptor expression in breast cancer in relation to molecular phenotype: results from the Nurses’ Health Study. Mod. Pathol. 24, 924–931 (2011).
Park, S., Koo, J. S., Kim, M. S., Park, H. S., Lee, J. S., Kim, S. I. et al. Androgen receptor expression is significantly associated with better outcomes in estrogen receptor-positive breast cancers. Ann. Oncol. 22, 1755–1762 (2011).
Castellano, I., Allia, E., Accortanzo, V., Vandone, A. M., Chiusa, L., Arisio, R. et al. Androgen receptor expression is a significant prognostic factor in estrogen receptor positive breast cancers. Breast Cancer Res. Treat. 124, 607–617 (2010).
Agoff, S. N., Swanson, P. E., Linden, H., Hawes, S. E., Lawton, T. J. Androgen receptor expression in estrogen receptor-negative breast cancer. Immunohistochemical, clinical, prognostic association. Am. J. Clin. Pathol. 120, 725–731 (2003).
Hu, R., Dawood, S., Holmes, M. D., Collins, L. C., Schnitt, S. J., Cole, K. et al. Androgen receptor expression and breast cancer survival in postmenopausal women. Clin. Cancer Res. 17, 1867–1874 (2011).
Rakha, E. A., El-Sayed, M. E., Green, A. R., Lee, A. H., Robertson, J. F. & Ellis, I. O. Prognostic markers in triple-negative breast cancer. Cancer 109, 25–32 (2007).
Bozovic-Spasojevic, I., Zardavas, D., Brohee, S., Ameye, L., Fumagalli, D., Ades, F. et al. The prognostic role of androgen receptor in patients with early-stage breast cancer: a meta-analysis of clinical and gene expression data. Clin. Cancer Res. 23, 2702–2712 (2017).
Yu, Q., Niu, Y., Liu, N., Zhang, J. Z., Liu, T. J., Zhang, R. J. et al. Expression of androgen receptor in breast cancer and its significance as a prognostic factor. Ann. Oncol. 22, 1288–1294 (2011).
Park, S., Koh, E., Koo, J. S., Kim, S. I., Park, B. W. & Kim, K. S. Lack of both androgen receptor and forkhead box A1 (FOXA1) expression is a poor prognostic factor in estrogen receptor-positive breast cancers. Oncotarget 8, 82940–82955 (2017).
Lundin, K. B., Henningson, M., Hietala, M., Ingvar, C., Rose, C. & Jernstrom, H. Androgen receptor genotypes predict response to endocrine treatment in breast cancer patients. Br. J. Cancer 105, 1676–1683 (2011).
Gonzalez-Angulo, A. M., Stemke-Hale, K., Palla, S. L., Carey, M., Agarwal, R., Meric-Berstam, F. et al. Androgen receptor levels and association with PIK3CA mutations and prognosis in breast cancer. Clin. Cancer Res. 15, 2472–2478 (2009).
Park, S., Park, H. S., Koo, J. S., Yang, W. I., Kim, S. I. & Park, B. W. Higher expression of androgen receptor is a significant predictor for better endocrine-responsiveness in estrogen receptor-positive breast cancers. Breast Cancer Res. Treat. 133, 311–320 (2012).
Witzel, I., Graeser, M., Karn, T., Schmidt, M., Wirtz, R., Schutze, D. et al. Androgen receptor expression is a predictive marker in chemotherapy-treated patients with endocrine receptor-positive primary breast cancers. J. Cancer Res. Clin. Oncol. 139, 809–816 (2013).
Loibl, S., Muller, B. M., von Minckwitz, G., Schwabe, M., Roller, M., Darb-Esfahani, S. et al. Androgen receptor expression in primary breast cancer and its predictive and prognostic value in patients treated with neoadjuvant chemotherapy. Breast Cancer Res. Treat. 130, 477–487 (2011).
Untch, M., Fasching, P. A., Konecny, G. E., Hasmuller, S., Lebeau, A., Kreienberg, R. et al. Pathologic complete response after neoadjuvant chemotherapy plus trastuzumab predicts favorable survival in human epidermal growth factor receptor 2-overexpressing breast cancer: results from the TECHNO trial of the AGO and GBG study groups. J. Clin. Oncol. 29, 3351–3357 (2011).
Untch, M., Mobus, V., Kuhn, W., Muck, B. R., Thomssen, C., Bauerfeind, I. et al. Intensive dose-dense compared with conventionally scheduled preoperative chemotherapy for high-risk primary breast cancer. J. Clin. Oncol. 27, 2938–2945 (2009).
Varga, Z., Lebeau, A., Bu, H., Hartmann, A., Penault-Llorca, F., Guerini-Rocco, E. et al. An international reproducibility study validating quantitative determination of ERBB2, ESR1, PGR, and MKI67 mRNA in breast cancer using MammaTyper(R). Breast Cancer Res. 19, 55 (2017).
Sikic, D., Wirtz, R. M., Wach, S., Dyrskjot, L., Erben, P., Bolenz, C. et al. Androgen receptor mRNA expression in urothelial carcinoma of the bladder: a retrospective analysis of two independent cohorts. Transl. Oncol. 12, 661–668 (2019).
McShane, L. M., Altman, D. G., Sauerbrei, W., Taube, S. E., Gion, M. & Clark, G. M. REporting recommendations for tumour MARKer prognostic studies (REMARK). Br. J. cancer 93, 387–391 (2005).
Rabiau, N., Dechelotte, P., Adjakly, M., Kemeny, J. L., Guy, L., Boiteux, J. P. et al. BRCA1, BRCA2, AR and IGF-I expression in prostate cancer: correlation between RT-qPCR and immunohistochemical detection. Oncol. Rep. 26, 695–702 (2011).
Mina, A., Yoder, R. & Sharma, P. Targeting the androgen receptor in triple-negative breast cancer: current perspectives. OncoTargets Ther. 10, 4675–4685 (2017).
Bonnefoi, H., Grellety, T., Tredan, O., Saghatchian, M., Dalenc, F., Mailliez, A. et al. A phase II trial of abiraterone acetate plus prednisone in patients with triple-negative androgen receptor positive locally advanced or metastatic breast cancer (UCBG 12-1). Ann. Oncol. 27, 812–818 (2016).
Traina, T. A., Miller, K., Yardley, D. A., Eakle, J., Schwartzberg, L. S., O’Shaughnessy, J. et al. Enzalutamide for the treatment of androgen receptor-expressing triple-negative breast cancer. J. Clin. Oncol. 36, 884–890 (2018).
Gucalp, A., Tolaney, S., Isakoff, S. J., Ingle, J. N., Liu, M. C., Carey, L. A. et al. Phase II trial of bicalutamide in patients with androgen receptor-positive, estrogen receptor-negative metastatic Breast Cancer. Clin. Cancer Res. 19, 5505–5512 (2013).
McNamara, K. M., Yoda, T., Takagi, K., Miki, Y., Suzuki, T. & Sasano, H. Androgen receptor in triple negative breast cancer. J. Steroid Biochem. Mol. Biol. 133C, 66–76 (2012).
Naderi, A., Chia, K. M. & Liu, J. Synergy between inhibitors of androgen receptor and MEK has therapeutic implications in estrogen receptor-negative breast cancer. Breast Cancer Res. 13, R36 (2011).
Mobus, V., von Minckwitz, G., Jackisch, C., Luck, H. J., Schneeweiss, A., Tesch, H. et al. German Adjuvant Intergroup Node-positive Study (GAIN): a phase III trial comparing two dose-dense regimens (iddEPC versus ddEC-PwX) in high-risk early breast cancer patients. Ann. Oncol. 28, 1803–1810 (2017).
Group EBCTC. Increasing the dose intensity of chemotherapy by more frequent administration or sequential scheduling: a patient-level meta-analysis of 37 298 women with early breast cancer in 26 randomised trials. Lancet 393, 1440–1452 (2019).
Author contributions
I.W., R.W., V.M. and M.U. conceived of the translational sub-study. P.F., C.D., H.L., J.H., T.K., M.M., F.M., C.S. and E.S. are part of the translational research board of the German Breast Group and approved the study concept. C.D. provided tumour tissue for the analysis. R.W. was responsible for the molecular analysis. K.W. performed the statistical analysis. I.W., K.W., S.L., R.W. and V.M. analysed and interpreted data. I.W., K.W. and V.M. prepared manuscript drafts. All authors commented on the manuscript drafts, and all authors read and approved the final manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
R.W. is employee of Stratifyer Molecular Oncology. V.M. received speaker honoraria from Amgen, Astra Zeneca, Celgene, Daiichi-Sankyo, Eisai, Pfizer, Novartis, Roche, Teva, and consultancy honoraria from Genomic Health, Hexal, Roche, Pierre Fabre, Amgen, Novartis, MSD, Daiichi-Sankyo and Eisai, Lilly, Tesaro and Nektar and research support from Novartis, Roche, Seattle Genetics, Genentech. All other authors declare that they have no conflict of interest.
Ethics approval and consent to participate
The clinical trials TECHNO and PREPARE received ethical approval (Ethics Committee, Medical Faculty of the Ludwig-Maximilians University, Munich, Germany, project no. 224/01 and 225/01, respectively) and the patient signed a consent to participate in the translational research project. The study was performed in accordance with the Declaration of Helsinki. The study was approved by each local institutional review board. A full list of the boards can be found in the supplementary material.
Funding
The AR analysis was supported by a donation of a private person to the University Medical Center Hamburg, Department of Gynecology.
Consent to publish
Not applicable.
Data availability
The datasets used during the present study are available from the German Breast Group, Neu-Isenburg, Germany on reasonable request.
Additional information
Note: This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution 4.0 International (CC BY 4.0).
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Witzel, I., Loibl, S., Wirtz, R. et al. Androgen receptor expression and response to chemotherapy in breast cancer patients treated in the neoadjuvant TECHNO and PREPARE trial. Br J Cancer 121, 1009–1015 (2019). https://doi.org/10.1038/s41416-019-0630-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41416-019-0630-3
This article is cited by
-
Classification of triple-negative breast cancer based on pathway enrichment levels
Medical Oncology (2023)
-
Development and validation of a nomogram based on pretreatment dynamic contrast-enhanced MRI for the prediction of pathologic response after neoadjuvant chemotherapy for triple-negative breast cancer
European Radiology (2022)