Abstract
Background
Tissue kallikrein-related peptidases 4, 5, 6 and 7 (KLK4–7) strongly increase the malignancy of ovarian cancer cells. Deciphering their downstream effectors, we aimed at finding new potential prognostic biomarkers and treatment targets for ovarian cancer patients. KLK4–7-transfected (OV-KLK4–7) and vector-control OV-MZ-6 (OV-VC) ovarian cancer cells were established to select differentially regulated factors.
Methods
With three independent approaches, PCR arrays, genome-wide microarray and proteome analyses, we identified 10 candidates (MSN, KRT19, COL5A2, COL1A2, BMP5, F10, KRT7, JUNB, BMP4, MMP1). To determine differential protein expression, we performed western blot analyses, immunofluorescence and immunohistochemistry for four candidates (MSN, KRT19, KRT7, JUNB) in cells, tumour xenograft and patient-derived tissues.
Results
We demonstrated that KLK4–7 clearly regulates expression of MSN, KRT19, KRT7 and JUNB at the mRNA and protein levels in ovarian cancer cells and tissues. Protein expression of the top-upregulated effectors, MSN and KRT19, was investigated by immunohistochemistry in patients afflicted with serous ovarian cancer and related to KLK4–7 immunoexpression. Significant positive associations were found for KRT19/KLK4, KRT19/KLK5 and MSN/KLK7.
Conclusion
These findings imply that KLK4–7 exert key modulatory effects on other cancer-related genes and proteins in ovarian cancer. These downstream effectors of KLK4–7, MSN and KRT19 may represent important therapeutic targets in serous ovarian cancer.
Similar content being viewed by others
Background
Ovarian cancer is the fifth leading cause of cancer-associated deaths in women in the United States, with 14,070 estimated deaths in 2018, and the most lethal gynaecological malignancy.1 The majority (>70%) of ovarian cancer patients are diagnosed at an advanced stage of the disease (FIGO III/IV). After standard treatment for advanced ovarian cancer, including cytoreductive surgery and platinum-containing chemotherapy, the cure rate is only 14% and thus has marginally changed in the last 25 years.2 Therefore, it is imperative to find new therapeutic targets to ameliorate these patients’ prognosis.
The tissue kallikrein-related peptidases (KLKs) are a family of 15 closely related serine proteases, KLK1–15,3 and show great promise as cancer biomarkers and therapeutic targets for ovarian cancer due to their biological substrates and signalling interactors present in the ovarian tumour microenvironment.4 At least 12 KLKs (KLK2-8, 10, 11, 13–15) are upregulated in ovarian cancer.3,5 In particular, elevated KLK4–7, 10 and 15 expression correlates with unfavourable prognosis and late-stage disease.6,7 KLK4 and KLK7 expression mediates resistance to the first-line chemotherapeutic paclitaxel in ovarian cancer.8,9 Accumulating evidence suggests that KLK4–7 together are part of an enzymatic cascade that promotes ovarian cancer.10 In fact, simultaneous overexpression of KLK4–7 in OV-MZ-6 ovarian cancer cells significantly increases cell invasion in vitro11 and tumour burden and metastatic spread in tumour xenograft models compared to KLK4–7-deficient control tumours.11,12 Moreover, KLK4–7-overexpressing ovarian cancer cells exhibit decreased cell adhesion and integrin expression as well as resistance to paclitaxel.13 Secreted proteome (secretome) and proteolytic profile (degradome) analyses revealed that KLK4–7 overexpression induced transforming growth factor beta-1 (TGFβ-1) signalling in ovarian cancer cells, suggesting a pronounced, yet non-degrading impact on the secretome and an association between KLK expression and the TGFβ-1 network in the tumour microenvironment.14
Thus, tumour-associated KLK4–7 overexpression significantly enhances ovarian cancer cell functions and the progression of this disease. However, the underlying mechanisms are not yet fully elucidated. KLK-mediated pericellular proteolysis occurs in various aspects of cancer cell proliferation, invasion and metastasis and in a complex microenvironment.15,16 The objective of this study was to determine genes and pathways regulated by KLK4–7 and to investigate the expression of these regulated factors in tumour tissues of ovarian cancer patients, potentially finding new biomarkers and therapeutic targets to treat the advanced stages of ovarian cancer.
Material and methods
Cell culture
Human OV-MZ-6 ovarian cancer cells were established from ascites of a patient with serous adenocarcinoma of the ovary and cultured as reported.17 OV-MZ-6 cells were stably co-transfected with an expression vector, pRcRSV (Invitrogen, Dreieich, Germany) harbouring human full-length cDNAs of KLK4–7, resulting in OV-KLK4–7 cells.11 Cells transfected with the vector only were used as controls (OV-VC cells).11
RT² Profiler PCR array
RNA was extracted from the OV-MZ-6-derived cell lines using the RNeasy Mini kit (Qiagen, Hilden, Germany) and the QIAcube fully automated spin column system (Qiagen), followed by DNase (Qiagen) treatment according to the manufacturer’s instructions. Reverse transcription was performed using a RT² First-Strand kit (Qiagen) and subsequent quantitative PCR analysis using a RT² SYBR Green PCR Mastermix (Qiagen) and primer pre-coated RT² Profiler PCR array plates (Qiagen), with 84 pathway- or disease-focused genes per plate. Three different human-specific PCR arrays were conducted: TGFβ/bone morphogenic protein (BMP), protease-activated receptor (PAR) and epithelial-to-mesenchymal transition (EMT) signalling pathways.
Quantitative reverse transcriptase-PCR
RNA was isolated from three biological replicates as described and reverse transcription was performed with the Cloned AMV First-Strand cDNA synthesis kit (Invitrogen). Quantitative reverse transcriptase-PCR analysis was performed using Universal ProbeLibray Probes (Roche, Mannheim, Germany), Brilliant III Ultra-Fast SYBR Green Low ROX Mastermix (Agilent Technologies, Waldbronn, Germany), 25 ng/well cDNA and 20 µM primers (Table S1).
Transcriptome-wide expression microarray
RNA was isolated as described above and a custom Agilent 4x180k oligo-array was performed using three biological replicates as reported.18 Microarray data were processed with Agilent’s Feature Extraction software (v10.7), and differential expression determined using a Bayesian adjusted t-statistic within the ‘Linear Models for Microarray Data’ (LIMMA) R-package. Normalised gene expression data of the experiment are Minimum Information About a Microarray Experiment (MIAME) compliant. Probes with a fold change (FC) of ≥2/≤−2 and false discovery rate (FDR) corrected P ≤ 0.05 between both groups were identified as significantly different. Pathway and gene enrichment analysis were used for functional gene annotations. Data have been deposited at the Gene Expression Omnibus (GEO) repository with the accession number GSE109261.
Secretome and degradome profiling
The secretome and degradome analysis has been performed previously.14 The fold changes were calculated for each replicate or averaged and included in Table 1.
Western blot analysis
Cell lysates of the OV-MZ-6-derived cell lines were generated using Tris-buffered saline containing 1% Triton X-100 and 0.1% complete proteases inhibitor cocktail (Roche). Protein concentrations were determined with the bicinchoninic acid protein assay (Thermo Fisher, Dreieich, Germany) and 40 µg/lane of protein was electrophoresed on a 12% sodium dodecyl sulfate (SDS) polyacrylamide gel and transferred onto polyvinylidene difluoride (PVDF) membranes (Millipore, Darmstadt, Germany) via semi-dry blotting. PVDF membranes were incubated with primary antibodies (Table S2) overnight at 4 °C and the secondary antibody (horseradish peroxidase (HRP)-conjugated anti-mouse/rabbit IgG, 1:5000; Millipore) 1 h at room temperature. Proteins were visualised using enhanced chemiluminescence substrate (Thermo Fisher) with a V3 Western Workflow (Bio-Rad, Dreieich, Germany). For Coomassie Blue staining, SDS polyacrylamide gels were fixed in 50% methanol plus 10% acetic acid, stained with 0.1% Coomassie R-250 blue (Thermo Fisher) solution (40% ethanol plus 10% acetic acid) and destained in 40% methanol plus 10% acetic acid.
Immunofluorescence
Immunofluorescence staining was performed as reported13 using primary (Table S2) and secondary antibodies (Alexa488-conjugated anti-mouse/rabbit IgG, 1:1000; Life Technologies, Melbourne, Australia), followed by DAPI counterstaining.
Immunohistochemistry using tumour xenograft tissues
Immunohistochemistry on tumour xenograft tissues was performed as reported14 using primary (Table S2) and HRP-conjugated anti-mouse/rabbit IgG (Dako, Hamburg, Germany). Antibody binding was visualised by 3,3′-diaminobenzidine (Dako), followed by Mayer’s hematoxylin counterstaining.
Immunohistochemistry using patient-derived tissues
Formalin-fixed, paraffin-embedded ovarian cancer tissues were obtained from the archives of the Department of Pathology of the Technical University of Munich. Tissue microarrays were produced as described previously.19 Briefly, samples were deparaffinised, blocked by 3% hydrogen peroxide and pressure-cooked in 10 mM citrate buffer, pH 6.0, for antigen retrieval. Samples were incubated with primary antibodies (Table S2) for 1 h and HRP-conjugated anti-mouse/rabbit IgG (Zytomed, Berlin, Germany) for 30 min at room temperature in a wet chamber. Antibody binding was visualised by 3,3′-diaminobenzidine (Dako), followed by Mayer’s hematoxylin counterstaining.
Statistical analysis
Data were analysed for statistical significance using SPSS 22.0. For comparison of mean values, the independent samples T-Test was used. Differences with a P-value ≤0.05 were considered statistically significant (P ≤ 0.05 is marked as *, P ≤ 0.01 as ** and P ≤ 0.001 as ***).
Results
KLK4–7 overexpression induces significant changes in gene expression in ovarian cancer cells
To identify genes and/or pathways regulated via simultaneous overexpression of KLK4–7 in ovarian cancer cells, three different PCR array analyses, targeting the TGFβ, PAR and EMT pathways, were performed. KLK4–7 gene expression was confirmed on the mRNA (Table S3) and protein levels via qPCR13 and enzyme-linked immunosorbent assay11 respectively. In total, 74 genes were found to be more than two-fold deregulated in OV-KLK4–7 compared to OV-VC cells (Table S4, Figure S1). In parallel, an independent transcriptome-wide RNA microarray (Figure S2) and a secretome14 analysis were performed, and 10 candidates were identified to be differentially expressed across all three approaches (Table 1). Pathway and gene enrichment analyses identified different molecular and cell functions, canonical pathways and upstream regulators being significantly regulated upon KLK4–7 expression (Table S5). To validate differential regulation of the candidate genes, qPCR analysis was performed using different probes and RNA samples compared to the PCR arrays. MSN, KRT19, COL5A2, COL1A2, BMP5 and F10 were significantly upregulated in OV-KLK4–7 compared to OV-VC cells in both PCR arrays and qPCR (Fig. 1a–f). KRT7, JUNB, BMP4 and MMP1 were significantly downregulated in OV-KLK4–7 compared to OV-VC cells in both PCR arrays and qPCR (Fig. 1g–k). Thus, at the mRNA level, all 10 candidate genes were found to be significantly differentially expressed following overexpression of KLK4–7 in ovarian cancer cells.
Analysis of mRNA expression in OV-KLK4–7 versus control OV-VC cells via PCR array and qPCR. Selected genes identified as being deregulated in OV-KLK4–7 (red columns) versus OV-VC (blue columns) cells in the PCR array were validated by qPCR using independently isolated RNA. OV-KLK4–7 cells displayed significant upregulation of MSN (a), KRT19 (b), COL5A2 (c), COL1A2 (d), BMP5 (e) and F10 (f) as well as downregulation of KRT7 (g), JUNB (h), BMP4 (j) and MMP1 (k) compared to OV-VC cells (*P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001)
KLK4–7 overexpression induces upregulation of MSN and KRT19 as well as downregulation of KLRT7 and JUNB proteins in ovarian cancer cells
Next, we sought to investigate the protein expression pattern of the highest deregulated candidate genes in all three independent analytical approaches. Thus, MSN, KRT19, KRT7 and JUNB were selected for analyses by western blotting (Fig. 2) and immunofluorescence (Fig. 3a) using the same human-specific monoclonal antibodies (Table S2). A strong upregulation of MSN and KRT19, together with downregulation of KRT7 and JUNB, was observed in OV-KLK4–7 compared to OV-VC cells. Downregulation of JUNB expression was more pronounced at the protein than at the mRNA level in OV-KLK4–7 cells, probably due to post-transcriptional regulation events. Immunofluorescence microscopy revealed specific staining patterns of the analysed proteins (Fig. 3a). While MSN showed a distinct cell membrane-associated staining, JUNB staining was associated with cell nuclei. Accordingly, MSN is a membrane-organising extension spike protein, whereas JUNB is a transcription factor. KRT19 and KRT7 are members of the large keratin gene family encoding typical intermediate filament proteins of epithelia. Thus, as expected, both KRT19 and KRT7 showed a cytoplasmic staining pattern, with a perinuclear staining for KRT7.
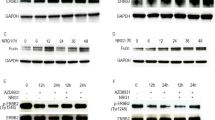
Western blot detection of differential protein expression of MSN, KRT19, KRT7 and JUNB in OV-KLK4–7 versus control OV-VC cells. Coomassie Blue staining (a) showed no obvious difference between OV-VC and OV-KLK4–7 protein patterns (40 µg protein/lane). Western blot analyses demonstrated upregulation of MSN (b) and KRT19 (c) and downregulation of KRT7 (d) and JUNB (e) in OV-KLK4–7 compared to OV-VC cells. GAPDH served as reference protein for equal loading and uniform transfer. Degradation products of the analysed proteins by limited proteolysis were co-visualised with MSN (b), KRT7 (d) and JUNB (e)
Visualisation of differential protein expression of MSN, KRT19, KRT7 and JUNB in OV-KLK4–7 versus control OV-VC cells and tumour xenograft tissues by immunofluorescence and immunohistochemistry. (a) An increased staining of MSN and KRT19 and a decreased staining of KRT7 and JUNB was observed in OV-KLK4–7 compared to OV-VC cells. MSN was cell membrane-associated, while JUNB was located in the nucleus. KRT19 and KRT7 showed a cytoplasmic staining pattern, with a perinuclear staining for KRT7 (scale bars, 25 µm). (b) In accordance with the immunofluorescence results, MSN and KRT19 showed increased and KRT7 and JUNB decreased immunohistochemical staining in OV-KLK4–7-derived tumour compared to OV-VC-derived tumour tissues (scale bars, 100 µm; insets: scale bars, 25 µm)
In a previous study, OV-KLK4–7 and OV-VC cells were encapsulated into hydrogels and implanted into NOD/SCID mice to form tumours in vivo.12 Formalin-fixed, paraffin-embedded tumour xenograft tissues from that study were used for immunohistochemical analysis of the four candidate proteins in this study using the same antibodies as above (Table S2). A strong staining of MSN and KRT19, together with a weak staining of KRT7 and JUNB was observed in OV-KLK4–7-derived tumour compared to OV-VC-derived tumour tissues (Fig. 3b). Overall, expression of MSN, KRT19, KRT7 and JUNB proteins was demonstrated to be clearly different in OV-KLK4–7 compared to OV-VC cells, as assayed via western blotting, immunofluorescence and immunohistochemistry.
Protein expression patterns of KLK4–7, MSN and KRT19 in tumour tissue of patients afflicted with advanced ovarian cancer
We sought to compare the relationship of KLK4–7 expression with that of MSN and KRT19, respectively, rather than the downregulated KRT7 and JUNB, on the protein level of all factors by immunohistochemistry in formalin-fixed, paraffin-embedded, patient-derived tissues. In previous studies, we have evaluated protein expression of KLK5 (ref. 19), KLK6 (ref. 20) and KLK7 (ref. 21) in ovarian cancer by immunohistochemistry. In the present study, we used a previously established protocol for KLK4, which has been used to assess KLK4 expression in breast cancer tissues.22 In addition, we established immunohistochemical protocols for MSN and KRT19, respectively. In Fig. 4, examples of MSN (Fig. 4a–c) and KRT19 (Fig. 4d–f) staining patterns are presented (for KLK4 see Figure S3), with an occasional distinct polar association of MSN with the cell membrane and KRT19 and KLK4 in the cytoplasm. For estimation of MSN, KRT19 or KLK4 immunoreactivity, a semi-quantitative score, based on staining intensity and percentage of positively stained cancer cells, was used. Scoring of KLK5, KLK6 and KLK7 immunostaining was previously done in a similar manner. In total, data for all factors were obtained from tumour tissues of 66 patients afflicted with advanced serous ovarian cancer (FIGO III/IV). The relationship of either KLK4, KLK5, KLK6 or KLK7 immuno-reactive score values with those of MSN and KRT19 is summarised in Table 2. A significant association was found for KLK7/MSN (P = 0.001), KLK4/KRT19 (P = 0.010) and KLK5/KRT19 (P = 0.030), while KLK6/MSN (P = 0.056) and KLK6/KRT19 (P = 0.075) showed a trend towards significance. In all cases, a lower KLK expression was associated with a lower expression of MSN or KRT19 and vice versa. In case of KLK4/MSN, in the KLK4 high-expressing group, a distinctly elevated number of MSN high-expressing cases (23 high versus 16 low) was observed. Similarly, KRT19 high-expressing cases were distinctly more frequent in the KLK7 high-expressing group (21 high versus 15 low). We also performed an in silico correlation analysis using the TCGA and ICGC high-grade serous ovarian cancer transcriptome data sets.23,24 Here, both MSN and KRT19 were found to be positively associated with KLK5, KLK6 and KLK7 (data not shown). Taken together, the immunohistochemistry data and the later in silico analyses are in line with the results obtained with the OV-MZ-6 ovarian cancer cell line and strongly indicate that overexpression of KLK4–7 in ovarian cancer patients is associated with an elevated MSN and KRT19 protein expression.
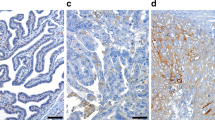
Immunohistochemical detection of MSN and KRT19 protein expression in high-grade serous ovarian cancer tissue samples. a–c Representative immunoexpression of MSN at low (a) and high (b) levels. Occasionally, a distinct polar association of MSN with the cell membrane was observed (c). d–f Representative micrographs corresponding to low (d), moderate (e) and high (f) cytoplasmic KRT19 immunoexpression (scale bar, 100 µm)
Discussion
Most members of the KLK serine protease family, including KLK4, 5, 6 and 7, are upregulated in ovarian cancer. Crosstalk between KLK4–7, possibly together with other proteases, may lead to modulation of diverse signalling pathways accompanied by complex interactions promoting both stimulatory and inhibitory signals. There may be a link between KLKs and the steroid hormone receptor pathway.25 These may contribute to tumorigenesis by affecting several tumour-associated processes, such as proliferation, survival, migration and invasion. In fact, our present results demonstrate that overexpression of KLK4–7 significantly regulates expression of other tumour-associated factors and pathways in OV-MZ-6 cells.
Using two different methods for mapping mRNA expression, qPCR arrays and a genome-wide RNA microarray, we identified several genes whose transcript might be affected by KLK4–7 overexpression. In addition to these RNA-based methods, we also considered data from our previous proteome study,14 in which the secretome profile of cell-conditioned medium of OV-KLK4–7 versus control OV-VC cells has been analysed by mass spectrometry. We selected 10 candidates for further analysis. Six of the 10 selected genes (MSN, KRT19, COL5A2, COL1A2, BMP5, F10) were upregulated upon KLK4–7 overexpression, while the other 4 genes were downregulated (KRT7, JUNB, BMP4, MMP1). MMP1 was only found significantly downregulated in the PCR array but not in the microarray and secretome analyses, thus was not further considered. Four of these genes (MSN, KRT19, KRT7, JUNB) are not annotated as secreted proteins. Therefore, one might have expected that these proteins would not be detectable in the secretome analysis. However, MSN and KRT19 were present in significantly higher amounts and KRT7 in significantly lower amounts in cell-conditioned medium on comparison of OV-KLK4–7 versus control OV-VC cells. This is in line with the detected differential mRNA expression. The presence of these proteins in cell-conditioned medium may result from exosomes and/or increased cell death during culture of the cells in serum-free medium to obtain the cell-conditioned medium. It should be noted that soluble protein fragments of several keratins, including KRT19, can be detected in the circulation of cancer patients and are, in fact, used to monitor tumour load and disease progression in certain tumour types.26
Differential mRNA expression of all 10 candidate genes was verified by qPCR, whereby different primers, different probes and also independently prepared mRNA for cDNA synthesis were used. We selected the two top-upregulated (MSN, KRT19) and two top-downregulated (KRT7, JUNB) genes to analyse whether this differential mRNA pattern in both cell lines is also reflected at the protein level in cell culture and tumour xenograft models. Indeed, we validated a strong increase of MSN and KRT19 and a clear decrease of KRT7 and JUNB in OV-KLK4–7 versus control OV-VC cells by western blot analysis, immunofluorescence and immunohistochemistry.
Jun proteins (c-Jun, JunB and JunD) are important in the regulation of cell proliferation, apoptosis and angiogenesis and associated with tumorigenesis and progression of several tumour types. Together with Fos proteins, they form different homo- and heterodimers, named AP-1 transcription factors.27 In combination with c-Fos, c-Jun expression supports tumorigenesis, whereas JunB is an inhibitor of cell division, an inducer of senescence and a tumour suppressor.28,29,30,31 In the present study, JunB showed downregulation upon overexpression of KLK4–7 in ovarian cancer cells, which may increase cell proliferation and promote tumorigenesis.
Among the top-regulated candidates, we identified a pair of genes, KRT7 and KRT19, encoding the intermediate filament proteins keratins 7 and 19, whose expression was inversely affected by overexpression of KLK4–7. The family of keratins consists of two major subgroups, the type I and type II keratins, that form obligate heterodimers by pairing of a type I with a type II keratin, which then assemble into filamentous polymers. Besides being important structural stabilisers of epithelial cells, keratins also have other cellular functions, including maintenance of cell integrity, positioning of subcellular organelles, signalling and protection from apoptosis.26 In malignant tumours, keratin 8/18 is the most prominent keratin pair. However, keratins 7 (type II) and 19 (type I) are frequently expressed and also occur as a keratin pair. Interestingly, in the absence of keratin 7, keratin 19 hetero-dimerises with keratin 8.26 Our findings may indicate that reduction of keratin 7 expression with concomitant increase of keratin 19 may induce a switch for keratin 7/19 (and 8/18) to a novel keratin dimer composed of keratin 8/19 (and 8/18). Besides ductal breast carcinomas and lung adenocarcinomas, non-mucinous ovarian carcinomas tend to lose keratin 7 expression upon metastasis.32
Keratin 19 lacks the carboxy-terminal tail domain typical for all other keratins and functionally overlaps with keratin 18. As stated above, cancer cells release soluble keratin 19 fragments. Serum levels of keratin 19 fragments (which are detected by the so-called Cyfra 21-1 assay) were found to be significantly higher in patients with ovarian cancer compared with those with benign ovarian disease.33,34 Furthermore, Cyfra 21-1 serum levels were significantly higher in serous versus non-serous types of ovarian cancer and higher in advanced (FIGO III/IV) versus early (FIGO I/II) stage disease. Interestingly, high pre-operative Cyfra 21-1 serum levels were associated with increased chemoresistance of ovarian cancer patients. Thus, increased release of keratin 19 fragments into the circulation may be an indicator of a more aggressive disease. In the study by Wu et al.34 Cyfra 21-1 was proposed to be of prognostic value. However, the analysed patient cohort was very heterogeneous encompassing different subtypes of ovarian cancer as well as early and advanced disease stages. To test whether increased serum levels of keratin 19 fragments are of prognostic relevance, homogeneous patient cohorts using only advanced high-grade serous ovarian cancer patients have to be analysed. In hepatocellular carcinoma, pre-operative Cyfra 21-1 serum levels were positively correlated with KRT19 expression. Additionally, KRT19 was linked to poor prognosis, cancer cell proliferation, invasion, TGFβ signalling and EMT. KRT19-positive cells showed sensitivity to TGFβ receptor inhibition, which suppressed TGFβ signalling and reduced tumour growth in a xenograft model.35
Our data strongly suggest an association between KLK4–7 and KRT19 expression in ovarian tumour tissues. As shown by immunohistochemical analysis, expression of both KLK4 and KLK5 was significantly and positively associated with KRT19 expression; KLK6 showed a trend towards significance. In the case of KLK7, the group of KLK7 high-expressing cases encompassed distinctly more high KRT19-expressing tumours (21 high versus 15 low). Taken together, the published data and our findings in the present study indicate that upregulation of KRT19, induced by KLK4–7 overexpression, may increase malignancy of ovarian cancer cells and furthermore be an explanation for our previous report from Loessner et al.13 that OV-KLK4–7 ovarian cancer cells were less sensitive to paclitaxel treatment compared to OV-VC cells.
Besides KRT19, MSN was identified as a strongly upregulated factor upon overexpression of KLK4–7. MSN is a membrane-organising extension spike protein, which belongs to the ERM family encompassing three homologous members: ezrin, radixin and moesin. MSN functions as a cross-linker between the plasma membrane and the cytoskeleton and is involved in cell–cell recognition, signalling and cell movement. High expression of MSN was positively correlated with disease progression in different tumour types.36,37 In ovarian cancer, ezrin and moesin showed strong expression in 49% and 48% of tumour tissues, respectively, and their expression correlated with reduced overall survival.38 Similar to KRT19, we observed distinct associations of MSN expression with KLK4–7 expression in tissues from serous ovarian cancer patients. MSN significantly and positively correlated to KLK7 expression; KLK6 showed a trend towards significance. In case of KLK4/MSN, a distinctly elevated number of MSN high-expressing cases (23 high versus 16 low) was observed in the KLK4 high-expressing group. This finding may indicate that upon overexpression of KLK4–7 an increased MSN expression promotes a malignant phenotype of ovarian cancer cells.
In muscular dystrophy, MSN, TGFβ and collagen type-I levels were found to be elevated and linked to cell motility and invasion. Enhanced immunoexpression of MSN and collagen type-I was observed in patient tissues, suggesting that MSN might be a potential disease-associated biomarker.39 Dislocation of ERM proteins into the cytoplasma alters cell-to-cell communications and induces cytoskeleton remodelling, subsequently promoting cancer progression and metastatic processes like EMT. Upregulation and phosphorylation of MSN, and other ERM proteins, are prerequisites for a malignant phenotype in other tumour types, for example in glioblastoma,40 melanoma,41 cervical cancer,42 oral squamous cell carcinoma,43 diffuse large B-cell lymphoma44 and breast cancer.45 MSN was positively correlated with tumour grade of breast cancers, depicting a cytoplasmic and membranous staining pattern mostly in normal adjacent tissues.45 Interaction of the tumour suppressor BRCA1 with ERM proteins reduced spreading and motility of breast cancer cells.46 ERM proteins also mediated multidrug resistance of breast cancer cells as they control both membrane insertion and functionality of P-glycoprotein that actively unloads chemotherapeutics from resistant cancer cells.47 The study by Ansa-Addo et al.48 showed that MSN is translationally regulated by TGFβ and required for induced regulatory T cell (iTreg) development and TGFβ signalling. Loss of MSN caused a significant reduction in the generation of iTregs due to inefficient TGFβ signalling. This reduced amount of iTregs resulted in a delayed tumour recurrence in a melanoma animal model.48 The findings from our study are in accordance with these above-mentioned reports and suggest that ERM proteins control various signalling pathways, including TGFβ and EMT cascades. Downregulation and/or inactivation of ERM proteins may be effective in abolishing tumour growth. Our results demonstrate that MSN is significantly upregulated upon KLK4–7 overexpression in ovarian cancer cells compared to controls. So far, upregulation of MSN has not been shown in ovarian cancer cells compared to normal ovarian tissue and if the upregulated MSN is in its phosphorylated form. This will be subject of future studies. Increased MSN levels may be responsible for tumour progression and metastasis in concert with elevated expression of KLK4–7 in ovarian tumours and thus may be combated by combination therapy regimes using MSN or KLK inhibitors. Reduction of MSN, as induced via inhibition of KLKs, may also lead to a decreased generation of iTregs delaying tumour recurrence and may reverse multidrug resistance of tumour cells.
The herein identified KLK4–7-related factors are key members of the TGFβ and BMP network that is a well-known modulator of EMT in cancer progression and metastasis. The TGFβ receptor activin receptor-like kinase 1 (ALK1) interacts with BMP9 and BMP10 and is therapeutically pursued to interfere with TGFβ and BMP binding and downstream signalling, thus inhibiting tumour angiogenesis. Combination of the ALK1 inhibitor, dalantarcept, with doxorubicin and cisplatin enhanced the efficacy of these chemotherapeutics and impaired growth of solid tumours in different animal models.49 The BMP–SMAD pathway and its activation status are associated with enhanced cell proliferation and poor prognosis in serous ovarian cancer, thus representing a potential therapeutic target for this disease.50 The multi-kinase inhibitors dorsomorphin and LDN-193189, which also interfere with BMP signalling, have shown efficacy in an intraperitoneal xenograft model and in resensitising platinum-resistant ovarian cancer cells, thus are promising alternatives for the adjuvant therapy of chemoresistant disease.51 The combination of TGFβ/BMP inhibitors with KLK-targeting agents may be an alternative to treat ovarian cancer. However, one needs to have in mind that different members of the TGFβ superfamily have overlapping and opposing functions in a tissue-specific context and TGFβ receptor signalling is controlled by a variety of extracellular factors, including proteases, growth factors, cytokines and ECM molecules.52 Their roles in cancer cell proliferation, metastatic and immuno-regulatory processes are complex and tightly regulated by the tumour microenvironment.
Future research will include a functional characterisation of the top-regulated factors in other KLK-expressing cell lines or treating OV-KLK4–7 cells with recombinant TGFβ. As we only focused on factors significantly regulated in all three approaches, future studies might validate other significant genes from the PCR array that are involved in KLK-mediated functions, e.g. cancer cell invasion and resistance to paclitaxel. In addition, some of the previous studies did not distinguish expression in individual ovarian cancer histotypes, which is an essential factor to understand the relevance of expression within different ovarian cancer histotypes. Herein, we evaluated the expression of KLK4–7 in high-grade serous ovarian cancer cells and tissues in a homogeneous patient cohort.
Conclusions
The association of the expression of different KLKs with those of MSN and KRT19 indicate that coordinate presence of KLK4–7 may be critical to induce the observed modulations of KLK-targeted genes. In fact, in contrast to KLK4–7 overexpression in ovarian cancer cells, overexpression of individual KLKs (e.g. KLK7) or other combinations (e.g. KLK5 + KLK7) does not lead to increased tumour growth and spread in an ovarian cancer xenograft model (P. Prezas, A. Krüger, V. Magdolen, unpublished results). KLK-mediated extracellular proteolysis is involved in many steps of cancer development and progression, including cell growth, angiogenesis, invasion and metastasis. In addition to degradation effects, KLKs can also activate signalling cascades, including TGFβ and EMT pathways, independent of their degrading function.14 Overexpression of KLK4–7 distinctly regulates the expression of other factors in OV-MZ-6 ovarian cancer cells, which are strongly associated with tumorigenesis. This may explain the findings in our previous publications.11,12,13,14,53 In an in vitro invasion assay,11 we observed a significantly increased invasive behaviour of OV-KLK4–7 versus control OV-VC cells. In tumour xenograft studies,11,12 we detected a significant role of KLK4–7 expression in tumour growth and spread. Our data imply that KLK4–7 exert key modulating effects on cancer-related targets and provide preclinical evidence for combination therapy regimens of MSN or TGFβ/BMP inhibitors with KLK-targeting agents for women afflicted with ovarian cancer.
Availability of data and material
All data generated or analysed during this study are included in this published article and its supplementary information files.
References
Siegel, R. L., Miller, K. D. & Jemal, A. Cancer statistics, 2018. CA Cancer J. Clin. 68, 7–30 (2018).
Wright, J. D. et al. Trends in relative survival for ovarian cancer from 1975 to 2011. Obstet. Gynecol. 125, 1345–1352 (2015).
Borgono, C. A. & Diamandis, E. P. The emerging roles of human tissue kallikreins in cancer. Nat. Rev. Cancer 4, 876–890 (2004).
Dorn, J. et al. Function and clinical relevance of kallikrein-related peptidases and other serine proteases in gynecological cancers. Crit. Rev. Clin. Lab. Sci. 51, 63–84 (2014).
Yousef, G. M. et al. Parallel overexpression of seven kallikrein genes in ovarian cancer. Cancer Res. 63, 2223–2227 (2003).
Yousef, G. M. & Diamandis, E. P. The human kallikrein gene family: new biomarkers for ovarian cancer. Cancer Treat. Res. 149, 165–187 (2009).
Yousef, G. M. et al. Prognostic value of the human kallikrein gene 15 expression in ovarian cancer. J. Clin. Oncol. 21, 3119–3126 (2003).
Dong, Y. et al. Paclitaxel resistance and multicellular spheroid formation are induced by kallikrein-related peptidase 4 in serous ovarian cancer cells in an ascites mimicking microenvironment. PLoS ONE 8, e57056 (2013).
Dong, Y. et al. Kallikrein-related peptidase 7 promotes multicellular aggregation via the alpha(5)beta(1) integrin pathway and paclitaxel chemoresistance in serous epithelial ovarian carcinoma. Cancer Res. 70, 2624–2633 (2010).
Dong, Y. et al. Metastasis of ovarian cancer is mediated by kallikrein related peptidases. Clin. Exp. Metastasis 31, 135–147 (2014).
Prezas, P. et al. Overexpression of the human tissue kallikrein genes KLK4, 5, 6, and 7 increases the malignant phenotype of ovarian cancer cells. Biol. Chem. 387, 807–811 (2006).
Loessner, D. et al. A bioengineered 3D ovarian cancer model for the assessment of peptidase-mediated enhancement of spheroid growth and intraperitoneal spread. Biomaterials 34, 7389–7400 (2013).
Loessner, D. et al. Combined expression of KLK4, KLK5, KLK6, and KLK7 by ovarian cancer cells leads to decreased adhesion and paclitaxel-induced chemoresistance. Gynecol. Oncol. 127, 569–578 (2012).
Shahinian, H. et al. Secretome and degradome profiling shows that Kallikrein-related peptidases 4, 5, 6, and 7 induce TGFbeta-1 signaling in ovarian cancer cells. Mol. Oncol. 8, 68–82 (2014).
Fuhrman-Luck, R. A. et al. Proteomic and other analyses to determine the functional consequences of deregulated kallikrein-related peptidase (KLK) expression in prostate and ovarian cancer. Proteomics Clin. Appl. 8, 403–415 (2014).
Silva, L. M. & Clements, J. A. Mass spectrometry based proteomics analyses in kallikrein-related peptidase research: implications for cancer research and therapy. Expert Rev. Proteomics 14, 1119–1130 (2017).
Mobus, V. et al. Morphological, immunohistochemical and biochemical characterization of 6 newly established human ovarian carcinoma cell lines. Int. J. Cancer 52, 76–84 (1992).
Sieh, S. et al. Phenotypic characterization of prostate cancer LNCaP cells cultured within a bioengineered microenvironment. PLoS ONE 7, e40217 (2012).
Dorn, J. et al. Assessment of kallikrein-related peptidase 5 (KLK5) protein expression in tumor tissue of advanced ovarian cancer patients by immunohistochemistry and ELISA: correlation with clinical outcome. Am. J. Cancer Res. 6, 61–70 (2016).
Seiz, L. et al. Stromal cell-associated expression of kallikrein-related peptidase 6 (KLK6) indicates poor prognosis of ovarian cancer patients. Biol. Chem. 393, 391–401 (2012).
Dorn, J. et al. Clinical value of protein expression of kallikrein-related peptidase 7 (KLK7) in ovarian cancer. Biol. Chem. 395, 95–107 (2014).
Yang F. et al. Tissue kallikrein-related peptidase 4 (KLK4), a novel biomarker in triple-negative breast cancer. Biol. Chem. 398, 1151–1164 (2017).
Cancer Genome Atlas Research N. Integrated genomic analyses of ovarian carcinoma. Nature 474, 609–615 (2011).
Patch, A. M. et al. Whole-genome characterization of chemoresistant ovarian cancer. Nature 521, 489–494 (2015).
Yousef, G. M. & Diamandis, E. P. Kallikreins, steroid hormones and ovarian cancer: is there a link? Minerva Endocrinol. 27, 157–166 (2002).
Moll, R., Divo, M. & Langbein, L. The human keratins: biology and pathology. Histochem. Cell Biol. 129, 705–733 (2008).
Ameyar, M., Wisniewska, M. & Weitzman, J. B. A role for AP-1 in apoptosis: the case for and against. Biochimie 85, 747–752 (2003).
Bakiri, L., Lallemand, D., Bossy-Wetzel, E. & Yaniv, M. Cell cycle-dependent variations in c-Jun and JunB phosphorylation: a role in the control of cyclin D1 expression. EMBO J. 19, 2056–2068 (2000).
Passegue, E. & Wagner, E. F. JunB suppresses cell proliferation by transcriptional activation ofp16(INK4a) expression. EMBO J. 19, 2969–2979 (2000).
Passegue, E., Jochum, W., Schorpp-Kistner, M., Mohle-Steinlein, U. & Wagner, E. F. Chronic myeloid leukemia with increased granulocyte progenitors in mice lacking junB expression in the myeloid lineage. Cell 104, 21–32 (2001).
Piechaczyk, M. & Farras, R. Regulation and function of JunB in cell proliferation. Biochem. Soc. Trans. 36(Pt 5), 864–867 (2008).
Tot, T. Cytokeratins 20 and 7 as biomarkers: usefulness in discriminating primary from metastatic adenocarcinoma. Eur. J. Cancer 38, 758–763 (2002).
Gadducci, A. et al. The clinical relevance of serum CYFRA 21-1 assay in patients with ovarian cancer. Int. J. Gynecol. Cancer 11, 277–282 (2001).
Wu, H. H. et al. Serum cytokeratin-19 fragment (Cyfra 21-1) is a prognostic indicator for epithelial ovarian cancer. Taiwan J. Obstet. Gynecol. 53, 30–34 (2014).
Kawai, T. et al. Identification of keratin 19-positive cancer stem cells associating human hepatocellular carcinoma using CYFRA 21-1. Cancer Med. 6, 2531–2540 (2017).
Clucas, J. & Valderrama, F. ERM proteins in cancer progression. J. Cell Sci. 127(Pt 2), 267–275 (2014).
Vitorino, P. et al. MAP4K4 regulates integrin-FERM binding to control endothelial cell motility. Nature 519, 425–430 (2015).
Kobel, M. et al. Ezrin promotes ovarian carcinoma cell invasion and its retained expression predicts poor prognosis in ovarian carcinoma. Int. J. Gynecol. Pathol. 25, 121–130 (2006).
Pines, M. et al. Elevated expression of Moesin in muscular dystrophies. Am. J. Pathol. 187, 654–664 (2017).
Qin, Y. et al. MiR-200c inhibits the tumor progression of glioma via targeting moesin. Theranostics 7, 1663–1673 (2017).
Riecken, L. B. et al. CPI-17 drives oncogenic Ras signaling in human melanomas via Ezrin-Radixin-Moesin family proteins. Oncotarget 7, 78242–78254 (2016).
Adada, M. M. et al. Intracellular sphingosine kinase 2-derived sphingosine-1-phosphate mediates epidermal growth factor-induced ezrin-radixin-moesin phosphorylation and cancer cell invasion. FASEB J. 29, 4654–4669 (2015).
Li, Y. Y., Zhou, C. X. & Gao, Y. Moesin regulates the motility of oral cancer cells via MT1-MMP and E-cadherin/p120-catenin adhesion complex. Oral Oncol. 51, 935–943 (2015).
Pore, D. et al. Identification of Ezrin-Radixin-Moesin proteins as novel regulators of pathogenic B-cell receptor signaling and tumor growth in diffuse large B-cell lymphoma. Leukemia 29, 1857–1867 (2015).
Bartova, M. et al. Expression of ezrin and moesin in primary breast carcinoma and matched lymph node metastases. Clin. Exp. Metastasis 34, 333–344 (2017).
Coene, E. D. et al. A novel role for BRCA1 in regulating breast cancer cell spreading and motility. J. Cell Biol. 192, 497–512 (2011).
Pokharel, D. et al. The role of CD44 and ERM proteins in expression and functionality of P-glycoprotein in breast cancer cells. Molecules 21, 290 (2016).
Ansa-Addo, E. A. et al. Membrane-organizing protein moesin controls Treg differentiation and antitumor immunity via TGF-beta signaling. J. Clin. Invest. 127, 1321–1337 (2017).
Hawinkels, L. J. et al. Activin receptor-like kinase 1 ligand trap reduces microvascular density and improves chemotherapy efficiency to various solid tumors. Clin. Cancer Res. 22, 96–106 (2016).
Peng, J. et al. The BMP signaling pathway leads to enhanced proliferation in serous ovarian cancer—a potential therapeutic target. Mol. Carcinog. 55, 335–345 (2016).
Ali, J. L. et al. Differential cellular responses induced by dorsomorphin and LDN-193189 in chemotherapy-sensitive and chemotherapy-resistant human epithelial ovarian cancer cells. Int. J. Cancer 136, E455–E469 (2015).
Chen, W. & Ten Dijke, P. Immunoregulation by members of the TGFbeta superfamily. Nat. Rev. Immunol. 16, 723–740 (2016).
Dorn, J. et al. Function and clinical relevance of kallikrein-related peptidases and other serine proteases in gynecological cancers. Crit. Rev. Clin. Lab. Sci. 51, 63–84 (2014).
Acknowledgements
The authors thank Melanie L. Lehman and Colleen C. Nelson for the design of the custom Agilent 4x180k oligo-array.
Author’s contributions
D.L. conceptualised and designed this study. V.M., M.S., J.A.C. and D.L. designed experiments and supervised this study. P.W., E.D., M.K., F.Y., O.S., A.R. and D.L. performed experiments and data analyses. J.D. and E.D. provided patient-derived samples. P.W., V.M., C.S. and D.L. wrote and reviewed the manuscript. All authors read and approved the final manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethics approval
Patient-derived tissue samples were collected between 1990 and 2012 at the Department of Obstetrics and Gynecology, Technical University of Munich, Klinikum rechts der Isar, Germany. The study was approved by the Ethics Committee of the Faculty of Medicine, Technical University Munich (1230/04) and written informed consent was obtained from all patients.
Competing interests
The authors declare no competing interests.
Funding
This study was supported in part by the German Research Foundation, the Wilhelm Sander-Stiftung, the National Health and Medical Research Council of Australia, Cancer Council Queensland, Movember Foundation and the Prostate Cancer Foundation of Australia through a Movember Revolutionary Team Award, Queensland University of Technology and the German Academic Exchange Service (German-Australian Network for Personalized Medicine).
Note
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 4.0 Unported License.
Electronic supplementary material
Rights and permissions
This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Wang, P., Magdolen, V., Seidl, C. et al. Kallikrein-related peptidases 4, 5, 6 and 7 regulate tumour-associated factors in serous ovarian cancer. Br J Cancer 119, 1–9 (2018). https://doi.org/10.1038/s41416-018-0260-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41416-018-0260-1
This article is cited by
-
A blood-based composite panel that screens Alzheimer’s disease
Biomarker Research (2023)
-
The long non-coding RNA keratin-7 antisense acts as a new tumor suppressor to inhibit tumorigenesis and enhance apoptosis in lung and breast cancers
Cell Death & Disease (2023)
-
Remodelling of the tumour microenvironment by the kallikrein-related peptidases
Nature Reviews Cancer (2022)
-
Benchmarking of analysis strategies for data-independent acquisition proteomics using a large-scale dataset comprising inter-patient heterogeneity
Nature Communications (2022)
-
Evaluation of hTERT, KRT7, and survivin in urine for noninvasive detection of bladder cancer using real‐time PCR
BMC Urology (2021)