Abstract
Cutaneous neurofibromas (cNF) are a nearly ubiquitous symptom of neurofibromatosis type 1 (NF1), a disorder with a broad phenotypic spectrum caused by germline mutation of the neurofibromatosis type 1 tumour suppressor gene (NF1). Symptoms of NF1 can include learning disabilities, bone abnormalities and predisposition to tumours such as cNFs, plexiform neurofibromas, malignant peripheral nerve sheath tumours and optic nerve tumours. There are no therapies currently approved for cNFs aside from elective surgery, and the molecular aetiology of cNF remains relatively uncharacterised. Furthermore, whereas the biallelic inactivation of NF1 in neoplastic Schwann cells is critical for cNF formation, it is still unclear which additional genetic, transcriptional, epigenetic, microenvironmental or endocrine changes are important. Significant inroads have been made into cNF understanding, including NF1 genotype–phenotype correlations in NF1 microdeletion patients, the identification of recurring somatic mutations, studies of cNF-invading mast cells and macrophages, and clinical trials of putative therapeutic targets such as mTOR, MEK and c-KIT. Despite these advances, several gaps remain in our knowledge of the associated pathogenesis, which is further hampered by a lack of translationally relevant animal models. Some of these questions may be addressed in part by the adoption of genomic analysis techniques. Understanding the aetiology of cNF at the genomic level may assist in the development of new therapies for cNF, and may also contribute to a greater understanding of NF1/RAS signalling in cancers beyond those associated with NF1. Here, we summarise the present understanding of cNF biology, including the pathogenesis, mutational landscape, contribution of the tumour microenvironment and endocrine signalling, and the historical and current state of clinical trials for cNF. We also highlight open access data resources and potential avenues for future research that leverage recently developed genomics-based methods in cancer research.
Similar content being viewed by others
Neurofibromatosis type 1: aetiology and symptoms
Neurofibromatosis type 1 (NF1) is a genetic disorder that affects 1:2600–1:4500 live births.1,2 The disease has nearly complete penetrance, and patients present with a diverse spectrum of manifestations. Hallmark traits of NF1 include café-au-lait spots, cutaneous neurofibromas (cNFs, or dermal neurofibromas), plexiform neurofibromas (pNFs) and malignant peripheral nerve sheath tumours (MPNSTs), among other symptoms.3,4 Although the disorder was first clinically described in the 1800s, it was not until much later that the neurofibromatosis type 1 gene (NF1) was identified.5,6,7,8 NF1 is caused by inherited or de novo germline mutations in the NF1 tumour suppressor gene, and it is thought that somatic loss-of-function of the second allele results in the development of tumours such as pNFs, MPNSTs and cNFs.
NF1-linked tumours present with differing frequencies across NF1 patients. PNFs are benign nerve sheath tumours that occur in ~40% of NF1 patients.9,10 Notably, pNFs have the capacity to develop into MPNSTs, which affect 6–13% of NF1 patients.11,12 Unlike pNFs, cNFs do not progress to malignancy, but they are observed in >99% of adult patients and can range widely in both size and number (Fig. 1).13,14,15,16 While these tumours are generally observed during puberty and pregnancy, cNFs are sometimes observed before 5 years of age.17,18 These benign cNF tumours can cause itching, pain and a cosmetic burden that has been linked with psychosocial challenges.15,19 Therefore, NF1 patients often identify the cNF tumours as their greatest burden. Additionally, treatment options for cNF are limited to elective surgical approaches, which presents a challenge for patients with thousands of tumours. The term 'cutaneous neurofibroma' (cNF) is used in this review to describe benign neurofibromas that are found exclusively within the cutaneous dermis layer; cNFs are therefore also often called dermal neurofibromas. In addition, 'benign neurofibroma' can also refer to non-cNFs including benign subcutaneous, internal, diffuse or plexiform neurofibromas. Currently, there are no well-defined subtypes of cNF that represent stages of tumour growth or phenotypically distinct cutaneous neurofibromas; elucidating these subtypes is an important challenge for the field.
Cutaneous neurofibromas in NF1 patients. Cutaneous neurofibromas occur in nearly all NF1 patients, but they present with great diversity in both tumour frequency and tumour size.13,14,15,16 These tumours represent one of the most challenging burdens for neurofibromatosis type 1 patients.15,16,19 Images are used with patient permission
While many studies have contributed to our understanding of cNF, there are still large gaps that remain to be explored. In this review, we summarise the present understanding of cNF pathogenesis including recent studies that propose a putative tumour cell of origin, the identification of mutations in NF1 and other genes in cNF, the role of the tumour microenvironment and endocrine signalling in cNF, and past, recent and ongoing clinical trials for cNF. We also propose the application of genomics-oriented approaches for future investigations of cNF.
Histology, origin and pathogenesis of cNF
cNFs manifest as small (2 mm–3 cm), circumscribed tumours that associate with nerves in the skin.14,20 Clinically, they can undergo a rapid initial proliferative phase but then quickly become quiescent with extremely slow to no growth.21 Their diverse composition is similar to that of the nerve sheath, consisting of Schwann cells, perineural cells, fibroblasts and a collagenous matrix.14,22,23 cNFs also contain an infiltrating immune cell population that is comprised of macrophages and mast cells.23,24,25 Studies in mouse models have identified a critical role for mast cells in pNF formation, but it is unknown whether mast cells and macrophages contribute to the growth of cNF tumours.23,24,26 Histologically, cNFs share similar markers to other NF1-associated nerve tumours such as pNFs, and can be identified by wavy nuclei, S100 positivity and the expression of collagen IV, Sox10, CD34 and CD44. Of note, S100 and Sox10 can be used as diagnostic markers.20,27,28
The cNF-tumour-initiating event in humans is currently unknown and the cell of origin has yet to be identified; however, studies in mice have led to the identification of Schwann-cell-like skin-derived precursor (SKP) cells as the likely murine cNF tumour cell of origin. Wu and colleagues observed cNF-like tumours in the Nf1fl/fl; DhhCre mouse model, and proposed that these tumours may originate from neural-crest-derived cells in the hair follicles.29 Shortly thereafter, Le et al. found that cNFs can originate from a population of neural-crest-derived progenitors residing in the dermis, identified as SKPs.30 These authors generated CMV-CreERT2;Nf1flox/-;ROSA26 mice and ablated Nf1 in the presumed cNF cell of origin in the skin; this was achieved with cutaneous application of tamoxifen.30 They observed the formation of cNFs at the tamoxifen application sites 6–7 months later, suggesting that the cNF cell of origin resides in the skin. Nf1-deficient SKPs also have the capacity to form pNFs or cNFs, which appears to be contingent on their local microenvironment, and these cells exhibit the same properties as the embryonic Schwann cell progenitors that give rise to pNFs.29,30 These observations revealed that loss of Nf1 gene expression in SKPs is required, but not sufficient, for neurofibroma development, and revealed critical roles for the tumour microenvironment. Consistently, when Nf1−/− SKPs were autologously implanted intradermally, they efficiently gave rise to cNFs in mice that were pregnant at the time of implantation.30 This suggested that the hormonal milieu can facilitate induction of cNF development from Nf1-deficient SKPs in the skin. More recent work demonstrated that implantation of Nf1−/− SKPs in sciatic nerve tissue, but not subcutaneous implantation in unprimed athymic mice, resulted in neurofibroma formation, further indicating that the microenvironment may be a key regulator of cNF development.24
It is not yet known whether human cNFs also originate from SKPs or other cell populations. Longitudinal natural history studies are currently underway that may address this gap.31,32 Considering this information, the natural history of cNF is poorly understood for multiple reasons: patients can develop many cNFs throughout life, cNF burden varies greatly among NF1 patients and the precise sequence and nature of all cNF-initiating events is unknown. This limited understanding of cNF natural history is one obstacle in generating models of the disease but there are several other challenges to developing translationally relevant models of cNF. cNFs are slow-growing benign tumours that may take more than a year to develop, possibly beyond the useful lifespan of commonly used experimental organisms. In addition, although the Nf1−/− Schwann cell is the neoplastic cell in cNFs, human cNFs involve multiple other cell types and a complex extracellular matrix, making it challenging to recapitulate the human disease in vivo as well as in vitro. Finally, in vivo and in vitro cNF models must originate from the same cell of origin as the human tumour, they must be driven to tumourigenesis by the same genetic drivers (intrinsic factors), undergo similar downstream transcription profiles, develop within a similar tumour microenvironment (extrinsic factors) and must be robust for preclinical screening. Many of these key elements of cNF remain to be clearly defined.
Role of NF1 mutations in the phenotype of cNF
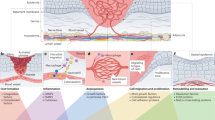
The NF1 gene product, neurofibromin, plays an important role in regulating cellular transcription, proliferation and survival (Fig. 2a). Ligand-activated receptor tyrosine kinases drive Ras activation by signalling via Ras guanidine exchange factors, such as SOS;33 however, NF1 is a Ras GTPase activating protein, and thus facilitates Ras inactivation.33,34 In cells lacking NF1, Ras activation is not inhibited by NF1, which in turn induces transcriptional changes and proliferative signalling via the RAF-MAP kinase pathway, and prosurvival signalling via the PI3K-mTOR axis.33,35 Consequently, loss or mutation of NF1 is an important step in NF1 patient tumourigenesis. Of note, next-generation sequencing has recently demonstrated that the NF1 gene is frequently mutated in sporadic cancers not usually observed in NF1 patients,36 such as conjunctival melanoma.37
The RAS signalling cascade and tumour microenvironment in cNF. a Loss of NF1 reduces the ability of Ras to hydrolyse GTP and shift from an active to a GDP-bound inactive state. Consequently, Schwann cells lacking NF1 have increased proliferation and altered transcription (RAF-MEK-ERK signalling) and increased prosurvival signalling (PI3K-mTOR). b The cNF microenvironment is composed of NF1-deficient Schwann cells, mast cells, fibroblasts, macrophages and neurons, among other cell types. While roles for some of these cell types have been studied or hypothesised, particularly with regard to mast cells, the bulk of these cellular interactions are poorly understood or unstudied
The mutational landscape of the NF1 gene, however, is more diverse in NF1 patients; germline mutations are observed across the entire 3.5 kb NF1 sequence.6,7,8,38,39 In contrast to oncogenic mutations, which often occur at specific loci and result in hyperactivation of the gene product, inactivating mutations observed in tumour suppressor genes such as NF1 are not site specific. For example, a survey of germline NF1 mutations in 189 NF1 patients determined that 45% of patients harboured one of 38 recurrent NF1 mutations detected in this study (for example, NF1 c.910C>T, NF1 c. 1885G>A and NF1 c.6792C>A), while 55% of patients had a unique NF1 mutation.38 Other studies have similarly identified many novel germline NF1 mutations across the length of the NF1 coding region with no evidence of mutational hotspots.40,41 One database, the Leiden Open Variation Database – NF1 (https://databases.lovd.nl/shared/genes/NF1), has identified over two thousand unique germline NF1 variants.39
In addition to point mutations, germline NF1 loss can be caused by microdeletion or pathogenic intronic mutations.42,43 The relationship between germline NF1 microdeletion and cNF is somewhat clearer, compared with our understanding of the role of germline and somatic intragenic NF1 mutations. Studies suggest that germline NF1 microdeletions guide cNF development; cNFs that form in NF1 microdeletion patients do not exhibit somatic loss of heterozygosity (LOH) of the second NF1 allele, but instead typically contain point mutations in NF1.44 NF1 microdeletion patients generally exhibit more severe symptoms than the general NF1 patient population, an effect attributed to the deletion of one or many modifying genes neighbouring NF1, such as CRLF3, ATAD5, OMG, RAB11FIP4, SUZ12, LRRC37B and several others.45,46 Generally, these NF1 microdeletions fall into one of four categories: type 1 (1.4 Mb, 70–80% of deletions), type 2 (1.2 Mb, >10% of deletions), type 3 (1.0 Mb, 1–4% of deletions) and atypical (non-standard, 8–10% of deletions).46 Type 1 and type 3 deletions are generally germline mutations, type 2 deletions are generally somatic and atypical microdeletions may be germline or somatic.46 Furthermore, the type of microdeletion present appears to impact cNF burden. For example, 50% of NF1 microdeletion syndrome patients with a type 1 deletion exhibit a high cNF burden (>1000 cNFs),43,47 while at least one atypical microdeletion (a 2.7 Mb region spanning from intron 21 of NF1 to intron 1 of ACCN1) is associated with a complete absence of cNFs, despite the fact that this microdeletion partially overlaps with the typical (type 1, 2 and 3) microdeletion regions.45,46 However, other patients with similar atypical deletions still present with cNF.45 Collectively, these observations suggest that critical genes or epigenetic regulatory elements exist in the NF1 microdeletion region, and that unresolved complexity remains, particularly regarding the impact of NF1 microdeletions on cNF presence.
NF1 function can also be impaired by post-transcriptional alterations such as alternative splicing, microRNA-mediated repression of the transcript or excessive proteasomal degradation of the protein.48,49,50 Consequently, detection of NF1 loss-of-function events and clarification of the NF1 genotype–phenotype relationship are existing challenges to the field.51 Some studies have started to characterise the genotype–phenotype relationship between NF1 and cNFs, and these further suggest that there are a wide range of somatic NF1 mutations associated with cNF.52,53,54 For example, Upadhyaya and colleagues identified 77 distinct somatic NF1 mutations (53 of which were not previously described) in a study of 109 cNFs from 46 NF1 patients, of which 25/109 (~23%) cNFs exhibited NF1 LOH.54 Another study by the same group identified NF1 LOH in 22/89 (25%) of cNFs tested, and somatic NF1 mutations in 57/89 (64%) of cNFs from three patients.53 The impact of most NF1 somatic intragenic mutations on cNF characteristics is currently unclear. Similarly, it is unknown what effect intragenic germline NF1 mutations have on cNF, with the exception of two mutations (NF1 c.2970-2972delAAT and NF1 c.5425C>T), which are associated with a lack of cNFs.55,56 NF1 c.5425C>T may disrupt the structure of the pleckstrin homology-like domain of NF1, whereas NF1 c.2970-2972delAAT is hypothesised to be a hypomorphic mutation.55,57 It is not currently understood why these mutations do not induce cNF formation.
Although it is not yet known for most clinical cases how NF1 genotype correlates with cNF phenotype, one possibility is a Ras-signalling-dependent growth mechanism. Mice with tissue-specific expression of oncogenic Ras (N-RasG12V) developed cNFs around 3 months post-birth, among other cutaneous symptoms of neurofibromatosis, suggesting that these phenotypes are caused by Ras activation.58 However, to the best of our knowledge, Ras mutations are not observed in either NF1-linked or sporadic cNFs. Considering this, experiments that examine the functional consequence of specific NF1 variants on Ras activity may help clarify the NF1 genotype–phenotype relationship.54,59 Perplexingly, germline inactivation of Spred1, which also results in Ras hyperactivation and the NF1-like disorder Legius syndrome, is not associated with the formation of cNFs.60 This indicates that there are unresolved mechanistic differences between NF1 and SPRED1 loss-of-function, and perhaps other modifiers that are requisite for cNF formation. Recent efforts using machine learning approaches, targeted sequencing panels for genes involved in RASopathies (diseases that are a consequence of Ras pathway dysregulation, such as NF1), and open access data resources are attempting to address these challenges.61,62,63 Specifically, these approaches are leveraging recent technological advances to assess NF1 loss-of-function and the relationship between germline or somatic NF1 mutations with the resulting phenotype. Way and colleagues trained a machine learning method to predict NF1 protein content, Bhoj et al. utilised a RASopathy sequencing panel in tandem with clinical examination to assess the presence of RASopathies, and Gosline et al. assembled the first public database of exome sequencing, RNA-seq and SNP array data for cNF.61,62,63 By integrating approaches such as these into cNF research, the field may be able to build upon prior studies to identify more accurate genotype–phenotype associations.
Genetic perturbation beyond the NF1 gene: impact on cNF development
Beyond the NF1 gene, cNFs are thought to have a low somatic mutation burden when compared with most cancers. Research comparing exome sequencing data from seven distinct cNF tumours to blood and skin samples obtained from a single patient did not identify somatic mutations in 5/7 cNFs, beyond a second-hit somatic NF1 mutation.52 The other two contained variants in HMCN1 and CEP131 (AZI1), respectively.52 A more recent study of exome data from three growing and three stable cNFs supports the hypothesis that these tumours are genomically quiet.64 Each tumour contained 1–11 somatic mutations, but no correlation was found between the type of variants and growth. Another study identified several recurrent somatic mutations in NF1-linked and sporadic cNFs, including MAML3, TAS2R30, DNAH3, KIAA0040, NDUFS7, SSPO and UBXN11.65 This study also suggested that NF1 cNFs, which can occur in the 10s–1000s in an individual patient, exhibit different critical molecular changes than sporadic NF1+/+ cNFs, which generally occur in isolation.65 Specifically, KIR2DL5 (a KIR-family receptor with no known ligand) mutation and/or loss of expression, which is only observed only in sporadic cNFs, serves to promote cellular proliferation in human Schwann cells.65 Other studies of cNF have observed signs of microsatellite instability with no accompanying somatic mismatch repair gene mutations in cNF tumour samples, and have observed only a few cNF tumours with somatic mutations in TP53 and RB1.53,54
Studies of gene copy number variation (CNV) in cNFs using array comparative genome hybridisation (aCGH) have observed that there is a scarcity of CNVs in cNF or other dermal (subcutaneous) neurofibromas.66,67,68,69 Beyond NF1 deletion, only one of these studies observed any CNVs in cNF, and this was a gain of CCND1.66 A more recent study identified a small number of regions that had chromosomal imbalances in nine cNFs from independent NF1 patients; 13 regions were decreased (33–76% of patients, depending on the region), and 3 were increased (44–76% of patients).70 Of the genes in these regions, the expression of UST and ARC were identified as significantly correlated with CNVs.70 In summary, very few non-NF1 genomic variants associated with cNF formation and growth are currently known, and future studies are needed to identify other non-NF1 variants that drive cNF formation. Furthermore, for the non-NF1 variants that have so far been observed in cNF, mechanistic studies may be valuable in determining their roles as potential drivers or functional modifiers in cNF development. For example, several mutations observed by Anastasaki et al. in NF1 cNFs such as MAML3 c.1513_1514del have also been observed in bona fide cancers, suggesting that they may be biologically meaningful mutations.65
A further gap in our knowledge of cNF development is a paucity of RNA expression data sets with matched normal nerve sheath tissue; however, generation of such a data set may suffer from logistic and ethical challenges. In this respect, animal models of cNF may provide an alternative source of tissue for genomic characterisation. Another approach may emulate the methodology currently employed by Steensma and colleagues. In this ongoing trial, the authors are obtaining cutaneous neurofibroma samples and patient-matched skin samples.71 Similarly controlled future translational studies may provide additional insight into novel genomic aspects of cNF biology. In designing such projects, it is important to consider the use of current genomics methodology such as single-cell RNA and whole-genome sequencing. These approaches may facilitate the exploration of the cellular and genomic heterogeneity of cNF, and this could provide a greater understanding of the relative contributions and roles of various cell types in cNF formation.
Another aspect of genomic perturbation in cNF that has not been well explored are mechanisms of genomic regulation such as DNA methylation, histone methylation and microRNA expression. Although there are currently no studies comparing gene/miRNA expression or methylation in cNF with unaffected normal tissue such as myelinated nerve tissue, several studies have compared methylation and RNA/miRNA expression patterns in cNF to other NF-related tumour types.72,73 These studies identified significant differences between cNFs, pNFs and MPNSTs, with respect to their DNA methylation, miRNA and gene expression profiles. Other genomics-driven studies have demonstrated a clear role for methylation and transcriptional dysregulation in NF1-linked tumours.74,75 Continued research in this area is critical to gain a comprehensive picture of the genomic landscape of cNF to guide future therapeutic development.
The microenvironment of cNF
Cutaneous neurofibromas have a complex and poorly understood microenvironment comprising multiple cell types, including NF1-deficient proliferative Schwann cells, macrophages, mast cells, fibroblasts and neurons, as well as blood vessels and a collagenous matrix (Fig. 2b). While many cellular components of cNF are not well studied, a role has been demonstrated for mast cells in neurofibroma growth/maintenance. These cells, which are a component of the innate immune system, are observed in several types of NF1-related tumours, including cNFs.25,76 Mast cells are thought to serve a complex pro-tumour role in these diseases.77 The presence of mast cells in cNFs was first described by Greggio in 1911, and it was later hypothesised that they may be an important element of the cNF microenvironment.25,78 Other studies have clarified the role of mast cells in cNF growth. Yang et al. demonstrated that homozygous deletion of Nf1 in Schwann cells caused excessive secretion of Kit ligand (SCF), a signalling molecule that activates mast cells. They also discovered that, in comparison to Nf1 wild-type mast cells, Nf1+/− mast cells are hyper-responsive to Kit ligand.79 This hyperactivation is associated with Ras-mediated PI3K signalling, indicating that Nf1+/− mast cells are uniquely affected by Nf1−/− Schwann cell signalling mechanisms.79 Mast cells may stimulate microenvironmental cNF changes by inducing the formation of a collagenous matrix by transforming growth factor beta (TGF-β)-mediated stimulation of cNF fibroblasts, and by secretion of other molecules such as platelet-derived growth factor (PDGF), fibroblast growth factor (FGF), vascular endothelial growth factor (VEGF) and histamine.25,80 In mouse models of cNF, studies demonstrated that cNFs contain infiltrating mast cells.24,30 A study of human cNF found that these tumours contain a greater abundance of mast cells and lymphatic vessels, as well as a larger lymphatic vessel diameter than several other NF-associated tumour types, thus suggesting that mast cells play a role in human cNF formation or growth.81
The relationship between other cell types and cNF growth is poorly understood. Macrophages are reported to be present in cNF and are abundant in pNFs.23,26,82,83 While the associations between macrophages and both NF1-associated mouse and human pNFs has been investigated, similar studies have not yet been performed in cNF.26,83 A genomics-focused approach may help to address the potential associations between the microenvironment, mutation burden and tumour growth. Genomic approaches to the deconvolution of microenvironmental cellular subtypes are increasingly utilised in oncology research; for example, methods such as CIBERSORT and TIMER have been used to characterise the diversity of tumour immune cell infiltrate populations across many cancer types.84,85 Applying these approaches to cNF data sets may uncover new details about these tumours and the role of the microenvironment.
Endocrine signalling and cNF
Endocrine signalling is thought to be an important factor in cNF growth and development.30,86,87 This hypothesis is underpinned by the observation of the rapid onset and growth of neurofibromas that can occur during puberty and pregnancy,17,86,88,89 and there are increasing amounts of molecular data to support this idea. The puberty-related increases in cNF growth may be due to changes in endocrine signalling;90 cNFs from NF1 patients have increased immunohistochemical staining for growth hormone receptor (GHR), compared with sporadic cNF and the ligand for GHR is increased during normal puberty.90 Growth hormone (GH) hypersecretion has been observed in some NF1 patients, but other studies have concluded that NF1 patients have GH deficiency,91,92,93 and thus the interplay between GH and cNF growth remains unclear at present.
It is well documented that NF1-associated cNFs are frequently progesterone receptor positive, although they are infrequently oestrogen receptor positive.87,94 McLaughlin and Jacks observed that the progesterone receptor is not expressed by NF1−/− Schwann cells in cNF, but rather by other cNF-associated cells that are S100-negative.94 One possibility proposed by the authors is that these cells are fibroblasts, perineurial cells, NF1+/− Schwann cells or NF1 wild-type Schwann cells.94 Others have found that neurofibroma-derived Schwann cells can proliferate in response to progesterone or oestrogen treatment in vitro.95,96 In vivo mouse xenograft experiments with cNF-derived Schwann cells demonstrated that one of four tumours treated with oestrogen grew, while another one of four tumours treated with progesterone grew, suggesting that the presence of oestrogen or progesterone can affect cNF growth.97 However, a study of 59 NF1 patients taking hormonal contraceptives observed that patients taking either oestrogen–progestogen or progestogen had no associated tumour growth;2 although, tumour growth was reported by two patients taking high-dose synthetic progesterone. Collectively, these data suggest that further research is needed to determine the role of progesterone on cNF tumour growth, and to elucidate the cNF cell population(s) most affected by progesterone receptor activation.2 Such future research could be enhanced with genomics approaches; for example, using a method such as single-sample gene-set enrichment analysis to detect oestrogen signalling pathways in cNFs, similar to a study that performed the same in colorectal cancer samples.98
Current and emerging treatment paradigms for cNF
There is currently no effective therapy for cNFs, and the mainstay of clinical management involves monitoring and informing the patients about their prognosis and the nature of their tumours. Treatment options for cNFs are limited to elective surgical approaches or laser-based ablation of cNFs.99,100 Other techniques have been applied for the removal of cNFs, including electrodessication and radiofrequency ablation.100,101,102 Surgical/ablative approaches involve a risk of scarring and skin discoloration,100,103 and surgical approaches are not practical in patients with numerous cNFs. These clinical deficiencies emphasise a need for novel, genomically guided therapies that are based on a greater understanding of the molecular and cellular mechanisms that underlie NF1-associated cNFs, and at the time of writing, nine interventional clinical trials were identified for cNFs (Table 1). Of these, five have been completed, three are active and not recruiting and one is currently active and recruiting.
Three cNF-specific clinical trials have evaluated the effect of topically applied treatments on cNF. Topical administration offers the benefit of limiting the systemic exposure; however, it can make effective drug delivery more difficult, as the skin acts as a barrier to most drugs.104 A second potential challenge for topical administration of cNF treatments concerns the potential impracticality for patients with extensive lesions, who may find it unfeasible to regularly apply a treatment to 40–70% of their body surface. Furthermore, no successful results have yet been reported for topically applied monotherapies in cNF (for example, imiquimod).105 Other reports have indicated that off-label topical application of ketotifen may be used to prevent cNF growth or reduce cNF burden by blocking the degranulation of mast cells in cNFs106,107,108; however, this drug has not seen widespread clinical adoption or success for treating cNF.
Beyond topical monotherapies, other trials have been initiated to study the safety and efficacy of photodynamic therapy (PDT) on cNFs. PDT involves pretreatment of a skin lesion with a photosensitising agent such aminolevulinic acid (ALA) followed by light exposure, to induce localised cell death. A current Phase 1 study is exploring the use of topical ALA and red light as a cNF intervention,109 and preliminary results have indicated that PDT may slow the growth of cNFs in adults. Applying PDT to developing cNFs during adolescence may be an effective preventative therapeutic measure, and a Phase 2 study will investigate whether this intervention will slow the growth of cNFs in adolescents.110 Other trials are investigating the direct injection of active agents into cNFs. One such study assessed diclofenac, a nonsteroidal anti-inflammatory drug, in cNF patients.111 The study did not show a conclusive change in neurofibroma size; in some cases, the neurofibromas grew, while in other cases the cNFs presented with necrosis and detached. These results appear to serve as a proof-of-concept for a study that will investigate topical diclofenac following laser microporation of cNFs.112
Since cNFs are reported to be composed of the same cell types found in the peripheral nerves, their biology may be similar to tumours like pNFs.113,114 Most of the interventional clinical trials described for cNFs stem from therapies under development for other manifestations of NF1 (Table 1). The use of targeted therapeutics such as VEGF, mTOR, c-kit or MEK inhibitors in cNF is likely based on data suggesting that these targets are important in NF1-deficient tumours.115,116,117,118 Additionally, the overlap between pathways dysregulated by NF1 loss and pathways dysregulated in other cancer types has also inspired clinical trials that reposition existing cancer treatment options. An example of such a target is vascular endothelial growth factor (VEGF), an angiogenic signalling molecule that is highly expressed in cNFs and other NF1-linked tumours.119,120 A trial investigated the potential for VEGF as a viable target by directly injecting the VEGF inhibitor ranibizumab into cNFs.121 The primary outcomes of the trial were changes to cNF volume and interstitial pressure. Patients served as their own controls, with three tumours treated with a single injection of ranibizumab and three tumours treated with saline controls. Results are not yet reported for this study. The mechanistic target of rapamycin (mTOR) is another target with relevance to NF1 tumour biology.115 This protein has previously been studied in pNF and MPNST-focussed clinical trials, using the mTOR inhibitor sirolimus as well as the mTOR inhibitor everolimus in combination with the VEGF inhibitor bevacizumab.122,123 However, the results from these studies indicated that these interventions were inactive in pNF and MPNST, respectively. In cNF, a single-arm interventional trial assessed the effect of everolimus on cNF growth.124 As with the studies of mTOR inhibitors in pNF and MPNST, this trial observed no change in cNF growth over the course of the intervention. It is possible that this is due to the stability of untreated cNFs over time, or due to the intervention preventing further growth of the cNFs. Additionally, there was no control arm for this study, so it remains to be confirmed whether mTOR inhibition is effective in reducing cNF burden.
Mast/stem cell growth factor receptor kit (c-kit), a cytokine receptor found on mast cells that is activated by SCF, was identified as a critical molecular component in NF1-deficient mast cells and tumours.25,76,125 One strategy for targeting tumours with mast cell components such as cNFs and pNFs is imatinib-mediated inhibition of c-kit. In NF1 patients with pNF, a Phase 2 clinical trial demonstrated response in 6/36 (17%) of participants.76 Changes in cNF size were not tracked in these patients, but this study may provide a model for designing a cNF-focussed clinical trial with imatinib. In a separate case report studying the effect of imatinib in an NF1 patient with cutaneous vasculopathy, the authors reported no change in the volume of the patient’s cNFs;126 however, the intervention was prematurely discontinued due to the development of adverse side effects.126 A clinical trial with multiple patients would be required to make any further conclusions about the effect of c-kit inhibition on cNFs.
Mitogen-activated protein kinase kinase (MAPK2/MEK), an integral signalling component of the Ras pathway, is a compelling target in many NF1-related tumours. The MEK inhibitor selumetinib has been reported as a low-toxicity and efficacious intervention in a trial for children with NF1 and inoperable pNFs.116,127 While trials of MEK inhibitors in cNF are yet to be conducted, the effect of selumetinib on cNF size as well as p-ERK and p-AKT levels will be explored in a recently initiated clinical trial.128 It remains to be seen whether selumetinib treatment is effective in cNF.
While MEK inhibition may be demonstrated to be a successful therapeutic approach for cNF, the development of new cNF trials is partially dependent on the identification of novel and pharmacologically tractable molecular targets with critical relevance to NF1 tumour biology. We propose that the application of modern oncogenomic approaches to cNF may yield valuable insights and allow the identification of putative drug targets to improve the therapeutic prospects of NF1 patients.
knowledge gaps and prospective avenues for future study
Cutaneous neurofibromas are a significant burden for individuals with NF1.15,16,19 As such, improved prevention and treatment of cNF is an important aspect in raising the quality of life for these patients. The observation that cNFs can undergo rapid proliferation followed by quiescence presents a challenge and an opportunity.21 The possibility that quiescent cNFs may not respond to anti-proliferative drugs is problematic; on the other hand, there is an opportunity to understand the mechanism underlying the quiescent process, and thus to identify new therapeutic targets. This may also inform the field of biological differences to consider when developing prophylactic therapeutic strategies, as opposed to cNF treatments. While the field has elucidated several aspects of cNF aetiology and pathology, most of the mechanisms involved in the evolution of these tumours remain unknown. Somatic and germline NF1 mutations are diverse, and for most cases the genotype–phenotype relationship, with respect to NF1 mutation and cNF burden, is not straightforward.
Further investigations of cNF biology are needed to facilitate the identification of novel therapeutic targets. It is not known which intracellular signalling mechanisms are most responsible for cNF formation, the specific mechanisms by which mast cells promote cNF formation, and the contribution, or lack thereof, of macrophages and other components of the tumour microenvironment to cNF growth and development. The question of whether pharmacologic targeting of these components in cNF is a viable therapeutic approach is currently being investigated, and may yield more effective or more convenient therapeutic interventions for patients with cNF.
It is also unclear which genetic factors, beyond NF1, are responsible for cNF growth and progression. Epigenetic regulatory mechanisms are also thought to be involved in the development of other NF1-linked tumours; these or other epigenetic mechanisms may also be dysregulated in cNF.74,75 While many large-scale resources exist for exploring the mutational landscape of malignant tumours (e.g., TCGA, cBioPortal, Project GENIE), there are limited genomic explorations of neurofibromas. To address this, the Children’s Tumour Foundation created a cutaneous neurofibroma data resource.63 This resource contains whole-genome sequencing and SNP microarray data from cNFs and patient-matched blood samples, as well as cNF RNA sequencing data.63 The data are open access and can be found on Sage Bionetworks’ Synapse platform (https://www.synapse.org/cutaneousNF). Other similar publicly available resources include microarray-based RNA expression data from cNF-derived Schwann cell cultures and cNF patient samples (Gene Expression Omnibus, GSE32029, GSE14038, GSE66743), and cNF aCGH data (GSE58000).
Progress is being made by the neurofibromatosis research community towards addressing the current knowledge gaps with different resources available for cNF research (Table 2). As part of these efforts, cNF research should leverage the power of current technologies developed for oncology research. Newer, faster and less costly techniques for genomic and transcriptomic profiling as well as sharing of the resulting data is an important approach to facilitating this type of research. Integrated genomics approaches, such as the simultaneous analysis of mutational and RNA expression data, can be used to identify novel cancer driver genes, or uncover critical cancer signalling mechanisms.129,130,131 In addition, the previously described immune characterisation techniques can be used for pan-cancer analysis to discover previously unknown features of the tumour microenvironment. By applying these methods in cNF research, we may be able to accelerate the pace with which novel therapies for cNFs are identified and address a major life-long burden for NF1 patients.
References
Evans, D. G. et al. Birth incidence and prevalence of tumor-prone syndromes: estimates from a UK family genetic register service. Am. J. Med. Genet. A 152A, 327–332 (2010).
Lammert, M., Friedman, J. M., Kluwe, L. & Mautner, V. F. Prevalence of neurofibromatosis 1 in German children at elementary school enrollment. Arch. Dermatol. 141, 71–74 (2005).
Boyd, K. P., Korf, B. R. & Theos, A. Neurofibromatosis type 1. J. Am. Acad. Dermatol. 61, 1–14 (2009).
Friedman, J. Neurofibromatosis 1 [Internet]. Gene reviews. University of Washington. http://www.ncbi.nlm.nih.gov/pubmed/20301288. Accessed 6 July 2016 (2014).
Recklinghausen, F. von. Ueber die multiplen Fibrome der Haut und ihre Beziehung zu den multiplen Neuromen [Internet]. https://wellcomelibrary.org. Accessed 8 August 2017 (1882).
Wallace, M. R. et al. A de novo Alu insertion results in neurofibromatosis type 1. Nature 353, 864–866 (1991).
Viskochil, D. et al. Deletions and a translocation interrupt a cloned gene at the neurofibromatosis type 1 locus. Cell 62, 187–192 (1990).
Cawthon, R. M. et al. A major segment of the neurofibromatosis type 1 gene: cDNA sequence, genomic structure, and point mutations. Cell 62, 193–201 (1990).
Plotkin, S. R. et al. Quantitative assessment of whole-body tumor burden in adult patients with neurofibromatosis. PLoS ONE 7, e35711 (2012).
Tonsgard, J. H., Kwak, S. M., Short, M. P. & Dachman, A. H. CT imaging in adults with neurofibromatosis-1: frequent asymptomatic plexiform lesions. Neurology 50, 1755–1760 (1998).
Evans, D. G. R. et al. Malignant peripheral nerve sheath tumours in neurofibromatosis 1. J. Med. Genet. 39, 311–314 (2002).
Ingham, S. et al. Malignant peripheral nerve sheath tumours in NF1: improved survival in women and in recent years. Eur. J. Cancer 47, 2723–2728 (2011).
Sabbagh, A. et al. Unravelling the genetic basis of variable clinical expression in neurofibromatosis 1. Hum. Mol. Genet. 18, 2768–2778 (2009).
Jouhilahti, E.-M. et al. The development of cutaneous neurofibromas. Am. J. Pathol. 178, 500–55 (2011).
Peltonen, J., Jouhilahti, E.-M. & Peltonen, S. in Neurofibromatosis Type 1 (eds Upadhyaya, M. & Cooper, D.) 393–403 (Springer, Berlin, Heidelberg, 2012).
Ferner, R. E. et al. Guidelines for the diagnosis and management of individuals with neurofibromatosis 1. J. Med. Genet. 44, 81–88 (2006).
Dugoff, L. & Sujansky, E. Neurofibromatosis type 1 and pregnancy. Am. J. Med. Genet. 66, 7–10 (1996).
Wu, B. L., Austin, M. A., Schneider, G. H., Boles, R. G. & Korf, B. R. Deletion of the entire NF1 gene detected by FISH: four deletion patients associated with severe manifestations. Am. J. Med. Genet. 59, 528–535 (1995).
Granström, S., Langenbruch, A., Augustin, M. & Mautner, V.-F. Psychological burden in adult neurofibromatosis type 1 patients: impact of disease visibility on body image. Dermatology 224, 160–167 (2012).
Rodriguez, F. J., Folpe, A. L., Giannini, C. & Perry, A. Pathology of peripheral nerve sheath tumors: diagnostic overview and update on selected diagnostic problems. Acta Neuropathol. 123, 295–319 (2012).
Ruggieri, M., Upadhyaya, M., Rocco, C. Di, Gabriele, A. & Pascual-Castroviejo, I. in Neurocutaneous Disorders Phakomatoses and Hamartoneoplastic Syndromes (eds Ruggieri, M., Castroviejo, I. P. & Rocco, C. D.) 51–151 (Springer, Vienna, 2008).
Ortiz-Hidalgo, C. & Weller, R. in Histology for Pathologists 2nd edn (ed. Sternberg, S.) 285–311 (Lippincott-Raven Publishers, Philadelphia, 1997).
Le, L. Q., Kesterson, R. A. & Gutmann, D. H. Defining the research landscape for dermal neurofibromas. Oncol. Times 38, 14–15 (2016).
Liao, C.-P. et al. The role of nerve microenvironment for neurofibroma development. Oncotarget 7, 61500–61508 (2016).
Yang, F. C. et al. Nf1+/- mast cells induce neurofibroma like phenotypes through secreted TGF-beta signaling. Hum. Mol. Genet. 15, 2421–2437 (2006).
Choi, K. et al. An inflammatory gene signature distinguishes neurofibroma Schwann cells and macrophages from cells in the normal peripheral nervous system. Sci. Rep. 7, 43315 (2017).
Hirose, T. et al. Immunohistochemical demonstration of EMA/Glut1-positive perineurial cells and CD34-positive fibroblastic cells in peripheral nerve sheath tumors. Mod. Pathol. 16, 293–298 (2003).
Riddle, N. D., Gorden, L., Rojiani, M. V., Hakam, A. & Rojiani, A. M. CD44 and p53 immunoexpression patterns in NF1 neoplasms - indicators of malignancy and infiltration. Int. J. Clin. Exp. Pathol. 3, 515–521 (2010).
Wu, J. et al. Plexiform and dermal neurofibromas and pigmentation are caused by Nf1 loss in desert hedgehog-expressing cells. Cancer Cell 13, 105–116 (2008).
Le, L. Q., Shipman, T., Burns, D. K. & Parada, L. F. Cell of origin and microenvironment contribution for NF1-associated dermal neurofibromas. Cell Stem Cell 4, 453–463 (2009).
(NHGRI) National Human Genome Research Institute. [NCT00314119] Natural History and Biology of Skin Neurofibromas in Neurofibromatosis Type 1. ClinicalTrials.gov. (2006).
National Cancer Institute (NCI). [NCT00924196] Natural History Study of Patients with Neurofibromatosis Type I. ClinicalTrials.gov. (2009).
Ratner, N. & Miller, S. J. A RASopathy gene commonly mutated in cancer: the neurofibromatosis type 1 tumour suppressor. Nat. Rev. Cancer 15, 290–301 (2015).
Eccleston, J. F., Moore, K. J., Morgan, L., Skinner, R. H. & Lowe, P. N. Kinetics of interaction between normal and proline 12 Ras and the GTPase-activating proteins, p120-GAP and neurofibromin. The significance of the intrinsic GTPase rate in determining the transforming ability of ras. J. Biol. Chem. 268, 27012–27019 (1993).
Downward, J. Targeting RAS signalling pathways in cancer therapy. Nat. Rev. Cancer 3, 11–22 (2003).
Philpott, C., Tovell, H., Frayling, I. M., Cooper, D. N. & Upadhyaya, M. The NF1 somatic mutational landscape in sporadic human cancers. Hum. Genomics 1, 13 (2017).
Scholz, S. et al. NF1 mutations in conjunctival melanoma. Br. J. Cancer. (2018). https://doi.org/10.1038/s41416-018-0046-5.
Ars, E. et al. Recurrent mutations in the NF1 gene are common among neurofibromatosis type 1 patients. J. Med. Genet. 40, e82 (2003).
van Minkelen, R. et al. A clinical and genetic overview of 18 years neurofibromatosis type 1 molecular diagnostics in the Netherlands. Clin. Genet. 85, 318–327 (2014).
Upadhyaya, M. et al. The heterogeneous nature of germline mutations in NF1 patients with malignant peripheral serve sheath tumours (MPNSTs). Hum. Mutat. 27, 716–716 (2006).
Upadhyaya, M. et al. Germline and somatic NF1 gene mutations in plexiform neurofibromas. Hum. Mutat. 29, E103–E111 (2008).
Raponi, M., Upadhyaya, M. & Baralle, D. Functional splicing assay shows a pathogenic intronic mutation in neurofibromatosis type 1 (NF1) due to intronic sequence exonization. Hum. Mutat. 27, 294–295 (2006).
Pasmant, E. et al. NF1 microdeletions in neurofibromatosis type 1: from genotype to phenotype. Hum. Mutat. 31, E1506–E1518 (2010).
De Raedt, T. et al. Somatic loss of wild type NF1 allele in neurofibromas: comparison of NF1 microdeletion and non-microdeletion patients. Genes Chromosomes Cancer 45, 893–904 (2006).
Kehrer-Sawatzki, H., Schmid, E., Fünsterer, C., Kluwe, L. & Mautner, V.-F. Absence of cutaneous neurofibromas in an NF1 patient with an atypical deletion partially overlapping the common 1.4 Mb microdeleted region. Am. J. Med. Genet. A 146A, 691–699 (2008).
Kehrer-Sawatzki, H., Mautner, V.-F. & Cooper, D. N. Emerging genotype-phenotype relationships in patients with large NF1 deletions. Hum. Genet. 136, 349–376 (2017).
Mautner, V.-F. et al. Clinical characterisation of 29 neurofibromatosis type-1 patients with molecularly ascertained 1.4 Mb type-1 NF1 deletions. J. Med. Genet. 47, 623–630 (2010).
Hinman, M. N., Sharma, A., Luo, G. & Lou, H. Neurofibromatosis type 1 alternative splicing is a key regulator of Ras signaling in neurons. Mol. Cell Biol. 34, 2188–2197 (2014).
Lenarduzzi, M. et al. MicroRNA-193b enhances tumor progression via down regulation of neurofibromin 1. PLoS ONE 8, e53765 (2013).
Cichowski, K., Santiago, S., Jardim, M., Johnson, B. W. & Jacks, T. Dynamic regulation of the Ras pathway via proteolysis of the NF1 tumor suppressor. Genes Dev. 17, 449–454 (2003).
Yap, Y.-S. et al. The NF1 gene revisited - from bench to bedside. Oncotarget 5, 5873–5892 (2014).
Emmerich, D. et al. Somatic neurofibromatosis type 1 (NF1) inactivation events in cutaneous neurofibromas of a single NF1 patient. Eur. J. Hum. Genet. 23, 870–873 (2015).
Thomas, L., Kluwe, L., Chuzhanova, N., Mautner, V. & Upadhyaya, M. Analysis of NF1 somatic mutations in cutaneous neurofibromas from patients with high tumor burden. Neurogenetics 11, 391–400 (2010).
Thomas, L. et al. Exploring the somatic NF1 mutational spectrum associated with NF1 cutaneous neurofibromas. Eur. J. Hum. Genet. 20, 411–419 (2012).
Upadhyaya, M. et al. An absence of cutaneous neurofibromas associated with a 3-bp inframe deletion in exon 17 of the NF1 gene (c.2970-2972 delAAT): evidence of a clinically significant NF1 genotype-phenotype correlation. Am. J. Hum. Genet. 80, 140–151 (2007).
Pinna, V. et al. p.Arg1809Cys substitution in neurofibromin is associated with a distinctive NF1 phenotype without neurofibromas. Eur. J. Hum. Genet. 23, 1–4 (2014).
Santoro, C. et al. Arg(1809) substitution in neurofibromin: further evidence of a genotype-phenotype correlation in neurofibromatosis type 1. Eur. J. Hum. Genet. 23, 1460–1461 (2015).
Saito, H., Yoshida, T., Yamazaki, H. & Suzuki, N. Conditional N-rasG12V expression promotes manifestations of neurofibromatosis in a mouse model. Oncogene 26, 4714–4719 (2007).
Sherman, L. S., Atit, R., Rosenbaum, T., Cox, A. D. & Ratner, N. Single cell Ras-GTP analysis reveals altered Ras activity in a subpopulation of neurofibroma Schwann cells but not fibroblasts. J. Biol. Chem. 275, 30740–30745 (2000).
Messiaen, L. et al. Clinical and mutational spectrum of neurofibromatosis type 1-like syndrome. JAMA 302, 2111 (2009).
Way, G. P. et al. A machine learning classifier trained on cancer transcriptomes detects NF1 inactivation signal in glioblastoma. BMC Genomics 18, 127 (2016).
Bhoj, E. J. et al. Phenotypic predictors and final diagnoses in patients referred for RASopathy testing by targeted next-generation sequencing. Genet. Med. 19, 715–718 (2016).
Gosline, S. J. C. et al. A high-throughput molecular data resource for cutaneous neurofibromas. Sci. Data 4, 170045 (2017).
Faden, D. L., Asthana, S., Tihan, T., De Risi, J. & Kliot, M. Whole exome sequencing of growing and non-growing cutaneous neurofibromas from a single patient with neurofibromatosis type 1. Gao J-X, editor. PLoS ONE 12, e0170348 (2017).
Anastasaki, C. et al. KIR2DL5 mutation and loss underlies sporadic dermal neurofibroma pathogenesis and growth. Oncotarget 8, 47574–47585 (2017).
Mantripragada, K. K. et al. High-resolution DNA copy number profiling of malignant peripheral nerve sheath tumors using targeted microarray-based comparative genomic hybridization. Clin. Cancer Res. 14, 1015–1024 (2008).
Mantripragada, K. K et al. Genome-wide high-resolution analysis of DNA copy number alterations in NF1-associated malignant peripheral nerve sheath tumors using 32K BAC array. Genes Chromosomes Cancer 48, 897–907 (2009).
Brekke, H. R. et al. Genomic changes in chromosomes 10, 16, and X in malignant peripheral nerve sheath tumors identify a high-risk patient group. J. Clin. Oncol. 28, 1573–1582 (2010).
Beert, E. et al. Atypical neurofibromas in neurofibromatosis type 1 are premalignant tumors. Genes Chromosomes Cancer 50,1021–1032 (2011).
Asai, A. et al. High-resolution 400K oligonucleotide array comparative genomic hybridization analysis of neurofibromatosis type 1-associated cutaneous neurofibromas. Gene 558, 220–226 (2015).
Steensma, M. [NCT02777775] Targeting the Mechanisms Underlying Cutaneous Neurofibroma Formation in NF1: A Clinical Translational Approach. ClinicalTrials.gov. (2016).
Masliah-Planchon, J. et al. MicroRNAome profiling in benign and malignant neurofibromatosis type 1-associated nerve sheath tumors: evidences of PTEN pathway alterations in early NF1 tumorigenesis. BMC Genomics 14, 473 (2013).
Röhrich, M. et al. Methylation-based classification of benign and malignant peripheral nerve sheath tumors. Acta Neuropathol. 131, 877–887 (2016).
De Raedt, T. et al. PRC2 loss amplifies Ras-driven transcription and confers sensitivity to BRD4-based therapies. Nature 514, 247–251 (2014).
Lee, W. et al. PRC2 is recurrently inactivated through EED or SUZ12 loss in malignant peripheral nerve sheath tumors. Nat. Genet. 46, 1227–1232 (2014).
Robertson, K. A. et al. Imatinib mesylate for plexiform neurofibromas in patients with neurofibromatosis type 1: a phase 2 trial. Lancet Oncol. 13, 1218–1224 (2012).
Maciel, T. T., Moura, I. C. & Hermine, O. The role of mast cells in cancers. F1000Prime Rep. 7, 9 (2015).
Greggio, H. Les cellules granuleuses (Mastzellen) dans les tissus normaux et dans certaines maladies chirurgicales. Arch. Méd. Exp. 23, 323–375 (1911).
Feng-Chun, Y. et al. Neurofibromin-deficient Schwann cells secrete a potent migratory stimulus for Nf1+/– mast cells. J. Clin. Invest. 112, 1851–1861 (2003).
Viskochil, D. H. It takes two to tango: mast cell and Schwann cell interactions in neurofibromas. J. Clin. Invest. 112, 1791–1793 (2003).
Friedrich, R. E., Naber, U., Glatzel, M. & Hagel, C. Vessel and mast cell densities in sporadic and syndrome-associated peripheral nerve sheath tumors. Anticancer Res. 35, 4713–4722 (2015).
Takata, M., Imai, T. & Hirone, T. Factor-XIIIa-positive cells in normal peripheral nerves and cutaneous neurofibromas of type-1 neurofibromatosis. Am. J. Dermatopathol. 16, 37–43 (1994).
Prada, C. E. et al. Neurofibroma-associated macrophages play roles in tumor growth and response to pharmacological inhibition. Acta Neuropathol. 125, 159–168 (2013).
Newman, A. M. et al. Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods 12, 1–10 (2015).
Li, B. et al. Comprehensive analyses of tumor immunity: implications for cancer immunotherapy. Genome Biol. 17, 174 (2016).
Sharpe, J. C. & Young, R. H. Neurofibromatosis: the effect of pregnancy on the skin manifestations. J. Am. Med. Assoc. 106, 682 (1936).
Geller, M. et al. Progesterone and estrogen receptors in neurofibromas of patients with NF1. Clin. Med. Pathol. 1, 93–97 (2008).
Cesaretti, C. et al. Neurofibromatosis type 1 and pregnancy: maternal complications and attitudes about prenatal diagnosis. Am. J. Med. Genet. A. 161, 386–388 (2013).
Rasmussen, S. A. & Friedman, J. M. NF1 gene and neurofibromatosis 1. Am. J. Epidemiol. 151, 33–40 (2000).
Cunha, K. S. G., Barboza, E. P. & Fonseca, E. C. Identification of growth hormone receptor in localized neurofibromas of patients with neurofibromatosis type 1. J. Clin. Pathol. 56, 758–763 (2003).
Bizzarri, C. & Bottaro, G. Endocrine implications of neurofibromatosis 1 in childhood. Horm. Res. Paediatr. 83, 232–241 (2015).
Howell, S. J., Wilton, P., Lindberg, A. & Shalet, S. M. Growth hormone and neurofibromatosis. Horm. Res. 53, 70–76 (2000).
Vassilopoulou-Sellin, R., Klein, M. J. & Slopis, J. K. Growth hormone deficiency in children with neurofibromatosis type 1 without suprasellar lesions. Pediatr. Neurol. 22, 355–358 (2000).
McLaughlin, M. E. & Jacks, T. Progesterone receptor expression in neurofibromas. Cancer Res. 63, 752–755 (2003).
Overdiek, A., Winner, U., Mayatepek, E. & Rosenbaum, T. Schwann cells from human neurofibromas show increased proliferation rates under the influence of progesterone. Pediatr. Res. 64, 40–43 (2008).
Fishbein, L. et al. In vitro studies of steroid hormones in neurofibromatosis 1 tumors and schwann cells. Mol. Carcinog. 46, 512–523 (2007).
Li, H. et al. Analysis of steroid hormone effects on xenografted human NF1 tumor schwann cells. Cancer Biol. Ther. 10, 758–764 (2010).
Liu, D. Gene signa tures of estrogen and progesterone receptor pathways predict the prognosis of colorectal cancer. FEBS J. 283, 3115–3133 (2016).
Chiang, Y. Z., Al-Niaimi, F., Ferguson, J., August, P. J. & Madan, V. Carbon dioxide laser treatment of cutaneous neurofibromas. Dermatol. Ther. 2, 7 (2012).
Kriechbaumer, L. K., Susani, M., Kircher, S. G., Distelmaier, K. & Happak, W. Comparative study of CO2- and Er:YAG laser ablation of multiple cutaneous neurofibromas in von Recklinghausen’s disease. Lasers Med. Sci. 29, 1083–1091 (2014).
Kim, D. H. et al. 27.12 MHz radiofrequency ablation for benign cutaneous lesions. Biomed. Res. Int. 2016, 1–6 (2016).
Lutterodt, C. G., Mohan, A. & Kirkpatrick, N. The use of electrodessication in the treatment of cutaneous neurofibromatosis: A retrospective patient satisfaction outcome assessment. J. Plast. Reconstr. Aesthetic Surg. 69, 765–769 (2016).
Kim, S.-H., Roh, S.-G., Lee, N.-H. & Yang, K.-M. Radiofrequency ablation and excision of multiple cutaneous lesions in neurofibromatosis type 1. Arch. Plast. Surg. 40, 57–61 (2013).
Wermeling, D. P. et al. Microneedles permit transdermal delivery of a skin-impermeant medication to humans. Proc. Natl Acad. Sci. USA 105, 2058–2063 (2008).
Massachusetts General Hospital. [NCT00865644] Topical Imiquimod 5% Cream for Treatment of Cutaneous Neurofibromas in Adults With Neurofibromatosis 1 No Title. ClinicalTrials.gov. (2009).
Riccardi, V. M. Mast-cell stabilization to decrease neurofibroma growth. Prelim. Exp. ketotifen. Arch. Dermatol. 123, 1011–1016 (1987).
Riccardi, V. M. A controlled multiphase trial of ketotifen to minimize neurofibroma-associated pain and itching. Arch. Dermatol. 129, 577–581 (1993).
Riccardi, V. M. Ketotifen suppression of NF1 neurofibroma growth over 30 years. Am. J. Med. Genet. A 167, 1570–1577 (2015).
Whelan, H. T. [NCT01682811] Photodynamic Therapy (PDT) for Benign Dermal Neurofibromas (NF1). ClinicalTrials.gov. (2012).
Whelan, H. T. [NCT02728388] Photodynamic Therapy for Benign Dermal Neurofibromas. ClinicalTrials.gov. (2016).
Geller, M. et al. A proof-of-concept assessment of the safety and efficacy of intralesional diclofenac in the treatment of cutaneous neurofibromas. Int. J. Clin. Med. 6, 975–983 (2015).
Fundação Educacional Serra dos Órgãos. [NCT03090971] Use of Topical Liquid Diclofenac Following Laser Microporation of Cutaneous Neurofibromas in Patients With NF1. ClinicalTrials.gov. (2017).
Williams, V. C. et al. Neurofibromatosis type 1 revisited. Pediatrics 123, 124–133 (2009).
Hirsch, N. P., Murphy, A. & Radcliffe, J. J. Neurofibromatosis: clinical presentations and anaesthetic implications [Internet]. Br. J. Anaesth. 86, 555–564 (2001).
Johannessen, C. M. et al. The NF1 tumor suppressor critically regulates TSC2 and mTOR. Proc. Natl Acad. Sci. USA 102, 8573–8578 (2005).
Dombi, E. et al. Activity of selumetinib in neurofibromatosis type 1–related plexiform neurofibromas. N. Engl. J. Med. 375, 2550–2560 (2016).
Kawachi, Y. et al. NF1 gene silencing induces upregulation of vascular endothelial growth factor expression in both Schwann and non-Schwann cells. Exp. Dermatol. 22, 262–265 (2013).
Yang, F.-C. et al. Nf1-dependent tumors require a microenvironment containing Nf1+/−- and c-kit-dependent bone marrow. Cell 135, 437–448 (2008).
Kawachi, Y., Xu, X., Ichikawa, E., Imakado, S. & Otsuka, F. Expression of angiogenic factors in neurofibromas. Exp. Dermatol. 12, 412–417 (2003).
Wasa, J. et al. Differential expression of angiogenic factors in peripheral nerve sheath tumors. Clin. Exp. Metastas. 25, 819–825 (2008).
Massachusetts General Hospital. [NCT00657202] Ranibizumab for Neurofibromas Associated With NF1. ClinicalTrials.gov. (2008).
Weiss, B. et al. Sirolimus for progressive neurofibromatosis type 1-associated plexiform neurofibromas: a neurofibromatosis clinical trials consortium phase II study. Neuro. Oncol. 17, 596–603 (2014).
Widemann, B. C. et al. SARC016: phase II study of everolimus in combination with bevacizumab in sporadic and neurofibromatosis type 1 (NF1) related refractory malignant peripheral nerve sheath tumors (MPNST). J. Clin. Oncol. 34, 11053 (2016).
The University of Texas Health Science Center - Houston. [NCT02332902] Everolimus for Treatment of Disfiguring Cutaneous Lesions in Neurofibromatosis1 CRAD001CUS232T (DCLNF1). ClinicalTrials.gov. (2014).
Ingram, D. A., Yang, F.-C., Travers, J. B. & Clapp, D. W. Nf1 modulates C-kit signaling in mast cells and neural crest derived melanocytes in vivo in a dose dependent fashion. Pediatr. Res. 45, 53A–53A (1999).
Khelifa, I., Saurat, J. H. & Prins, C. Use of imatinib in a patient with cutaneous vasculopathy in the context of von Recklinghausen disease/neurofibromatosis. Br. J. Dermatol. 172, 253–256 (2015).
National Cancer Institute (NCI). [NCT01362803] AZD6244 Hydrogen Sulfate for Children With Nervous System Tumors. ClinicalTrials.gov. (2011).
National Cancer Institute (NCI). [NCT02839720] Selumetinib in Treating Patients With Neurofibromatosis Type 1 and Dermal Neurofibroma. ClinicalTrials.gov. (2016).
Peifer, M. et al. Integrative genome analyses identify key somatic driver mutations of small-cell lung cancer. Nat. Genet. 44, 1104–1110 (2012).
Wang, K. et al. Whole-genome sequencing and comprehensive molecular profiling identify new driver mutations in gastric cancer. Nat. Genet. 46, 573–582 (2014).
Sivakumar, S. et al. Master regulators of oncogenic KRAS response in pancreatic cancer: an integrative network biology analysis. PLoS Med. 14, e1002223 (2017).
Assistance Publique - Hôpitaux de Paris. [NCT01412892] Use of RAD001 as Monotherapy in the Treatment of Neurofibromatosis 1 Related Internal Plexiform Neurofibromas (NFitor). ClinicalTrials.gov. (2011).
The University of Texas Health Science Center - Houston. [NCT01031901] Topical Rapamycin Therapy to Alleviate Cutaneous Manifestations of Tuberous Sclerosis Complex (TSC) and Neurofibromatosis I (NF1). ClinicalTrials.gov. (2009).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
S.L.R., P.K. and A.B. are employees of The Children’s Tumor Foundation, which is a prominent non-profit funding organisation within the neurofibromatosis research community.
Consent for publication
All patients provided consent for publication.
Funding
L.Q.L. holds a Career Award for Medical Scientists from the Burroughs Wellcome Fund, the Thomas L. Shield, M.D. Professorship in Dermatology and receives funding from the Giorgio Foundation, the National Cancer Institute of the NIH (grant number R01 CA166593), the Neurofibromatosis Therapeutic Acceleration Program and the NF1 Research Consortium Fund.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Allaway, R.J., Gosline, S.J.C., La Rosa, S. et al. Cutaneous neurofibromas in the genomics era: current understanding and open questions. Br J Cancer 118, 1539–1548 (2018). https://doi.org/10.1038/s41416-018-0073-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41416-018-0073-2
This article is cited by
-
Metalloproteinase 1 downregulation in neurofibromatosis 1: Therapeutic potential of antimalarial hydroxychloroquine and chloroquine
Cell Death & Disease (2021)
-
NF1 patient missense variants predict a role for ATM in modifying neurofibroma initiation
Acta Neuropathologica (2020)
-
Available Therapies for Patients with Neurofibromatosis-Related Nervous System Tumors
Current Treatment Options in Oncology (2020)
-
Cutaneous neurofibromas: patients’ medical burden, current management and therapeutic expectations: results from an online European patient community survey
Orphanet Journal of Rare Diseases (2019)