Abstract
Background
Cancer cells must alter their metabolism to support proliferation. Immune evasion also plays a role in supporting tumour progression. This study aimed to find whether enhanced glutamine uptake in breast cancer (BC) can derive the existence of specific immune cell subtypes, including the subsequent impact on patient outcome.
Methods
SLC1A5, SLC7A5, SLC3A2 and immune cell markers CD3, CD8, FOXP3, CD20 and CD68, in addition to PD1 and PDL1, were assessed by using immunohistochemistry on TMAs constructed from a large BC cohort (n = 803). Patients were stratified based on SLC protein expression into accredited clusters and correlated with immune cell infiltrates and patient outcome. The effect of transient siRNA knockdown of SLC7A5 and SLC1A5 on PDL1 expression was evaluated in MDA-MB-231 cells.
Results
High SLCs were significantly associated with PDL1 and PD1 +, FOXP3 +, CD68 + and CD20 + cells (p < 0.001). Triple negative (TN), HER2 + and luminal B tumours showed variable associations between SLCs and immune cell types (p ≤ 0.04). The expression of SLCs and PDL1, PD1 +, FOXP3 + and CD68 + cells was associated with poor patient outcome (p < 0.001). Knockdown of SLC7A5 significantly reduced PDL1 expression.
Conclusion
This study provides data that altered glutamine pathways in BC that appears to play a role in deriving specific subtypes of immune cell infiltrates, which either support or counteract its progression.
Similar content being viewed by others
Background
Altered metabolic pathways are readily accepted as part of the revised hallmarks of cancer where cancer cells adapt their metabolism in order to resist the unfavourable, nutrient-deprived conditions and to respond to the increased energy demands required by their unremitting proliferation.1 Many cancer cells are highly reliant on amino acids for their growth, not only because they are precursors for nucleotide and protein synthesis, but also because they activate mammalian target of rapamycin complex1 (mTORC1) through nutrient-signalling pathways, which in turn regulates protein translation and cell growth.2,3
Solute carrier family 1 member 5 (SLC1A5) and solute carrier family 7 member 5 (SLC7A5) are two key amino acid transporters that have been attracting attention due to their role in supporting tumour metabolism. Primarily, SLC1A5 maintains the sodium‐coupled influx of glutamine, whereas SLC7A5 mediates the efflux of this amino acid in exchange with the influx of leucine, an essential amino acid and potent activator of mTORC1.4,5 SLC7A5 requires a covalent association with the heavy chain of SLC3A2, for its functional expression in plasma membrane.6 We have previously described the potential utility of these transporters as prognostic factors in certain BC subtypes.7,8 Further, we have recently stratified BC patients into three accredited clusters based on the protein expression of these three solute carriers.9
The role of the tumour microenvironment (TME) is well known with respect to disease development and progression. One of the important components of the TME is immune cells, including the regulatory T cells (Treg) and tumour-associated macrophages (TAM), which gain pro-tumoural functions stimulating tumour growth, progression, invasion and metastasis. Conversely, other immune cells such as CD8 + and CD20 + lymphocytes are responsible for anti-tumoural responses by activating host defence mechanisms preventing immune evasion.10,11 Immune evasion is a strategy used by tumours to evade a host’s immune response in an attempt to maximise their probability to continue surviving and growing. Tumour immune evasion includes several mechanisms such as progressive formation of an immune-suppressive environment within the tumour and the selection of tumour variants resistant to immune effectors (immunoediting).12
Composition of the inflammatory cell infiltrates in BC also correlates with clinical outcome, where an abundant infiltration of pro-tumorigenic cells is associated with poor outcome, while TME enriched in cells with anti-tumorigenic functions has a favourable effect on patient survival.13,14,15 Programmed cell death 1 (PD1) and its activating ligand, programmed death ligand 1 (PDL1) act in attenuating the anticancer immune response and promoting T-regulatory cell development and function. Indeed, PDL1 is expressed by tumour cells of several cancer types, with evidence of an association with aggressive tumour behaviour and poor prognosis.16,17,18,19,20
Previous studies show that the cellular contents of the TME change in parallel with tumour growth/progression, and the accompanied alterations in glucose metabolism in cancer cells are tightly linked to the composition of the surrounding immune cells.21,22 We therefore hypothesise that the reprogramming of glutamine metabolism will have an impact on the structure of the immune cells. This study aimed to determine whether overexpression of the key glutamine solute carriers can derive the existence of specific subtypes of immune cells, in addition to their supportive role in estimating the clinical outcome.
Methods
Patient cohort
This study evaluated a well-characterised cohort of early-stage, primary operable, invasive BC patients aged ≤70 years. Patients (n = 803) presented at Nottingham City Hospital from 1989 to 1998. Patient management was uniform and based on tumour characteristics by Nottingham Prognostic Index (NPI) and hormone receptor status. Clinical history, tumour characteristics, information on therapy and outcomes are prospectively maintained. Outcome data included development and time to distant metastasis (DM) and breast cancer-specific survival (BCSS), defined as the time (in months) from the date of primary surgical treatment to the time of death from BC. The clinicopathological parameters for the BC series are summarised in Supplementary Table 1.
Tissue microarrays (TMAs) and immunohistochemistry
TMAs consisting of 0.6-mm tumour tissue cores were arrayed and immunohistochemically profiled for SLC1A5, SLC7A5, SLC3A2, PD1, PDL1, CD3, CD8, CD68, CD20 and FOXP3, as previously described.23,9,13,24
TMA sections stained with SLC1A5, SLC7A5, SLC3A2, PDL1 and PD1 were scanned by using high-resolution digital images (NanoZoomer; Hamamatsu Photonics, Welwyn Garden City, UK), at ×20 magnification. Evaluation of staining was based on a semi-quantitative assessment by using a modified histochemical score (H-score)25 as previously described.9 Clustering analysis of SLC1A5, SLC7A5 and SLC3A2 protein expression was previously performed by using two algorithms, partitioning around medoids (PAM) and K-means, to stratify tumours into the optimal number of clusters based on their H-score. The three clusters were characterised as follows: low SLCs (SLC1A5–/SLC7A5–/SLC3A2–), high SLC1A5 (SLC1A5 +/SLC7A5–/SLC3A2–) and high SLCs (SLC1A5 +/SLC7A5 +/SLC3A2 + ).9
Immunohistochemical detection of a panel of lymphocyte markers including pan-T-cell CD3, cytotoxic T-cell CD8, T-reg FOXP3, B-cell CD20 and histiocytic cell marker CD68 was previously determined, and the total number of each immune cell type was counted in each tumour core by using a Nikon Eclipse 80i microscope (Nikon, Tokyo, Japan) as described.13,14,15 BC molecular subtypes were defined, based on tumour IHC profile and the Elston–Ellis26 mitotic score as ER +/HER2– low proliferation (mitotic score 1), ER +/HER2– high proliferation (mitotic scores 2 and 3) and HER2-positive class: HER2 + regardless of ER status, triple negative (TN): ER–, PgR– and HER2–.27
siRNA transfection of SLC7A5 and SLC1A5
The TN cell line, MDA-MB-231, was obtained from American Type Culture Collection; Rockville, MD, USA and cultured in Roswell Park Memorial Institute (RPMI-1640) medium (Sigma-Aldrich, UK) supplemented with 10% foetal bovine serum (Sigma-Aldrich, UK). Mycoplasma testing was carried out on a routine basis by using the MycoAlert Detection kit (R&D Systems). In total, 5 × 104 cells were seeded per well in a 24-well plate and transfected by using the reverse transfection method with 25 and 100 pmol siRNA (ThermoFisher Scientific), for SLC7A5 and SLC1A5, respectively, and lipofectamine (RNAiMAX) according to the manufacturer’s protocol.
siRNA antisense sequences were as follows: 5′-UUGGGAUCUAGAUUGGACAca-3′ (for SLC7A5) and 5′-AAAGAGUAAACCCACAUCCtc-3′ (for SLC1A5). Untransfected cells were carried out alongside the experiment as controls. SLC7A5 and SLC1A5 expression of transfected cells was performed in duplicate and determined by Western blotting analysis (Supplementary Fig. 2A–B).
Statistical analysis
Statistical analysis was performed by using SPSS 24.0 statistical software (SPSS Inc., Chicago, IL, USA). The chi-square test was performed for interrelationships between categorical variables. Differences between two groups of normalised data were assessed by using t test. Survival curves were analysed by Kaplan–Meier with log-rank test. This was performed with BC- specific death; those who died of other causes, alive and lost to follow-up were censored. P-values were adjusted by using Bonferroni correction for multiple testing. A p-value < 0.05 was considered significant. The study endpoints were 10-year breast cancer-specific survival (BCSS) or distant metastasis-free survival (DMFS).
This study was approved by the Nottingham Research Ethics Committee 2 under the title ‘Development of a molecular genetic classification of breast cancer’ and the North West– Greater Manchester Central Research Ethics Committee under the title ‘Nottingham Health Science Biobank (NHSB)’ reference number 15/NW/0685.
Results
Expression of SLC1A5, SLC7A5, SLC3A2 and immune cell markers in BC
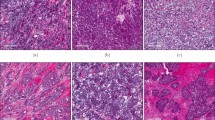
Expression of the three solute carriers was predominantly in the membrane of the invasive BC cells, with intensity levels varying from absent to high. High expression of the solute carriers was also observed in lymphocytic infiltrates, which were in the stroma adjacent to the tumour cells (Fig. 1a–c). Immunohistochemical expression of solute carriers and immune cell markers in invasive BC cores is illustrated in Supplementary Fig. 1.
Association of SLCs with immune cell infiltrates
Tumour-infiltrating FOXP3 + lymphocytes and CD68 + macrophages were both predominant in tumours with high SLCs and to a lesser extent in those tumours with high SLC1A5 expression (Table 1, p < 0.0001). CD20 + lymphocytes were mainly observed in tumours with high SLC expression (Table 1, p < 0.0001). While PDL1 was highly expressed in tumours with high SLCs and high SLC1A5 expression, PD1 + cells were mainly expressed with tumours with high SLC expression (Table 1, p < 0.0001). In contrast, there was no significant association between the SLC clusters and CD3 + or CD8 + T lymphocytes (Table 1, p = 0.84 and p = 0.24), respectively.
Association between SLC clusters and immune cell infiltrates varied among BC molecular subtypes
CD68 + cells were significantly associated with high SLC expression in both ER + high proliferative/luminal B and TN subtypes (Tables 2, 3, p = 0.02 and p = 0.03), respectively. In contrast, FOXP3 + cells were only associated within TN tumours showing high SLC expression (Table 3, p = 0.02). CD20 + cells were only observed in ER + high proliferative/luminal B tumours with high SLCs (Table 2, p = 0.04). PD1 and PDL1 were also significantly expressed in HER2 + tumours with high SLC expression (Table 3, p = 0.03 and p = 0.04), respectively. However, within TN tumours, only PDL1 expression was associated with high SLC expression (Table 3, p = 0.04). There were no significant associations between the SLC clusters and immune cell infiltrates in ER + low proliferative/luminal A tumours (Table 2).
The co-occurrence of SLCs and immune cell infiltrates correlates with patient outcome
Variable associations with patient outcome were observed when investigating the co-occurrence of the SLC clusters with the immune cell markers. The coexistence of high SLCs with FOXP3 + T lymphocytes was predictive of a shorter BCSS (Fig. 2a, p < 0.001). Similar associations were observed when CD68 + macrophages, PD1 + cells and PDL1 expression were considered (Fig. 2b–d, p < 0.001). However, patients with tumours showing both high SLCs and CD20 + lymphocytes, showed better BCSS (Fig. 2e, p < 0.001).
There was a comparable observation regarding the association of SLCs and immune cell markers with DMFS, where high SLC expression accompanied by the presence of FOXP3 +, CD68 + and PD1 + cells or PDL1 expression showed significantly shorter DMFS (Supplementary Fig. 3A–D, all p < 0.001). In contrast, the presence of CD20 + lymphocytes and high SLCs conferred a longer DMFS (Supplementary Fig. 3E, p = 0.002).
SLC7A5 plays a role in PDL1 expression in TNBC
Functional analysis was carried out by using the TNBC cell line, MDA-MB-231, due to the high expression of PDL1 together with SLC7A5 and/or SLC1A5 expression, and also based on the significant associations found between SLCs and immune cell markers, including PDL1, within the TNBC subtype (Fig. 3b, c).
PDL1 protein expression in western blotting. a Western blot analysis of PDL1 protein expression in MDA-MB-231 cells transfected with SLC7A5 and/or SLC1A5 SiRNA. Western blot results in different BC cell lysates for b SLC7A5 and c SLC1A5. The bar graph summarises the expression levels of PDL1 protein, by using β-actin as normalised control, upon d SLC7A5 SiRNA transfection. e SLC1A5 SiRNA transfection. f SLC7A5 and SLC1A5 SiRNA transfection. Data represent the mean and error bars of three independent experiments
siRNA knockdown of SLC1A5 or SLC7A5 in MDA-MB-231 reduced the protein expression of PDL1. However, this observation was only significant upon targeting SLC7A5 (Fig. 3d, p = 0.01) but not SLC1A5 (Fig. 3e, p = 0.13). Significant reduction of PDL1 protein expression was also observed in cells transfected with both SLC1A5 and SLC7A5 siRNAs (Fig. 3f, p = 0.02).
Discussion
Breast cancer is a heterogeneous disease with various subtypes28 that are different in terms of morphology, molecular profiles, response to therapy and clinical behaviour. Breast cancer also shows heterogeneity in metabolic reprogramming, where highly proliferative tumours are distinguished based on their metabolic signatures.29,30,31
Cancer cells undergo metabolic changes in order to satisfy the demands of necessary energy and cellular building blocks. One of the most prominent is the increase in glutamine consumption, which is reflected by the upregulation of the key glutamine transporters (SLC1A5 and the SLC7A5–SLC3A2 dimeric complex) at the surface of the tumour cells. We have recently demonstrated that the combined expression of the three solute carriers (SLC1A5, SLC7A5 and SLC3A2) is associated with poor prognosis and short BCSS, particularly in the highly proliferative BC subtypes.9
Besides metabolic reprogramming, immune evasion is also considered as an emerging hallmark of cancer.1 The role of immune cells in tumour evasion is increasingly barbed, as many tumours not only escape recognition by the adaptive immune response but also sometimes cooperate with the pro-tumorigenic immune cells to become invasive and more aggressive. Furthermore, there is a link between the two mentioned hallmarks, as changes in the tumour cell metabolism can influence the component and function of the inflammatory infiltrates.21,32 This study showed that altered glutamine metabolism, which was detected by the overexpression of the key glutamine transporters (SLC1A5, SLC7A5 and SLC3A2) was significantly associated with the existence of specific subtypes of immune cells, namely CD68 + macrophages, FOXP3 + regulatory T cells (Tregs), CD20 + B lymphocytes and PD1 + lymphocytes along with its tumour-expressing ligand (PDL1). However, no association was observed between the SLCs and CD3 + or CD8 + T lymphocytes.
Our previous study on the same BC cohort showed that the main component of the inflammatory infiltrates is the pan-T-lymphocyte population (CD3 + cells) along with CD8 + cells being more frequent than FOXP3 + cells. The CD68 + macrophages were more frequent while CD20 + B lymphocytes were the least.33 In this study, however, we observed that changes in the metabolic activity of the cancer cells, which is reflected by an increase in glutamine transport, derive specific components of immune cells, which were restricted to CD68 +, FOXP3 + and CD20 + along with PD1 + cells. This indicates that in these circumstances, the antigen- presenting cells (APC), CD68 + cells, are recognised only by specific subpopulations of T- and B lymphocytes.
When different BC subtypes were examined, a significant association was observed in ER + highly proliferative/luminal B, TN and HER2 + tumours, but not the ER + low proliferative/luminal A subtype. Both luminal B and TN tumours showed associations with CD68 + macrophages. These two subtypes are aggressive, highly proliferative and exhibit high metabolic activity. Consequently, aggressive cancer cells secrete high levels of reactive oxygen species (ROS) in their microenvironment.34 The latter can cause a state of pseudohypoxia in the adjacent stromal compartment with concomitant upregulation of hypoxia-inducible factor 1α (HIF1α), known to induce the pro-tumorigenic CD68 + macrophages.35,36 The same scenario can be applied when amino acids, particularly leucine, activate mTORC1 that upregulates HIF1α.37 Previous studies have shown that PD1/PDL1 are mainly expressed in HER2 + and TN subtypes.20,38,39 This study further shows that the expression of PD1 and/or PDL1 is mainly associated with high SLC expression but restricted to HER2 + and TN tumours.
In this study, we observed high expression of the solute carriers in the stromal lymphocytes. This is expected as glutamine transporters are not only necessary for cancer cells, but they are also important for optimal lymphocyte proliferation and differentiation.40,41,42,43 In addition, macrophages may require glutamine as it is the main precursor for arginine.44 The latter can be catalysed by Arginase 1 to support cell proliferation and tissue remodelling.45
Indeed, the upregulation of glutamine transporters in the cancer cells and their neighbouring immune cells, might indicate that both cell types are substantially comparable in their requirements of amino acids, which can be obtained from the TME, to support their survival and proliferation. Furthermore, TME might be a source of stromal glutamine, as it has been found that the metabolic stress in TME triggers genomic instability, which subsequently acquires the non-malignant cancer-associated fibroblasts (CAF) a catabolic phenotype with enhanced macroautophagy. This catabolic state produces a nutrient-rich environment, with increased amounts of pyruvate, lactate, ketone bodies and glutamine.46 This phenomenon also substantiates the cancer-stromal symbiosis that subsequently supports cancer growth and progression.
We and others revealed that high expression of glutamine solute carriers is associated with poor patient outcome.7,8,47 Similarly, the presence of FOXP3 + and CD68 + cells also correlates with shorter survival.33 However, CD20 + cells tend to be associated with better survival.33 In this study, we showed that the co-occurrence of SLCs with FOXP3 + and CD68 + cells can predict shorter survival compared with the presence of one without the other, whereas the combination of the SLCs with CD20 + cells derives better patient outcome.
The association between PD1/PDL1 and survival in BC is controversial.20,48,49,50 This study, however, showed that the co-expression of SLCs with PD1 or PDL1 was associated with shorter distant metastasis-free survival and breast cancer-specific survival, indicating that patients with BC show an increase in their amino acid metabolic activity that might influence poor outcome in PD1/PDL1 + tumours.
We observed that targeting SLC7A5 by transient siRNA significantly reduced the expression of PDL1 in TN cells. This can be attributed to the role played by this protein in activating the mTORC1 pathway, through importing essential amino acids, such as leucine. This might take place in parallel with the activation of mTORC1 through the AKT–mTOR signalling pathway, which is previously identified as a tight regulator of PDL1 expression in several cancers, including TNBC.38,51,52
Although clinical trials with monoclonal antibodies targeting PD1/PDL1 interaction have shown promising results, with durable responses, in several human cancers,53,54,55 not all patients respond to this targeted therapy. Therefore, it is critical to find effective approaches that could allow personalisation of treatment of PD1/PDL1 + tumours. This study not only provides clinical evidence that SLCs in BC could aid the personalisation of anti-PD1/PDL1 inhibition therapies, but it also emphasises that targeting the amino acid transporter, SLC7A5, along with the anti-PDL1 immunotherapy could be considered as a novel approach to synergistically enhance the therapeutic effect.
Conclusion
This study revealed that there are associations between the two cancer hallmarks, metabolic reprogramming and immune evasion. Altered glutamine pathways in cancer cells can derive specific subtypes of inflammatory infiltrates, which act either with or against the aggressiveness and progression of the BC cells. Targeting both SLC7A5 and PD1/PDL1 can be a new approach that will counteract the highly proliferative and aggressive BC subtypes.
References
Hanahan, D. & Weinberg, R. A. Hallmarks of cancer: the next generation. Cell 144, 646–674 (2011).
Bar-Peled, L. & Sabatini, D. M. Regulation of mTORC1 by amino acids. Trends Cell Biol. 24, 400–406 (2014).
Bond, P. Regulation of mTORC1 by growth factors, energy status, amino acids and mechanical stimuli at a glance. J. Int. Soc. Sports Nutr. 13, 8 (2016).
Bhutia, Y. D., Babu, E., Ramachandran, S. & Ganapathy, V. Amino acid transporters in cancer and their relevance to “glutamine addiction”: novel targets for the design of a new class of anticancer drugs. Cancer Res. 75, 1782–1788 (2015).
Fuchs, B. C. & Bode, B. P. Amino acid transporters ASCT2 and LAT1 in cancer: partners in crime? Semin. Cancer Biol. 15, 254–266 (2005).
Yanagida, O., Kanai, Y., Chairoungdua, A., Kim, D. K., Segawa, H., Nii, T. et al. Human L-type amino acid transporter 1 (LAT1): characterization of function and expression in tumor cell lines. Biochim, Biophys, Acta. 1514, 291–302 (2001).
El Ansari, R., Craze, M. L., Miligy, I., Diez-Rodriguez, M., Nolan, C. C., Ellis, I. O. et al. The amino acid transporter SLC7A5 confers a poor prognosis in the highly proliferative breast cancer subtypes and is a key therapeutic target in luminal B tumours. Breast Cancer Res. 20, 21 (2018).
El Ansari, R., Craze, M. L., Diez-Rodriguez, M., Nolan, C. C., Ellis, I. O., Rakha, E. A. et al. The multifunctional solute carrier 3A2 (SLC3A2) confers a poor prognosis in the highly proliferative breast cancer subtypes. Br. J. Cancer 118, 1115 (2018).
El-Ansari, R., Craze, M. L., Alfarsi, L., Soria, D., Diez-Rodriguez, M., Nolan, C. C. et al. The combined expression of solute carriers is associated with a poor prognosis in highly proliferative ER+ breast cancer. Breast Cancer Res. Tr. 175, 27–38 (2019).
Monjazeb, A. M., Zamora, A. E., Grossenbacher, S. K., Mirsoian, A., Sckisel, G. D. & Murphy, W. J. Immunoediting and antigen loss: overcoming the achilles heel of immunotherapy with antigen non-specific therapies. Front. Oncol. 3, 197 (2013).
Qian, B. Z. & Pollard, J. W. Macrophage diversity enhances tumor progression and metastasis. Cell 141, 39–51 (2010).
Vinay, D. S., Ryan, E. P., Pawelec, G., Talib, W. H., Stagg, J., Elkord, E. et al. Immune evasion in cancer: mechanistic basis and therapeutic strategies. Semin. Cancer Biol. 35(Suppl), S185–S98. (2015).
Mahmoud, S. M., Paish, E. C., Powe, D. G., Macmillan, R. D., Grainge, M. J., Lee, A. H. et al. Tumor-infiltrating CD8+ lymphocytes predict clinical outcome in breast cancer. J. Clin. Oncol. 29, 1949–1955 (2011).
Mahmoud, S. M., Paish, E. C., Powe, D. G., Macmillan, R. D., Lee, A. H., Ellis, I. O. et al. An evaluation of the clinical significance of FOXP3+ infiltrating cells in human breast cancer. Breast Cancer Res. Tr. 127, 99–108 (2011).
Mahmoud, S. M., Lee, A. H., Paish, E. C., Macmillan, R. D., Ellis, I. O. & Green, A. R. The prognostic significance of B lymphocytes in invasive carcinoma of the breast. Breast Cancer Res. Tr. 132, 545–553 (2012).
Konishi, J., Yamazaki, K., Azuma, M., Kinoshita, I., Dosaka-Akita, H. & Nishimura, M. B7-H1 expression on non-small cell lung cancer cells and its relationship with tumor-infiltrating lymphocytes and their PD-1 expression. Clinical Cancer Res. 10, 5094–5100 (2004).
Nomi, T., Sho, M., Akahori, T., Hamada, K., Kubo, A., Kanehiro, H. et al. Clinical significance and therapeutic potential of the programmed death-1 ligand/programmed death-1 pathway in human pancreatic cancer. Clinical Cancer Res. 13, 2151–2157 (2007).
Hamanishi, J., Mandai, M., Iwasaki, M., Okazaki, T., Tanaka, Y., Yamaguchi, K. et al. Programmed cell death 1 ligand 1 and tumor-infiltrating CD8+ T lymphocytes are prognostic factors of human ovarian cancer. Proc. Natl Acad. Sci. USA 104, 3360–3365 (2007).
Badoual, C., Hans, S., Merillon, N., Van Ryswick, C., Ravel, P., Benhamouda, N. et al. PD-1-expressing tumor-infiltrating T cells are a favorable prognostic biomarker in HPV-associated head and neck cancer. Cancer Res. 73, 128–138 (2013).
Sabatier, R., Finetti, P., Mamessier, E., Adelaide, J., Chaffanet, M., Ali, H. R. et al. Prognostic and predictive value of PDL1 expression in breast cancer. Oncotarget. 6, 5449–5464 (2015).
Kareva, I. & Hahnfeldt, P. The emerging “hallmarks” of metabolic reprogramming and immune evasion: distinct or linked? Cancer Res. 73, 2737–2742 (2013).
Carmona-Fontaine, C., Bucci, V., Akkari, L., Deforet, M., Joyce, J. A. & Xavier, J. B. Emergence of spatial structure in the tumor microenvironment due to the Warburg effect. Proc. Natl Acad. Sci. USA 110, 19402–19407 (2013).
Abd El-Rehim, D. M., Ball, G., Pinder, S. E., Rakha, E., Paish, C., Robertson, J. F. et al. High-throughput protein expression analysis using tissue microarray technology of a large well-characterised series identifies biologically distinct classes of breast cancer confirming recent cDNA expression analyses. Int. J. Cancer 116, 340–350 (2005).
Green, A. R., Aleskandarany, M. A., Ali, R., Hodgson, E. G., Atabani, S., De Souza, K. et al. Clinical impact of tumor DNA repair expression and T-cell infiltration in breast cancers. Cancer Immunol. Res. 5, 292–299 (2017).
McCarty, K. S. Jr. & KS, Mc. Carty Sr. Histochemical approaches to steroid receptor analyses. Semin. Diagn. Pathol. 1, 297–308 (1984).
Elston, C. W. & Ellis, I. O. Pathological prognostic factors in breast cancer. I. The value of histological grade in breast cancer: experience from a large study with long-term follow-up. C. W. Elston & I. O. Ellis. Histopathology 19, 403–410 (1991). Histopathology 41,151–152 (2002), discussion 2–3.
Senkus, E., Kyriakides, S., Ohno, S., Penault-Llorca, F., Poortmans, P., Rutgers, E. et al. Primary breast cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 26(Suppl 5), v8–v30 (2015).
Perou, C. M., Sorlie, T., Eisen, M. B., van de Rijn, M., Jeffrey, S. S., Rees, C. A. et al. Molecular portraits of human breast tumours. Nature 406, 747–752 (2000).
Kim, S., Kim, D. H., Jung, W. H. & Koo, J. S. Expression of glutamine metabolism-related proteins according to molecular subtype of breast cancer. Endocr. Relat. Cancer 20, 339–348 (2013).
Hilvo, M., Denkert, C., Lehtinen, L., Muller, B., Brockmoller, S., Seppanen-Laakso, T. et al. Novel theranostic opportunities offered by characterization of altered membrane lipid metabolism in breast cancer progression. Cancer Res. 71, 3236–3245 (2011).
Gatenby, R. A. & Gillies, R. J. Why do cancers have high aerobic glycolysis? Nat. Rev. Cancer. 4, 891–899 (2004).
Netea-Maier, R. T., Smit, J. W. A. & Netea, M. G. Metabolic changes in tumor cells and tumor-associated macrophages: A mutual relationship. Cancer Lett. 413, 102–109 (2018).
Althobiti, M., Aleskandarany, M. A., Joseph, C., Toss, M., Mongan, N., Diez-Rodriguez, M. et al. Heterogeneity of tumour infiltrating lymphocytes (TILs) in breast cancer and its prognostic significance. Histopathology 73, 887–896 (2018).
Martinez-Outschoorn, U. E., Lin, Z., Ko, Y. H., Goldberg, A. F., Flomenberg, N., Wang, C. et al. Understanding the metabolic basis of drug resistance: therapeutic induction of the Warburg effect kills cancer cells. Cell Cycle 10, 2521–2528 (2011).
Zhang, W. J., Chen, C., Zhou, Z. H., Gao, S. T., Tee, T. J., Yang, L. Q. et al. Hypoxia-inducible factor-1 alpha correlates with tumor-associated macrophages infiltration, influences survival of gastric cancer patients. J. Cancer 8, 1818–1825 (2017).
Li, N., Li, Y., Li, Z., Huang, C., Yang, Y., Lang, M. et al. Hypoxia inducible factor 1 (HIF-1) recruits macrophage to activate pancreatic stellate cells in pancreatic ductal adenocarcinoma. Int. J. Mol. Sci. 17, 799–811 (2016).
Land, S. C. & Tee, A. R. Hypoxia-inducible factor 1alpha is regulated by the mammalian target of rapamycin (mTOR) via an mTOR signaling motif. J. Biol. Chem. 282, 20534–20543 (2007).
Mittendorf, E. A., Philips, A. V., Meric-Bernstam, F., Qiao, N., Wu, Y., Harrington, S. et al. PD-L1 expression in triple-negative breast cancer. Cancer Immunol. Res. 2, 361–370 (2014).
Tsang, J. Y., Au, W. L., Lo, K. Y., Ni, Y. B., Hlaing, T., Hu, J. et al. PD-L1 expression and tumor infiltrating PD-1+ lymphocytes associated with outcome in HER2+ breast cancer patients. Breast Cancer Res. Tr. 162, 19–30 (2017).
Rohde, T., MacLean, D. A. & Klarlund Pedersen, B. Glutamine, lymphocyte proliferation and cytokine production. Scand J. Immunol. 44, 648–650 (1996).
Nakaya, M., Xiao, Y., Zhou, X., Chang, J. H., Chang, M., Cheng, X. et al. Inflammatory T cell responses rely on amino acid transporter ASCT2 facilitation of glutamine uptake and mTORC1 kinase activation. Immunity. 40, 692–705 (2014).
Sinclair, L. V., Rolf, J., Emslie, E., Shi, Y. B., Taylor, P. M. & Cantrell, D. A. Control of amino-acid transport by antigen receptors coordinates the metabolic reprogramming essential for T cell differentiation. Nat. Immunol. 14, 500–508 (2013).
Ren, W., Liu, G., Yin, J., Tan, B., Wu, G., Bazer, F. W. et al. Amino-acid transporters in T-cell activation and differentiation. Cell Death Dis. 8, e2757 (2017).
Ligthart-Melis, G. C., van de Poll, M. C., Boelens, P. G., Dejong, C. H., Deutz, N. E. & van Leeuwen, P. A. Glutamine is an important precursor for de novo synthesis of arginine in humans. Am. J. Clin. Nutr. 87, 1282–1289 (2008).
Biswas, S. K. & Mantovani, A. Orchestration of metabolism by macrophages. Cell Metab. 15, 432–437 (2012).
Penkert, J., Ripperger, T., Schieck, M., Schlegelberger, B., Steinemann, D. & Illig, T. On metabolic reprogramming and tumor biology: A comprehensive survey of metabolism in breast cancer. Oncotarget 7, 67626 (2016).
Liu, Y., Yang, L., An, H., Chang, Y., Zhang, W., Zhu, Y. et al. High expression of solute carrier family 1, member 5 (SLC1A5) is associated with poor prognosis in clear-cell renal cell carcinoma. Sci. Rep. 5, 16954 (2015).
Muenst, S., Soysal, S. D., Gao, F., Obermann, E. C., Oertli, D. & Gillanders, W. E. The presence of programmed death 1 (PD-1)-positive tumor-infiltrating lymphocytes is associated with poor prognosis in human breast cancer. Breast Cancer Res. Tr. 139, 667–676 (2013).
Muenst, S., Schaerli, A. R., Gao, F., Daster, S., Trella, E., Droeser, R. A. et al. Expression of programmed death ligand 1 (PD-L1) is associated with poor prognosis in human breast cancer. Breast Cancer Res. Tr. 146, 15–24 (2014).
Baptista, M. Z., Sarian, L. O., Derchain, S. F., Pinto, G. A. & Vassallo, J. Prognostic significance of PD-L1 and PD-L2 in breast cancer. Hum. Pathol. 47, 78–84 (2016).
Parsa, A. T., Waldron, J. S., Panner, A., Crane, C. A., Parney, I. F., Barry, J. J. et al. Loss of tumor suppressor PTEN function increases B7-H1 expression and immunoresistance in glioma. Nat. Med. 13, 84–88 (2007).
Lastwika, K. J., Wilson, W. 3rd, Li, Q. K., Norris, J., Xu, H., Ghazarian, S. R. et al. Control of PD-L1 expression by oncogenic activation of the AKT-mTOR pathway in non-small cell lung cancer. Cancer Res. 76, 227–238 (2016).
Brahmer, J. R., Tykodi, S. S., Chow, L. Q., Hwu, W. J., Topalian, S. L., Hwu, P. et al. Safety and activity of anti-PD-L1 antibody in patients with advanced cancer. N. Engl. J. Med. 366, 2455–2465 (2012).
Topalian, S. L., Hodi, F. S., Brahmer, J. R., Gettinger, S. N., Smith, D. C., McDermott, D. F. et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N. Engl. J. Med. 366, 2443–2454 (2012).
Powles, T., Eder, J. P., Fine, G. D., Braiteh, F. S., Loriot, Y., Cruz, C. et al. MPDL3280A (anti-PD-L1) treatment leads to clinical activity in metastatic bladder cancer. Nature 515, 558–562 (2014).
Acknowledgements
We thank the Nottingham Health Science Biobank and Breast Cancer Now Tissue Bank for the provision of tissue samples. We thank the University of Nottingham (Nottingham Life Cycle 6 and Cancer Research Priority Area) for funding.
Author contributions
R.E. contributed to writing, IHC staining, scoring, data analysis and interpretation; M.L.C. contributed to writing and reviewing the paper; M.A. and L.A. contributed to analysis and reviewing the paper; I.O.E. and E.A.R. contributed to writing and reviewing the paper; A.R.G. contributed to study design, data analysis and interpretation, writing and reviewing the paper.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
This study was approved by the Nottingham Research Ethics Committee 2 under the title ‘Development of a molecular genetic classification of breast cancer’ and the North West–Greater Manchester Central Research Ethics Committee under the title ‘Nottingham Health Science Biobank (NHSB)’ reference number 15/NW/0685. The study was performed in accordance with the Declaration of Helsinki.
Funding
Sources of study funding are the University of Nottingham (Nottingham Life Cycle 6 and Cancer Research Priority Area).
Consent to publish
Not applicable.
Data availability
The data sets generated during this study are available from the corresponding author on reasonable request.
Additional information
Note: This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution 4.0 International (CC BY 4.0).
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Ansari, R.E., Craze, M.L., Althobiti, M. et al. Enhanced glutamine uptake influences composition of immune cell infiltrates in breast cancer. Br J Cancer 122, 94–101 (2020). https://doi.org/10.1038/s41416-019-0626-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41416-019-0626-z
This article is cited by
-
Nutrient transporters: connecting cancer metabolism to therapeutic opportunities
Oncogene (2023)
-
Simultaneous glutamine metabolism and PD-L1 inhibition to enhance suppression of triple-negative breast cancer
Journal of Nanobiotechnology (2022)
-
Association of L-type amino acid transporter 1 (LAT1) with the immune system and prognosis in invasive breast cancer
Scientific Reports (2022)
-
Co-expression effect of LLGL2 and SLC7A5 to predict prognosis in ERα-positive breast cancer
Scientific Reports (2022)
-
Development of cancer metabolism as a therapeutic target: new pathways, patient studies, stratification and combination therapy
British Journal of Cancer (2020)