Abstract
Tissue engineering is rapidly progressing toward clinical application. In the musculoskeletal field, there has been an increasing necessity for bone and cartilage replacement. Despite the promising translational potential of tissue engineering approaches, careful attention should be given to the quality of developed constructs to increase the real applicability to patients. After a general introduction to musculoskeletal tissue engineering, this narrative review aims to offer an overview of methods, starting from classical techniques, such as gene expression analysis and histology, to less common methods, such as Raman spectroscopy, microcomputed tomography, and biosensors, that can be employed to assess the quality of constructs in terms of viability, morphology, or matrix deposition. A particular emphasis is given to standards and good practices (GXP), which can be applicable in different sectors. Moreover, a classification of the methods into destructive, noninvasive, or conservative based on the possible further development of a preimplant quality monitoring system is proposed. Biosensors in musculoskeletal tissue engineering have not yet been used but have been proposed as a novel technology that can be exploited with numerous advantages, including minimal invasiveness, making them suitable for the development of preimplant quality control systems.
Similar content being viewed by others
Introduction
Tissue engineering aims to study and develop new methods for the replacement of damaged or diseased tissue. The fundamental idea is to use the appropriate type of cells, seed them on suitable support materials, induce controlled cell differentiation by the addition of specific growth factors and use the mature construct for replacement of the lost tissue. Mesenchymal stromal cells (MSCs) are at the root of this technology due to their regenerative potential and capability to differentiate into osteogenic, chondrogenic and adipogenic lineages,1 which make them particularly attractive for musculoskeletal tissue engineering. To improve in vitro models and generate tissues more similar to native tissue, traditional two-dimensional (2D) cell culture methods have been translated toward establishing three-dimensional (3D) constructs. In the past 20 years, considerable progress has been made in moving tissue engineering2 closer to clinics, which opens the opportunity to treat several musculoskeletal diseases or injuries in the near future. Although research in this field has been rapidly growing, progress needs to be made toward translation from the bench to the bedside.3 The translation of a tissue engineered product, defined by the regulatory framework as an advanced therapy medicinal product (ATMP), includes standardization of protocols for the fabrication of the implants that meets good laboratory manufacturing and practice as well as regulatory aspects. Standardization and quality control are important aspects that can ensure a boost in clinical translation. These criteria require modern methods allowing for standardized quality control in a 3D setup. The development of standard protocols will help to ensure the reproducibility of experiments and quality in manufacturing products in the clinic and consequently can ensure safe translation to patients.
Each analytical technique has advantages and disadvantages and can be used at a specific stage during the development of engineered tissues. This review article aims to provide an overview of the main methods used for quality control in tissue engineering of the musculoskeletal system, with a focus on cartilage and bone regeneration, using emerging techniques in the field. The techniques described herein are divided into 3 groups depending on the possibility of a future implant of engineered tissues. According to this principle, destructive methods result in the samples being destroyed, so they cannot be used to perform further analyses or for clinical purposes. Examples of such methods include classical techniques such as gene expression analysis, immunohistological staining, and some imaging techniques. Nondestructive or noninvasive techniques maintain sample integrity; therefore, the same specimen can be used for further analyses with destructive techniques. However, the samples might not be suitable for clinical use, as the technique might affect cell behavior; for example, X-ray doses for microCT require extra attention for their mutagenic properties. Finally, conservative methods keep the sample intact and do not affect cell biology, ideally enabling use for clinical implantation. Some types of biosensors can be included in this group. However, there is still no reported use of biosensors in musculoskeletal tissue engineering. Therefore, the potential of sensors for quality monitoring is introduced by discussing meaningful studies from other fields, such as neurosciences, as biosensors may represent a potential tool for the advancement of tissue engineered products both in the preclinical and clinical settings. Furthermore, a selection of the main regulatory standards that can be applied in the field is discussed in the respective paragraphs. To conclude, standard procedures, together with innovative methods of quality control, can contribute to the identification of key factors that influence the safety and quality of products4 with the overall aim of quality control, thereby achieving full translational clinical potential.
Methods for musculoskeletal tissue engineering
Cell sources for musculoskeletal tissue engineering
The ultimate goal of tissue engineering is the restoration of the structure and function of tissues lost due to trauma or disease, including musculoskeletal tissues such as bone or cartilage. Despite the tremendous effort and the number of clinical and preclinical studies in diverse fields, there is still a paucity of clinically approved solutions including cells with or without materials.3,5 A successful tissue engineering approach is complex, with many factors to be tuned to recreate functional tissue that can integrate with the native environment. Among these factors, the cells used represent the building block, and their careful selection and management is of critical importance. For a recent overview, see the TERMIS book series (https://www.springer.com/series/13441). For more than a decade, mesenchymal stromal cells have been indicated as the most promising source in regenerative medicine/tissue engineering of the musculoskeletal system because of their regenerative potential and their physiological tendency to differentiate toward cells of the mesodermal lineage, such as osteoblasts and chondrocytes.6
There are several active clinical trials involving MSCs (clinicaltrials.gov, n = 237 actively recruiting studies as of August 2020), with more than 50 studies focused on the treatment of musculoskeletal disorders and osteoarthritis in primis. Bone marrow, adipose tissue and umbilical cords are the major sources of stem cells in current trials, probably owing to tissue accessibility, the number of cells that can be obtained and the large number of preclinical studies conducted. Even though biobanking of bone marrow-derived MSCs is possible, they have mainly been studied for autologous use in musculoskeletal applications7. Analogous considerations can be made for adipose tissue-derived stromal cells (ADSCs) and stromal vascular fractions (SVFs).
Umbilical cord-derived stem cells are promising, especially for allogeneic use, since cord blood or Wharton Jelly can be collected from term births with no invasive procedure.8 Allogeneic transplantation relies on the use of biobanks, with higher requirements in terms of manipulation and regulations (see9,10,11,12 for reference on production and cryopreservation of cells).
Induced stem cells are a highly promising cell type in the field of regenerative medicine. In 2006, Takahashi and Yamanaka, by screening 24 different genes, concluded that adult fibroblasts could be reversed to totipotent cells by upregulating Oct3/4, Sox2, c‐Myc, and Klf4. These reprogrammed cells, designated as induced pluripotent stem cells (iPSCs), displayed a typical embryonic stem (ES) cell‐like morphology and growth physiology.13
On the other hand, the use of cells with a more mature phenotype (i.e., differentiated cells) has been more commonly employed for cartilage tissue engineering than for bone. Indeed, autologous chondrocyte implantation (ACI) has already been used in clinical practice, although with variable results,14 donor site morbidity and cell dedifferentiation during expansion and after implantation. Cells for ACI can be derived from low-weight bearing areas of the joint,15,16 auricular cartilage17 or nasal chondrocytes,18 the latter showing promising results for the development of hyaline cartilage. For bone tissue engineering applications, rather than mature cells, potential, autologous bone grafts are usually employed with good results and still represent the gold standard in the treatment of large bone defects.19,20
Regulatory standards for cell isolation and manipulation
Interest in MSCs for the treatment of several diseases has constantly increased in recent years due to their ability to improve tissue regeneration in patients who are refractory to other therapies. Minimal criteria for the identification and characterization have been proposed for both MSCs1 and ADSCs.21
Most of these cell preparations are autologous; thus, the cells have to be isolated, expanded, and manipulated before being reinjected into the patient. In November 2017, the European Commission published a set of new guidelines on good manufacturing practices (GMPs) specific to ATMPs. The guidelines for GMP 2003/94/EC6 highlighted three important pillars for patient and product standardization: (1) Standardization of quality control methods; (2) Integration of procedures for product tracking; and (3) Establishment of self-inspection analysis to monitor quality standards. Later, these principles became an integral part of legislative redlines concerning TE products 2004/23/EC and 2006/17/EC. Furthermore, ATMPs have mainly focused on the establishment of regulations for somatic cell therapy and gene therapy medicinal products (European Directive 2001/83/EC), which regulate the use of standard operating procedures (SOPs), guidelines, reference manuals, donor records, and reporting forms for tissue or cell quality for their quality management systems (QMSs)22.
All stages of cell processing and storage must comply with strict GMP guidelines and must be accompanied by accurate documentation.23,24 These guidelines define general measures to ensure that the processes required for production and validation are clearly defined, validated, reviewed and documented and that personnel, premises and materials are suitable for the production of pharmaceutical and biological products.25 The quality and safety of the product must be guaranteed, from the first phases of production, to minimize microbiological and cross-contamination through continuous environmental monitoring26. The first World Health Organization (WHO) draft text on GMPs was adopted in 1968. A Supplementary Annex on Biological Standardization (ECBS) of biological medicinal products was adopted in 1991, setting out the general approach to quality control of biological medicinal products such as vaccines, blood and blood products, antigens, cell and tissue therapies, biopharmaceuticals and others.27 Worldwide, more than 100 countries have incorporated WHO GMP requirements into their national medicine laws. Grade A facilities are essential for production and packaging of the product under aseptic conditions, which are normalized by specific quality controls to produce a safe ATMP for direct human application. The International Standards Organization (ISO) provides requirements, specifications or guidelines that can be used to ensure that materials, products, processes and services are safe, efficient and environmentally friendly.28 Table 1 summarizes applicable standards and relevant GMP literature for cell isolation and manipulation.
Development of 3D constructs and 3D bioprinting
Tissues of the musculoskeletal system are characterized by their complex 3D environment and hierarchical nature of the matrix component on the microscale, such as the proteoglycan gradient29,30 and collagen fiber orientation in articular cartilage.31,32 To develop advanced biologically functional in vitro models, special emphasis is placed on 3D culture setups to better mimic the 3D environment in the target tissue. Among additive manufacturing techniques, 3D bioprinting is the most prominent approach to fabricate patient-specific and thus anatomically shaped 3D biological functional scaffolds combining cells, supporting biomaterial and bioactive molecules for implantation, drug screening or in vitro models.33
The ultimate aim of biofabrication technologies is to produce patient-specific tissues or anatomically shaped organs by using autologous or allogeneic cells combined with a biomaterial to replicate the original geometry of the diseased or damaged tissue.34,35 There are promising results in bioprinting implants for bone repair36,37,38,39,40 or complex multidimensional structures to produce (cell-free) scaffolds mimicking the osteochondral environment for full cartilage defects.41,42,43,44,45,46,47 However, major limitations of these studies are the missing tools to characterize the printing quality and reproducibility postprinting and to investigate cell viability, process-induced cell stress, senescence, apoptosis or related cell phenotypic changes within the 3D constructs.48 In the literature, some basic experimental works identifying cell death induced by the 3D printing process49,50 and different behaviors of cell differentiation comparing casted vs. printed constructs51,52 have been published. This topic needs to be studied more in-depth to better understand the impact of the printing process on cells and evaluate whether printed constructs outweigh simple casted implants with regard to their biological performance. In addition to biological evaluation, functional characterization, such as histological or mechanical characterization, of printed engineered constructs is essential. For example, mechanical tests of both cartilage-like and bone-like constructs are needed to quantify their mechanical strength, elasticity and absorption at the material interface and therefore to understand their interaction with the tissues.53 However, while the mechanical characterization of bone has been well established for preclinical studies, functional testing of cartilage has yet to be fully characterized.54 In fact, although it is well known that the mechanical properties of engineered cartilage constructs have to match those of native cartilage at load-bearing joints, there is still a lack of standard procedures for testing the mechanical properties of engineered articular cartilage, which often leads to an inability to compare the results of different studies. A recent review by Patel and colleagues55 discusses mechanical testing of articular cartilage and underlines that compression testing (various modes such as ramp, stress relaxation, creep, dynamic and testing configurations such as unconfined, confined, in situ) is the most common test performed, followed by an evaluation of frictional properties. Similarly, a systematic review published by Marchiori et al.56 discussed cartilage mechanical characterization, showing that unconfined compression (both dynamic and static conditions) is the most diffuse configuration, although it does not have a standard reference. Additionally, confined compression tests are more common and standardized than indentation tests, probably due to the more straightforward experimental and analytical configuration. Mechanical tests of cartilage devices are not reported by ISO, whereas the FDA has clear guidelines for these devices,57 specifically for knees, where they recommend the major evaluations that have to be carried out, such as the ability of an implant to withstand expected in vivo static and dynamic loading (e.g., compression, shear and tension) and the most relevant measurements, such as maximum recoverable compressive strain, aggregate modulus, shear modulus, permeability and complex shear modulus G* measurements.
Furthermore, more specifically for 3D printed constructs, GLPs that follow quality criteria and standards for characterization of the printed samples need to be defined to bring the field closer toward clinical translation and application.
To date, there are no regulatory guidelines of the US FDA or other institutions available for 3D bioprinting cellular or acellular constructs for application in clinical studies, which can be explained by the infancy of the technologies. In contrast to living 3D bioprinted products, nonliving and thus cell-free printed devices or surgical tools made of medical-grade plastics (e.g., polyether ketone), which are tolerant for subsequent sterilization procedures,58 are already in clinical use. Medical applications of these products include the use of anatomical models based on patient radiological data serving as intraoperative surgical guides, biomodels to plan surgical approaches or education tools59,60,61,62,63 and constructs serving as space fillers or bridges for dental use.64 To produce the abovementioned biomedical products, the FDA released guidance covering device design, software workflow, material controls, postprocessing and process validation (Technical Considerations for Additive Manufactured Medical Devices Guidance for Industry and Food and Drug Administration Staff, FDA-2016-D-1210). ISO provides a similar document, the ISO/ASTM 52900:2015 (ASTM F2792) Additive manufacturing—General principles—Terminology (2015).
Quality monitoring of tissue engineering constructs
Destructive methods for quality analysis
Analysis of cellular toxicity
Cytotoxicity testing is a first step toward ensuring the biocompatibility of a biomaterial that might be used as a medical device. Cytotoxicity testing is reported in the ISO 10993-5: “Tests for Cytotoxicity—In Vitro Methods” This ISO standard describes a set of rapid, sensitive, and inexpensive tests designed to standardize cellular toxicity and quantify the effect of biologically harmful products. These tests are widely used for the assessment of cell viability in 3D tissue engineered constructs.
It is crucial to follow specific assay conditions. Details on the procedures in the preparation of samples are reported in part ISO 10993-12:2012(E). For example, in the extract test method, extracting conditions should simulate the clinical use conditions without altering the chemical properties of the samples.
According to ISO 10993, a reduction in cell viability by more than 30% is considered to be a cytotoxic effect. This cellular toxicity can be determined following different evaluation categories: assessment of cell damage by morphological means, measurement of cell growth, and measurement of specific aspects of cellular metabolism. Additional qualitative morphological scoring is available to assess changes in the general morphology, vacuolization, detachment, cell lysis and membrane integrity.
One of the most common cytotoxicity tests is the (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazoliumbromid) (MTT) assay (ISO 10993, Annex C), based on an optical density measurement.
In addition to the evaluation of the cytotoxicity of the compounds connected to ISO 10993, there are various other methodologies to monitor cell viability and metabolic activity in 2D and 3D culture setups, including live-dead staining and lactate dehydrogenase (LDH) assays (Fig. 1).
Analysis of cell viability in 2D and 3D cultures. a Comparison of viability and metabolic activity assessment via spectrophotometric readouts or (confocal) microscopy in 2D and 3D in vitro culture setups. b 2D cultures are shown as monolayers in well plates or tissue culture flasks where reagents have direct contact with the cells. c 3D samples are characterized by the presence of a biomaterial as a supportive structure (e.g., fabricated via 3D bioprinting) in which the cells are embedded. Reagents need to diffuse through the material to stain or be metabolized by the cells, and thus, a limitation in penetration depth may result in low or no positive staining toward the center of the 3D construct, as illustrated for live-dead staining. Furthermore, many biomaterials or compounds present in the materials can interfere and show an autofluorescent signal that, in the worst case, results in higher readout values or background staining than that derived from the cells only
To date, the abovementioned methodologies, based on spectrophotometric or imaged-based readouts, have been established and are routinely used in 2D cultures where the reagents are applied on cell monolayers and are thus in direct contact with the cells. The main difference between 2D and 3D cultures is that in 3D cultures, the cells are embedded within a biomaterial or seeded onto a 3D scaffold. Reagent-based assays to assess cell viability or differentiation have some limitations when cells are embedded in a 3D material: on the one hand, materials may absorb dyes due to electrostatic interactions and thus interfere with subsequent absorbance or fluorescence measurements. Furthermore, many biomaterials or compounds present in the materials interfere with an autofluorescent signal that, in the worst case, result in higher readout values or background staining compared to the signal derived from the cells.
Another issue addresses the diffusion of the reagents into the 3D construct to assess the viability or metabolic activity of the full construct. In 3D cultures, the reagents need to diffuse through the material to stain the cells or be metabolized by the cells. The presence of a biomaterial reduces the diffusion of the reagents, and there is the risk of a penetration depth limit. Thus, gradients with high concentrations will occur at the periphery of the large, centimeter-scale samples with decreasing concentrations toward the center unless the incubation times are optimized to the specific biomaterials and geometry (shape and size).65
Analysis of molecular markers
The analysis of gene expression is a powerful tool for the evaluation of tissue engineering constructs since it provides information about the differentiation status and maturation of a construct, allows control of the status of cell differentiation pathways, and enables monitoring of the effectiveness of drug treatments.
There are different techniques that can be used for the analysis of gene expression (as schematically depicted in Fig. 2), each with specific advantages and disadvantages. Among those, the most useful in the field of tissue engineering are quantitative real-time polymerase chain reaction (qPCR), probe-based microarrays, RNA sequencing (RNAseq), or RNA imaging of fixed or live cells using specific probes.
Quality monitoring via gene expression analysis. a Gene expression analysis starts with RNA isolation, which implies the destruction of the sample. Different techniques can then be employed to assess gene expression levels, including qPCR analysis, microarrays, and RNA sequencing. b Intracellular RNA can be imaged using fluorescent nucleic acid-based probes. The probe should be transfected into the cells; once inside, the probe anneals to the target sequence, leading to an increase in fluorescence, which can then be analyzed via confocal microscopy
qPCR-based approaches represent the method of choice for targeted gene expression analysis. The main pros of qPCR are specificity, sensitivity, customizability, flexibility, robustness, constant technology and reagent development, and availability of validated assays to cover the whole human transcriptome and a large number of noncoding (nc)RNAs. The main disadvantage of qPCR is that it is a destructive technique, and even though a good number of targets can be analyzed in the same sample, it does not allow for a transcriptome-wide analysis and requires prior knowledge of targets to be investigated.
A different approach is based on gene expression microarrays that can analyze thousands of genes in the same sample. The advantage of microarray technology over qPCR is undoubtedly the possibility of analyzing a larger number of targets in the same sample, on the order of thousands of genes, therefore covering a larger portion of the transcriptome and allowing for pathway analysis.66 The main disadvantage is that microarrays are probe-based, thus requiring prior knowledge of the sequences to be analyzed.67
Some of these limitations can be overcome by RNAseq methods. The main advantage of RNAseq is that its approach is unbiased and allows for a transcriptome-wide analysis. RNAseq disadvantages are mainly destructivity and high costs, although they decrease with time.68,69 Moreover, the analysis of data is complex and requires specialized bioinformatics skills.
More recently, imaging techniques based on RNA probe hybridization have been developed. These techniques are less commonly used in the field of musculoskeletal tissue engineering. RNA probe-based imaging has the great advantage of being potentially adapted for live cell imaging, as described by some authors with molecular beacons.70,71,72,73 Its main limitation is the low multiplexing capability and a high level of optimization required for signal detection and maintenance of cell viability, especially in 3D constructs.
Regarding quality guidelines, there are a few documents applicable to gene expression analysis, but standards are under development for sequencing. Table 2 summarizes standards and literature on quality relevant for gene expression analysis.
Histological assays
One of the main purposes in tissue engineering is to create organized and functional tissues that are as close as possible to physiological tissues.74 Therefore, a morphological assessment is necessary to evaluate the quality of the in vitro matured tissues through histological and histochemical techniques.75
One of the principal tissue stains used in histology is hematoxylin-eosin staining (H&E), which gives a general overview of cell distribution and tissue organization. However, this type of assay does not provide much information regarding the quality of the extracellular matrix produced, i.e., of the presence of proteoglycans and of cartilage-specific collagens. For this purpose, extracellular matrix analysis is commonly performed by staining with Safranin-O,76 a dye that highlights the presence of proteoglycans and glycosaminoglycans. Counterstaining is usually achieved with Fast Green and Weigert’s hematoxylin for cell nuclei. The staining is characterized by a red appearance in the presence of negatively charged glycans, with a green color indicating other cells and collagen. Other types of staining, such as Masson’s trichrome, are used for the visualization of collagenous connective tissue fibers in tissue sections.77 Tolonium chloride (also known as toluidine blue) and Alcian blue staining can indicate the presence of acidic polysaccharides such as glycosaminoglycans in cartilage and other body structures.78 Another method is to use dimethyl methylene blue (DMB or DMMB) for cartilage detection.79 These techniques, although qualitative, do not allow us to discriminate which specific molecule is expressed in the construct. Especially during stem cell commitment toward chondrocytes, it is extremely necessary to understand the quality and stage of differentiation. The main protein markers associated with cartilage are type II collagen, which constitutes the articular and hyaline cartilage; type I collagen, conventionally associated with fibrous cartilage; and type X collagen and matrix metallopeptidases (MMPs), usually expressed during hypertrophic differentiation, when cells commit to osteochondral and bone formation. The expression and distribution of those proteins can be analyzed by immunohistochemical staining.
In light of bone tissue engineering, the main characteristic is the production and thus presence of a mineralized matrix, which can be analyzed in different ways. Both Alizarin Red and Von Kossa staining can be performed on histological sections and in monolayer cultures to visualize deposited bone-like matrix. Other methods to stain calcium can include fluorochrome labels, such as calcein green, xylenol orange or tetracyclines, although their use is more applied to assess new bone formation in in vivo studies.80 Other fluorescent dyes with the ability to bind to hydroxyapatite (HA) have been developed and can be used in cell cultures and tissue sections, as in the case of Giemsa81 or fluorescein-bisphosphonate conjugates.82 In addition to mineralization, immunohistochemical detection of bone-specific proteins can be of course envisaged to assess the quality of bone.83,84
Although a first analysis through histological techniques can provide an idea about the morphology of the construct and its quality, this approach still has some limitations. The main advantage of using classical histological staining is its low cost, direct visualization where tissue-specific extracellular matrix is deposited, and high throughput. However, the qualitative evaluation of specimens should be performed by immunohistochemical assays in which specific antibodies bind proteins of interest, allowing effective assessment. Although this type of technique appears simple, it can be affected by several variables, including temperature, antibody concentration, fixation, demasking protocols, and binding to biomaterials, which can easily lead to false-positive or negative results. Thus, the parallel staining of internal and external controls, e.g., native tissue specimens or nondifferentiated cells, is of great importance for the evaluation of the staining results.
The main limitations of this technique already arise from the fixing method, which can radically change the recognition of the antibody used. The inclusion phase represents another limiting step, which can totally change the interpretation of the result; indeed, paraffin or resin embedding can generate artifacts in the results by modifying 3D constructs if mainly constituted by hydrogels.85,86 However, the use of snap freezing and subsequent cutting with cryotome could partly overcome this limit. Finally, the extracellular matrix can often interfere with the detection of fluorescent antibodies because of its intrinsic autofluorescence, which could easily lead to a false interpretation. Furthermore, histological and immunohistological analyses involve the destruction of the sample and the use of expensive reagents such as antibodies.
Laser confocal microscopy
Laser confocal scanning microscopy (LSCM) (Fig. 3) has become an important instrument that has been widely used in materials science,87 cytobiology, morphology,88 pharmacology,89 neuroscience90 and tissue engineering.91,92
Summary of microscopy techniques for qualitative and quantitative assessment of cells, scaffolds and tissue engineered constructs. a With laser scanning confocal microscopy (LSCM), it is possible to analyze the expression of specific proteins and nucleic acids and to assess the biological responses and morphological organization of cells using fluorescent dyes. b With scanning electron microscopy (SEM), it is possible to gather high-resolution structural and analytical information about tissues and constructs. c Atomic force microscopy (AFM) allows us to analyze the surface and mechanical properties of constructs
LSCM can acquire fluorescence from only one focal point at a time, allowing the generation of high-quality images. Compared with a traditional optical microscope, a confocal microscope can obtain extremely precise 3D images and measure subcellular structure.93
Similar to classical histology, cell characteristics, cell organization within a scaffold, and production of extracellular matrix proteins can be analyzed by using specific fluorescently labeled antibodies or other molecules that target cellular structures such as nucleic acids, membranes, and mitochondria (Table 3).
Confocal microscopy can be used both for semiquantitative studies94,95,96 and for qualitative evaluation of samples, with the possibility of reconstructing the 3D organization.95
Labeling of cells can be performed in fixed or living constructs, depending on the outcome needed, and quality assessments can be performed with different methods. The tracking of cells in a tissue engineering study is of paramount importance; for this reason, common techniques aim to label cell components with or without the use of antibodies.
Frequently, transfection of cells by using genes that encode fluorescent proteins, such as eGFP, tdTomato, or mCherry, is used to study cell morphology and organization; the use of vital dyes that label membranes is of considerable interest for the study of cell migration inside 3D tissues or in scaffolds. PKH dyes are used for monitoring cell trafficking and function, allowing the analysis of live tissues over time.97,98 This method is usually stable for some cellular doublings, but the signal can be diluted or lost in long-term experiments with high cell proliferation, leading to a decrease in quality or discontinuation of the analysis. Labeling of cytoskeletal components with phalloidin or Taxol derivative conjugates allows the visualization of fixed cells without the possibility of longitudinal application.
The labeling of DNA by DAPI, Hoechst dyes, or ethidium homodimers can facilitate the identification of cells into constructs, and higher concentrations of dyes can also penetrate and stain nuclei in live, intact cells.
Other molecules such as tetramethylrhodamine ethyl ester (TMRE) and tetramethylrhodamine methyl ester (TMRM) are membrane potential-sensitive, cationic fluorophores99 that stain mitochondria for oxidative stress and damage.
The study of cell viability, proliferation, and adhesion into the scaffold can also be performed using different methods. One possible approach uses phosphor dots inserted into the scaffold that become imperceptible when cells grow over them.100 Calcein-AM and ethidium homodimers are used for live/dead staining, allowing the study of cell viability in the construct. Nonfluorescent calcein-AM freely enters live cells where the acetoxymethyl ester is removed by hydrolysis by intracellular esterases; low concentrations of ethidium homodimer can cross the cell membrane, only in case of cell damage or death, and can bind to the DNA minor grove.101 The main advantage of using the live/dead method in confocal microscopy is the opportunity to evaluate cell viability in a 3D setting. However, one of the fundamental limitations of this technique is that it is necessary to acquire information in a short time once the sample has been stained, as calcein becomes toxic and can easily give false results. Furthermore, it is no longer possible to use and culture the constructs after staining.
Recently, a new method for the quantification of specific mRNA in living cells has been proposed with molecular beacons.70,71,72,73 These are hairpin-shaped nucleic acids, with a fluorescent reporter in one end and quencher on the other. Once the molecular beacon recognizes and anneals to the cytoplasmic target RNA, the structure opens, allowing the fluorophore to emit fluorescence. This conformation allows the real-time detection of the presence of a specific RNA target, making semiquantification possible.
The main limitation in LSCM depends on the optics, where a z-stack acquisition of 200 µm can restrict deeper analysis. Nonetheless, this method can be used for the observation of living cells in a complex 3D environment. Sample preparation can involve the use of antibodies and dedicated optical supports that could be quite expensive on a large scale. The use of dyes for scanning acquisition can interfere with further culturing and use. However, the broad use of confocal microscopy has some disadvantages. For example, the use of powerful lasers on living cells over a long period of time can create certain phototoxicity that can damage samples, while for fixed samples, photobleaching can wreck the fluorescent signal. However, the introduction of two-photon LSCM, where two low-energy photons enable stimulation of a fluorochrome instead of a single strong photon, partially solved these problems, reducing phototoxicity and photobleaching in samples.102
Regarding the guidelines proposed by the International Organization for Standardization, ISO 21073:2019 is applied only to confocal single point scanners using single photon excitation procedures, and this document referred to more general documents as ISO 8039, ISO 10934-1, and ISO 10934-2 (Table 4).
ISO 21073:2019 can be applied to LSCM to image fluorescent biological specimens with high performance, increasing the repeatability, reliability, consistency, and overall quality of output. ISO 21073:2019 includes mechanisms for the resolution and strength of optical sectioning, uniformity of field and centering accuracy, coregistration accuracy, stability of illumination power, field number of the confocal scan optic and scanning frequency. In addition, ISO 21073:2019 includes other information regarding the specific wavelength of light required to excite a fluorescent molecule, such as a fluorescent antibody or fluorescent protein, the emission of light at emission and wavelength range of light collected by the photodetector, the pinhole diameter in terms of the low numerical aperture approximation, the ratio of signal to noise, and the glycerol air and water immersion refractive index.
Light-sheet microscopy
In addition to conventional high-resolution microscopy, optical imaging solutions exist that allow for the label-based and label-free analyses of small to larger tissues (cm-range). Light-sheet microscopy (LSM) is essentially a nondestructive microtome that provides 3D images of tissue constructs. While it is still based on the detection of fluorescent labels, it is also typically referred to as light-sheet fluorescence microscopy (LSFM). Although LSFM is not label-free, it has advantages over conventional microscopy and even conventional histology/pathology, especially in rapid volumetric microscopy; that is, it serves end-point analyses as well as monitors live tissue at high-resolution and thick tissue constructs.103,104 Images are obtained by illuminating a focal plane at an angle of 45° relative to the vertical direction, and the fluorescent signal is imaged orthogonally to the incoming light. LSFM can easily reach intracellular resolutions of <1.5 µm and extreme tissue penetration depths of >200 µm.105,106 The disadvantages of LSFM for the nondestructive monitoring of tissue samples are label-dependent detection and a limited penetration depth. Furthermore, this method can be used to image only transparent samples but not calcified tissue, such as bone.107
Scanning electron microscopy
Scanning electron microscopy (SEM) uses high-energy electron beams to image the surface of samples and provides information about sample morphology, chemical composition, and crystalline structure. The signals produced by the interaction between accelerated electrons and the sample comprise secondary electrons and backscattered electrons used for sample imaging. X-rays, specifically emitted by each element, are used to determine chemical compositions. In particular, secondary electrons have very low energies on the order of 50 eV, so imaging is limited to the superficial nanometers of the sample, whereas backscattered electrons are reflected from deeper regions (<100 nm) of the sample by elastic scattering. The spatial resolution of SEM ranges between 1 nm and 20 nm, depending on the size of the electron spot, which in turn depends on both the wavelength of the electrons and the electron-optical system producing the scanning electron beam.
SEM has been mainly employed for materials characterization, e.g., nanoparticle characterization or surface topography;108,109 however, it has demonstrated great potential in imaging biological samples, such as tissue fragments and cells.110
To image a biological sample with a conventional SEM, vacuum is required, and therefore, the sample must be dried, frozen and coated with a metallic layer. The development of the environmental SEM (ESEM) in the late ‘80 s has allowed the analysis of samples containing water or other volatile substances because of differential pressure-limiting apertures in combination with a pumping system in the path of the electron beam that maintain the gaseous environment around the sample. However, the electron gun itself is kept at standard pressures (10–6 to 10–7 torr).
One application that involves the use of SEM is immunocytochemistry on biological samples.111,112 The antigen/antibody complex is labeled with a probe that produces a high secondary or backscattered electron signal. Colloidal gold probes are the selected probes due to their low background, difference from biological tissue components, and distinctive X-ray signal. Controls should be carefully chosen to check the performance of individual reagents or sample contaminations: a known positive control avoids false negatives and tests the effectiveness of the labeling procedure (reactivity of antigen, antibody, and markers), whereas a negative control assesses methodological nonspecificity of the technique, such as the secondary antibody labeling capacity (incubation with the secondary antibody alone).
In addition, SEM can be exploited to obtain both structural and analytical information regarding bone tissue, e.g., bone remodeling and bone pathology.113 In the field of bone research, the backscattered electron imaging mode is more useful than the secondary electron mode because it allows the determination of the mineral-density distribution patterns within bone.114 SEM can be employed to study bone resorption with the quantification of the resorption pits formed by osteoclasts in vitro.115 Another application is a comparison of human osteoblast growth on allografts and synthetic and xenogeneic bone grafts for bone lesion treatments.116
On the other hand, SEM imaging of cartilage is more challenging than that of bone due to its high water content and the need for dehydration steps during sample preparation that can lead to artifacts when analyzing the matrix structure. However, microwave fixation and cryogenic methods (e.g., freeze fracture) have been used in cartilage surface studies to overcome this limitation.117,118 In the work of Suso et al., environmental SEM helped obtain high-resolution images of fresh articular cartilage surfaces without sample fixation, thus minimizing the risk of creating artifacts in the structure.119
Overall, because of its user-friendly interface, rapid image acquisition, and ease of use, SEM is a powerful tool for imaging both materials and biological samples. The few limitations of SEM are represented by the limited size of the samples that can fit in the microscope chamber, the analyses of wet samples with conventional SEM (low vacuum and environmental SEM) and the use of conductive coating. Additionally, some solid-state X-ray detectors are not sensitive to low represented elements and cannot detect very light elements. Another limitation is the imaging of ultrafine but large structures due to the limited acquisition throughput of standard SEM.
Recently, a novel multibeam scanning electron microscope (mSEM) was developed for rapid-throughput scanning of large sample areas (e.g., a 2 mm-tissue block).120,121 Originally, mSEM was optimized for the quality control of semiconductor wafers at the nanometer-length scale, and afterward, in combination with sectioning and volume-rendering methods, it was used to reconstruct macroscopic volumes of murine brain tissue.122
Another recent advancement in imaging cellular and subcellular structures in 3D is represented by serial block face SEM (SBF-SEM) and focused ion beam SEM (FIB-SEM), more commonly used in the materials science and semiconductor fields. Although SBF-SEM can generate a stack of up to thousands of 2D images containing ultrastructural information for the bulk of the volume, it still has some limitations in terms of artifacts due to the use of a mechanical device to slice the sample and poor control of the thickness of each slide. Instead, in FIB-SEM, an ion beam is applied to carefully remove ultrathin layers of tissue, allowing for reconstruction of z-stacks capable of resolving intracellular organization in fine detail.123 For example, in the work of Hasegawa et al., the ultrastructure of cartilaginous extracellular fibrils and osteoblastic cytoplasmic processes were imaged via FIB-SEM. The results showed that osteoblasts not only extend their cytoplasmic processes to the bone matrix but also stack these cell processes on the osteoid of the primary trabeculae.124
There are no specific ISO procedures that apply to SEM technology; however, some ISOs should be followed to validate methods of sample processing and analysis and increase the repeatability, reliability, consistency, and overall quality of the output (Table 4). One of these documents is ISO 17025, which includes mechanisms for quality control, document control, analysis quality, and the trending of data. A more specific standard, ISO 13322-1:2014, can be applied to determine the particle size distribution of acquired SEM images. This ISO does not address the sample preparation (this is central to ISO 14887 and ISO 14488); however, a correct particle dispersion ensures accuracy of the final results.
Atomic force microscopy
Atomic force microscopy (AFM) belongs to the family of scanning probe microscopy (SPM) technologies and is used to image surfaces by mechanically scanning a probe over a surface and sensing the surface properties at the nanoscale or atomic scale. Specifically, a sharp probe with a nanometric tip is attached to a flexible cantilever; this probe is available with different stiffnesses and tip shapes. The cantilever deflection is recorded and provides information on the surface force. The standard geometry of the tip is a pyramid with a radius curvature of 20–30 nm.
AFM can provide not only information on the topography of the surfaces but also on mechanical, chemical, electric and magnetic properties and many others depending on the mode of operation and the property of the probe tip. Some examples of operation modes are contact mode (actual contact between the tip material and the surface), tapping mode (intermittent contact between the tip and surface, preferred for the imaging of soft biological materials), and pulsed force mode (sinusoidal movement of the cantilever for quantitative mapping of surface mechanical properties and acquisition of the surface topography in tapping mode).
AFM has been used in materials science and nanotechnology applications (e.g., imaging of polymers), biochemistry applications (nanostructural details and biomechanical properties of biomolecules, cellular components, cells or tissues), and chemistry, physics and biophysics applications.125 In the biological field, AFM allows us to measure the mechanical properties of the cell membrane, cell stiffness, and cell viscoelasticity and to assess cell adhesion.126 AFM has become an essential technique in biomedical applications,127 especially in the study of drug targets128 and has been gaining increasing interest in the field of bone and cartilage tissue engineering.
In particular, the mechanical properties of cells can be quantified from measurements of cell deformability, exploiting the AFM feature to acquire high-resolution measurements in a liquid environment.129,130 The resulting measure of cell elasticity (Young’s modulus) mainly reflects the deformability of the cell cytoskeleton and can be used as a marker for stem cell differentiation, as demonstrated in a study by Maloney et al.131. In particular, an inverse correlation between the actin fiber thickness and cell elasticity during the in vitro expansion of MSCs was demonstrated. In a recent study from Szydlak and coworkers, AFM was employed to determine the elasticity of Wharton’s jelly mesenchymal stem cells during in vitro culture.132 Generally, the amplitude and strength of local rupture events are not detectable by techniques performed at the level of the whole cell. Therefore, a constant velocity indentation AFM mode has been specifically used in the work from Streppa and colleagues to collect mechanical parameters of living adherent C2C12 myoblasts and myotubes.133
AFM is conceptually a simple technique that offers several advantages over electron microscopy, especially for imaging biological materials. This method provides a 3D surface profile at high resolutions (the z resolution is ~1 Å), does not require an expensive vacuum system and allows investigators to image the sample directly in its natural environment with minimal sample preparation (no coating, fixation) and fewer artifacts. There is generally no limitation in the medium selection, sample temperature, or chemical composition of the sample. AFM can give true atomic resolution in ultrahigh vacuum (UHV) as well as in liquid environments, and it can also be combined with a variety of optical microscopy and spectroscopy techniques, such as fluorescence microscopy or infrared spectroscopy. To avoid surface damage to biological samples, the spring constant of the cantilever must be kept at a low level (<0.2 N/m), whereas stiffer cantilevers are required to reduce noise when operating in tapping mode in air.
In contrast, one of the disadvantages of AFM is the scanning speed, which causes image distortions induced by thermal drift, followed by limitations in the scanning area (150 × 150 μm and a maximum height on the order of 10–20 μm); however, this issue can be overcome by the use of parallel probes. There is the possibility of image artifacts, which could be induced by an unsuitable tip, a poor operating environment, or scanning acquisition that is too fast. AFM images can also be affected by the nonlinearity, hysteresis, and creep of the piezoelectric material and crosstalk among the x-, y-, and z-axes. To overcome this crosstalk limitation, newer AFMs have introduced real-time correction software, closed loop scanners, or the use of separated orthogonal scanners.
There is a set of ISO guidelines available for scanning probe microscopy (Table 4). Most of these guidelines are of general relevance and focus, for example, the measurement of drift data (ISO 11039:2012), the determination of the cantilever normal spring (ISO 11775:2015), the vocabulary (ISO 18115:2013) and dimensional calibration (ISO 11952:2014). More detailed information can be found in the book “Quantitative Data Processing in Scanning Probe Microscopy: SPM Applications”.134 Additional standards, instead, are addressed to specific scanning probe microscopy techniques. For instance, ISO-27911 is specific for near-field optical scanning microscopes, and ISO 13095:2014, ISO 21222:2020 and ISO/DTR 13096 are specific for AFM. Since both the shape and the size of the AFM probe as well as the mechanism used to control the probe-sample distance can strongly affect AFM imaging, a quantitative expression of the probe tip shape is required to ensure reproducibility. Therefore, ISO 13095 specifies two methods for characterizing the shape of an AFM probe tip: a projection of the probe profile (PPP) that provides a continuous profile giving information regarding the quality of the probe, and the effective probe shape characteristic (EPSC) that provides few discrete points underlying the usefulness of a probe for depth measurements in narrow trenches. ISO 21222 describes a procedure to determine the elastic moduli of materials starting from force-distance curves acquired at the surface using AFM. The Johnson-Kendall-Roberts (JKR) two-point method is used to model the tip and surface contact in which adhesion forces outside the contact area are ignored and elastic stresses at the edge of the contact area are infinite; it applies to highly adhesive systems with low stiffnesses (elastic moduli ranging from 100 kPa to 1 GPa) and high radii of curvature.
A future improvement of high-speed atomic force microscopy is its combination with optical tweezers, which will allow the observation of a protein molecule responding to an external force applied to a specific locus in a given direction. To improve the investigation of biological samples, AFM can also be combined with standard and high-resolution fluorescence microscopy as well as Raman spectroscopy. This combination for cell imaging leads to high-resolution images that simultaneously display the 3D topography of the cytoskeleton and its stiffness and correlate them to its structural elements (e.g., distribution of actin filaments). Similarly, AFM can be coupled with a near-field scanning optical microscope to obtain simultaneous information on both the localization of molecular nanoclusters in cell membranes and protrusions. All these recent advancements are pushing AFM toward new developments in the pharmaceutical industry and clinical medicine.
Noninvasive methods
Raman spectroscopy
In the forties, Raman spectroscopy (RS) became one of the most commonly used vibrational spectroscopy techniques to analyze pure chemicals. As the H2O Raman spectrum is very weak, RS soon developed to be a powerful tool for the analysis of biological samples.135,136
In the early seventies, RS was used for the characterization of native, denatured or synthetized biomolecules137,138 and later for the study of proteins and lysosomes from isolated animal tissues.139 Since then, rapid innovation in microscopy and laser technologies has made it possible to discriminate living140 from dead cells141,142,143 and to identify cell cycle or differentiation stages.141,144,145,146,147 Being label-free, chemically selective, and minimally invasive, RS rapidly caught the interest of the medical field, where it may greatly impact diagnostics or monitoring methods.
A basic RS setup (Fig. 4) comprises a laser source typically in the near infrared range, a series of mirrors and a microscope directing the light toward the sample. After interaction with the sample, only a small portion of the scattered photons change their frequency. Following the filtration of nonshifted photons, photons with modified frequency are directed toward a spectrometer linked to a detector system (CCD), from which the data are transferred to a computer. However, as the Raman signal is very weak, in recent decades substantial effort has been devoted to finding ways to increase it.136 To date, there have been more than twenty enhanced RS techniques characterized by an amplified signal, and here, some of them are summarized in Fig. 5.
Simplified schematic of a Raman system. Reprinted by permission from Springer Nature Customer Service Center GmbH: Springer Nature. Springer eBook, Raman Micro-Spectroscopy as a Noninvasive Cell Viability Test, Verrier S. et al. © 2011143
Summary of Raman spectroscopy techniques: polarized Raman spectroscopy (PRS), suitable for highly oriented systems; Raman microscopy, able to couple chemical and morphological information; spatially offset Raman spectroscopy (SORS), used for collecting Raman signal from deep regions; coherent Raman spectroscopy (SRS & CARS), able to improve Raman signal intensity up to 105 factor; and surface enhanced Raman spectroscopy, (SERS) able to enhance signal intensity up to 1010 factor, exploiting the plasmonic effect between the sample and metal particles. The processing of Raman data collected from biological samples is accomplished by spectral analysis and statistical treatments
In spontaneous RS, the Stokes signal is detected under continuous radiation, typically from a diode laser with high spectral stability. For in vivo clinical RS applications, it is most convenient to deliver light to a tissue and collect it from the tissue by means of specifically designed fiber probes.148,149 However, one of the largest challenges of fiber probe design is to minimize tissue fluorescence.
Confocal RS implements a confocal microscope configuration to provide optical depth sectioning by spatial filtering of the collected RS light with a pinhole or an optical fiber to block out-of-focus signals. To date, confocal Raman probes have been used mainly for ex vivo and in vitro studies.150,151
Spatially offset RS (SORS) is similar to RS but collects Raman signals from deeper regions of the tissue by spatially offsetting the detection and excitation fibers. Collecting Raman signals at different offsets effectively samples different layers in the tissue. SORS typically uses a probe with an illumination fiber surrounded by detections fibers offset of 1–5 mm,152,153 but an offset as high as 16 mm has been used to perform Raman tomographic imaging in bone.154
Coherent RS uses two light fields (referred to as the pump and Stokes beam) so that the difference corresponds to a vibrational mode frequency of a molecular bond of interest. The coherent addition of the Raman signal from different molecules improves the signal compared to spontaneous Raman, typically by up to ~105.155 Coherent RS techniques include stimulated RS (SRS) and coherent anti-Stokes RS (CARS). Both SRS and CARS can be performed in highly fluorescent media, which is usually a rather limiting factor for Raman imaging in tissues.
Polarized RS (PRS) provides information regarding both the chemical composition and anisotropic directions of highly oriented systems, such as the amide I band and alpha helical conformation, representative of collagen.156
In the field of regenerative medicine and tissue engineering, Dooley et al.157 used phantom samples based on 3D-printed polycaprolactone (PCL):HA scaffolds to investigate the feasibility of SORS for monitoring the mineralization of bone tissue engineering scaffolds in large animal models. It was demonstrated that SORS was able to detect HA concentrations that were an order of magnitude lower than those found in living bones, even when a 4 mm thick layer of skin (mimicking in vivo transcutaneous measurements) was present.
Liao et al.158 disclosed the possibility of applying SORS for nondestructive analysis of bone tissue engineering scaffolds. A multilayered scaffold composed of bioactive glass foams (IEIC16), a 3D-printed biodegradable poly(lactic-coglycolic acid) scaffold and HA powder was used to model 3D tissue engineering constructs at a real scale for nondestructive investigation of the biomineralization process. The authors were able to evaluate spectral depth profiles at high speed (5 s for each spectrum) and the simplicity of SORS application as a promising noninvasive approach to study cell/tissue growth and for in vitro and in vivo monitoring of the 3D scaffold long-lasting biomineralization process.
With regard to Raman databases and standards, to date, available and free Raman spectra databases can be found only for minerals, inorganic materials, or simple organic molecules. For example, Bio–Rad’s SpectraBase (https://spectrabase.com) has over 24 000 spectra available, although most are basic organic compounds. Unfortunately, comprehensive databases of Raman spectra of biological compounds and tissues are still not available due to the complexity of biological systems and the necessity of standardization of Raman data. Nevertheless, some review articles reported an extensive collection of the most relevant Raman bands that can be found in a Raman tissue investigation. The review of Talari et al.159 represents today the most complete list of assigned Raman peaks from biological specimens. Talari reported in total more than 1 000 assigned bands extrapolated from Raman spectra belonging to both healthy and pathological tissues.
To meet the requirements of standardization, multidisciplinary and multicenter networks, such as Raman4Clinics (EU COST Action BM1401) and the International Society for Clinical Spectroscopy (ClirSpec), have recently been established to compare 35 Raman spectroscopic instruments and setups in 7 European countries.160
In addition, in a very recent study, Power et al. first proposed RS as a quality control tool for GMP-compliant manufacturing of tissue engineered cartilage.161 Current GMP-compliant quality controls for tissue engineering procedures include sterility testing and required cell number and viability testing. In addition to the current requirements, the authors propose to use RS to assess the quality of the donor site tissue (e.g., purity) and of the resulting tissue engineered construct prior to implantation.
RS analysis of biological samples contains much information, but once measurement procedures and data analysis protocols have been standardized and validated with other classical methods, Raman spectra can be produced and analyzed in a quick, automated, and noninvasive manner.
Microcomputed tomography
The application of imaging techniques based on microcomputed tomography (microCT) in the field of tissue engineering has been steadily increasing in importance for over a decade (Fig. 6). MicroCT can be used in different contexts (in vitro, in vivo and ex vivo) for the 3D and even 4D qualitative and quantitative evaluation of engineered materials, both in terms of optimization of the design and measurement of their regenerative capacity.
Schematic representation of the potential of microCT analysis for musculoskeletal tissue engineering. This nondestructive technique can be used for different applications for biomaterials per se or in combination with cells and tissue (both ex vivo and in vivo). The produced image dataset can be compared with that from different microscopy techniques, combined with biomechanical tests or used for 3D modeling
The technique is nondestructive and is based on the interaction of X-rays with matter. A series of radiographic projections taken at different angles is captured by a detector generating bidimensional images. Using computer algorithms, these projections are used to reconstruct the internal and external structure of the sample.162,163
The most common method of scanning is based on the attenuation of X-rays when they pass through matter. This makes the technique largely used for studying many aspects related to bone biology. In the field of tissue engineering, a wide range of materials can be directly examined, such as metals, ceramics, some polymers and mineralized tissues.162 The study of the morphological characteristics that are closely related to certain quality requirements, such as their regenerative properties, is crucial for improving existing materials or creating new products.164,165 In this context, the ability to obtain 3D datasets using microCT provides more accurate information on the structure of the sample than complementary two-dimensional (2D) methods, such as histology and scanning electron microscopy.166,167 Therefore, microCT is commonly used in the morphological and structural analysis of bone tissue engineering scaffolds prior to cell culture or implantation168,169,170 and in the in vitro evaluation of the influence of porosity on cell seeding.171 The nondestructive quantification of characteristics such as porosity, pore size distribution and interconnectivity is a key factor in terms of planning and optimization of scaffold designs because a defined porosity allows cell migration, improves proliferation and production of extracellular matrices and facilitates tissue growth and the invasion of blood vessels.172,173 Moreover, microCT is widely used in the evaluation of the extracellular matrix growth process within scaffolds.174 MicroCT provides a noninvasive and nondestructive tool for the evaluation of mineralization based on in vitro culture conditions,175,176,177 the osteoinductive and osteoconductive properties of scaffolds through the 3D quantification of newly formed bone in preclinical models ex vivo178,179,180 or longitudinally in vivo,181,182 and the resorption and degradation of biomaterials in vitro and in vivo.183,184,185
The morphological analysis of scaffolds can be coupled with mechanical tests (e.g., porosity versus mechanical load resistance), providing an additional rational basis for the design and optimization of scaffolds. For example, microCT imaging can act as a source for finite element (FE) analysis, increasing the simulation modeling accuracy,186 and it can be used for the dynamic monitoring of compressive strength.187,188 In addition, the combination of microCT imaging and mechanical tests allows a digital volume correlation (DVC) approach to study the micromechanics of bone-biomaterial systems.189
Although microCT is mainly used for mineralized tissues, currently the imaging of materials or tissues with a lower attenuation coefficient, such as hydrogels, natural polymers, cells, and soft tissues, is possible due to the increasingly widespread implementation of contrast agents.166,167 For example, osmium tetroxide (OsO4) has been used to increase the contrast of some polymers scanned in an aqueous medium166,167 or to evaluate 3D cell colonization inside scaffolds in combination with DNA quantification.190,191 Mixtures of osmium tetroxide and uranyl acetate or uranyl acetate and lead citrate were used for the study of collagen-based scaffolds.192 Iodine-based or phosphotungstic acid (PTA) contrast agents have been used for in vitro extracellular matrix visualization inside engineered scaffolds.193,194
However, these contrast agents are often toxic, making longitudinal and nondestructive evaluation impossible.
In addition to absorption, it is possible to use approaches based on other aspects of the X-ray/matter interaction. X-ray imaging techniques based on phase contrast (PC) use X-ray refraction and phase shifting. PC microCT has great potential in biomedical applications because variations in refractive indices are generally higher than variations in X-ray attenuation coefficients. Therefore, information on the structure of soft tissues or materials with low absorption can be theoretically obtained without contrast agents.195,196 PC techniques have been applied in the field of tissue engineering for the visualization of polymeric scaffolds under cell culture conditions,197,198 for the characterization of polydimethylsiloxane scaffold deformation caused by ultrasounds,199 and for the detection of 3D cell organization in polyglycolic acid—polylactic acid (PLGA) scaffolds.200
MicroCT images can also be used as valid input for 3D bioprinting, improving the manufacturing of increasingly complex and customized scaffolds.201
The advantages of the microCT imaging technique can therefore be summarized in its noninvasive and nondestructive intrinsic characteristics, the possibility of high-resolution 3D visualization, the possibility to perform longitudinal studies on the same sample, and digital-based information that can be used to mathematically describe a 3D object. MicroCT can be used both to integrate existing analysis techniques and in combination with them because the analyzed samples can then be histologically assessed or biomechanically tested.202 In addition, continuous hardware and software innovations increase the 3D rendering capabilities up to virtual reality and augmented reality applications.
The main limitations of the technique lie in the significant effect of acquisition settings (e.g., nominal resolution and the rotation step) on image quality,203 in the difficult identification of correct thresholding values for materials or tissues with similar absorption coefficients,183 and in the radiation dose in in vivo longitudinal studies.204,205 In addition, microCT imaging acquisition and analysis are not standardized. However, the coordination of the results can be maximized using references that describe the validation of the methods through a comparison and correlation with other traditional techniques such as histomorphometry.206,207
In the context of good laboratory practice (GLP) compliance, specific applications can be validated through SOPs, which mainly take into account the generation, integrity and monitoring of electronic data, calibration of the systems and validation of associated software. Nevertheless, nonclinical imaging data can be incorporated into regulatory communications even if they are not GLP compliant. In this respect, it is preferable to describe in detail (1) the imaging procedure enclosing complete descriptions of hardware and software; (2) the degree of reliability, quality, and integrity of the electronic data to be able to track and reconstruct the data processing; and (3) the data management and archiving procedures. Measures to prevent unintentional image changes and backup and restore procedures should also be considered208.
Some international standards in the field of computed tomography have been published, but none cover the validation and calibration of CT systems. The most important published standards for CT are summarized in Table 5.
Optical coherence tomography
A technology to overcome the label-dependent nature of optical tissue microscopy is optical coherence tomography/microscopy (OCT/OCM). This is an imaging technique with a resolution of >10 µm and penetration depths of up to 2 mm. In OCT, the reflected light of a broadband, low-coherence light source is detected by an interferometer, which results in clear 3D images. Despite its limitations in resolution, it can display macrostructural tissue morphologies with a label-free method and is even compatible with calcified scaffolds,209,210,211 making it particularly relevant for monitoring cell proliferation,212,213 musculoskeletal structures209 and diseases.210
Special modes of OCT can disclose more information with respect to a tissue construct than only its still complex morphology. Optical coherence elastography (OCE), for instance, essentially images a tissue undergoing mechanical deformation.214 Local strain distributions are then estimated based on cross-sectional maps of the mechanically induced (cell) displacement. OCE has thus the potential to mechanically characterize engineered tissue, essentially contributing to final quality control. However, more than the tissue itself can be analyzed by different modes of OCT; for example, Ghosn et al. reported the use of OCT for label-free imaging of glucose diffusion within tissues, thereby underpinning the versatility and potential of OCT for analytical tissue engineering imaging.215
Conservative methods
Electrochemical sensors and biosensors
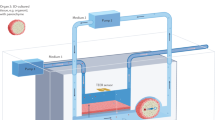
In disciplines such as neurosciences, for more than a decade, sophisticated analytical monitoring techniques have been in use for the in vitro and in vivo monitoring of chemical processes in real time. The use of these techniques is still limited in the fields that are the focus of this review. However, it is envisaged that the use of sensors, particularly miniaturized microsensors, can be developed, implemented, and adapted ad hoc for the study and monitoring of the cellular quality of 3D constructs for musculoskeletal tissue engineering and biofabrication (Fig. 7). From the technological, qualitative, and economic point of view, the translation of expertise and sensors from neuroscience to in vitro applications and tissue engineering would bring quality control of implants to the next level and thus accelerate translation to clinical applications.
Overview of the applicability of sensors to the field of tissue engineering. Critical parameters for construct quality control (e.g., glucose, lactate, and oxygen consumption) could be monitored in different types of in vitro culture systems (a) by using electrochemical (b) and optical (c) sensors/biosensors for real-time quality monitoring (d). These methods are conservative, allowing for longitudinal studies and further applicability of tissue engineered constructs
During the last few decades, biosensing technology has attracted much attention due to the numerous features that make it applicable in several fields, such as environmental, agri-food, and biomedical, both in vivo and in vitro.216,217,218,219,220 Sensors and biosensors are widespread not only in diverse fields of application but also because they offer the possibility of being miniaturized. This characteristic allows real-time monitoring of several compounds (e.g., glucose, lactate, glutamate) at very low concentrations depending on the sensor used, thus representing a noninvasive measurement for direct analysis of dissolved molecules. In addition, the low production cost in manufacturing makes these biosensors particularly appealing, as they are economically sustainable for researchers. Indeed, in recent years, electrochemical sensors and biosensors have become interesting tools for application in tissue engineering and regenerative medicine.216,217,218,219,220,221
The characteristics of an electrochemical sensor can be modified for interaction with one or more analytes, transforming the obtained chemical signal into a quantifiable electrical signal. Depending on the electrochemical technique used (voltammetry, amperometry, conductometry or potentiometry), this signal can be linearly or logarithmically proportional to the analyte concentration. Amperometric microsensors are valued devices for both in vivo and in vitro detection of different compounds, such as dopamine,222,223,224 ascorbic acid,222,225,226 nitric oxide (NO)227, and oxygen (O2).225,228 In addition to the high sensitivity and quite high spatial and temporal resolution, microsensors are very interesting because they allow the real-time monitoring of the compounds under examination.
Amperometric devices can work in both oxidation and reduction modes. Among the monitored reducible compounds, O2 is one of the most important because of its physiological and biochemical implications. Oxygen monitoring occurs when a cathodic potential is used.225,228,229 O2 reduction arises by means of a two-step reaction, leading to H2O production (Fig. 8).
A biosensor is used when the compounds dissolved in aqueous matrices cannot be directly oxidized or reduced on the surface of the transducer, as in the case of glucose, lactate, glutamate, ethanol, or polyphenols.219,220,230,231 The term biosensor indicates a chemical sensor in which the recognition system uses a biochemical or biological sensing element. Depending on the technology, biosensors can be divided into two main categories: biocatalytic devices, where enzymes, tissues or cells are used, and bioaffinity sensors, when antibodies, nucleic acids, or receptors are employed. In particular, amperometric biosensors have become very attractive because of their sensitivity, fast response, and high spatial resolution. The first amperometric sensor was developed for the measurement of oxygen tension.232 Since then, amperometric sensors have evolved and have become important tools in research due to their numerous technical advantages. In neuroscience, biosensors have been integrated into telemetry systems, allowing direct signal transmission and the simultaneous monitoring of different analytes. In the biomedical field, biosensors are commonly employed for the real-time monitoring of different important compounds, such as neurotransmitters (e.g., glutamate),233,234,235 glucose and lactate.235,236,237
The most common amperometric biosensors are enzyme-based. These devices exploit the capability of some enzymes of the oxidase class to convert the compound of interest into an amperometrically detectable analyte. Indeed, in a biosensor for glucose, glucose oxidase (GOx) allows its conversion in the presence of oxygen, as depicted in Fig. 9. The concentration of H2O2 produced from the GOx reaction is directly proportional to the glucose concentration in the matrix, and H2O2 is easily oxidized on a platinum surface when an anodic potential is applied.220,230,231,238
Enzyme-based biosensors, based on platinum wire modifications, have proven to be particularly useful when implanted in brain tissues in preclinical models due to their biocompatibility and to their high spatial and temporal resolution. Depending on the application, such as the study of the cellular microenvironment in 2D cell cultures,239,240 sensors and biosensors can be developed with different shapes, such as cylindrical and conical. The main advantage is related to the small dimensions (50–500 μm), which allow them to be inserted inside tissues and organs. Planar sensors have been developed as well241,242 since these types of sensors and biosensors also offer a notable detection capability, simplicity, and low cost.
Although the application of biosensors in tissue engineering and regenerative medicine is still limited, these tools can bring great possibilities in the field.221 Through these tools and these techniques, it is possible to check many critical parameters to ensure proper tissue growth,243 such as oxygen and nutrient uptake or the release of metabolites. In addition, monitoring metabolic parameters can contribute to the standardization of experiments. Sensor-based systems for cell metabolism monitoring have been recently reviewed by Kieninger and coworkers.240 The investigation showed an overview of the possibility of using sensor systems to track cell metabolism even in 3D cultures. It is possible to directly measure extracellular substances in the medium (e.g., glucose, lactate, oxygen),244,245 but matrix interference is an important issue to address217 to make the monitoring of biosensors reliable and reproducible. Several studies have been performed by means of integrated microfluidics techniques246,247 and through modification of flasks and plates. Compared to batch systems, microfluidics techniques allow important advantages, such as dimensions or sample dilution.248
The materials used for sensor construction need to be biocompatible, nontoxic and sterilizable. Different sterilization strategies can be successfully followed,249 especially in combination, but any procedure has to be adapted to each sensor depending on the design chosen. Accordingly, inactivation and loss of the recognition element must be considered, particularly with regard to enzyme-based biosensors. However, several development approaches, such as enzyme immobilization or biosensor storage, can be used to maintain and improve the analytical performances of these tools.217
Future prospectives on amperometric microsensors and biosensors are focused on the development of nanostructured transducers because of new and improved functionalities that cover a wide variety of applications in the biomedical field and on the improvement of the duration of analytical performance in terms of enzyme activity.
Optical tissue environment monitoring
An easy and nondestructive way to assess cell metabolic activity or physiology is the analysis of the cell/tissue environment, namely, the cell culture medium. Sampling of small volumes of the medium allows the quantification of a multitude of metabolites and nutrients; typically, even less than 5 µL is sufficient for the analysis of glucose, cholesterol, glutamate, lactate dehydrogenase, ions, or total protein content, among others.250 Such assay-based kits are available in small size for use in multiwell plates with optical plate readers or even at industrial scale-throughput packs to be used in conjunction with specialized spectrometers. While metabolite quantification is not a new concept, it is still not widely used in tissue engineering applications. However, as process analytical technologies (PAT) originating from the field of pharmaceuticals (U.S. Food and Drug Administration. Guidance for Industry: PAT—A Framework for Innovative Pharmaceutical Development, Manufacturing, and Quality Assurance) become more relevant for transplantation of engineered tissue constructs, they in turn will await their standard employment in modern tissue engineering.
Such in-depth quantifications of cell/tissue environmental parameters are already well-established solutions – as long as enough liquid is available for sampling. When the total amount of available liquid does not allow for sampling or sampling is technically not feasible, optical probes are available for a small range of cell environmental parameter analyses. These probes are typically based on parameter-sensitive fluorescent dyes: for instance, a fluorophore that changes its fluorescence intensity and fluorescent lifetime upon a change in pH or oxygen concentration. Examples of fluorophores are ruthenium- and metallo-porphyrin-based molecules. Due to their cytotoxic nature, they are typically encapsulated into polymers, sol-gels, or silica matrices. The encapsulated formats can be beads, rods, flat pads, and other geometrical objects to either flow through an entire fluidic system or be located at a region of interest. Since the parameter sensitivity of the currently available fluorophores is still limited, most commercially available systems offer probes for pH and oxygen only. Nevertheless, pH and oxygen are two essential parameters for the development of tissue engineering modules and reactors, which could benefit from the implementation of probes that pose a good alternative to electrochemical sensors. The implementation does, however, have direct implications on the module/reactor design, as the probes must be optically accessible and the optical penetration depth into a tissue construct is very limited. Given that the implementation does not affect the cell/tissue culture and a USP class-VI polymer is chosen for fluorophore encapsulation, these optical probes can pose a noncytotoxic sensor for tissue constructs engineered for transplantation.
Summary and prospectives
The main purpose of tissue engineering is to create functional and implantable constructs able to restore damaged or lost tissue. In recent years, while the acceleration of technological development has fostered great improvements in the field, the translation of tissue engineered constructs into the clinic is still limited. In the biological field, a variety of methods have been proposed to improve the reproducibility of protocols in terms of cell isolation and differentiation toward targeted tissues. The process of cell embedding and maturation in biomaterials has also been developed further to move toward functional cell-laden implants. With the recent advances of technologies, skills, and methodologies to produce complex constructs that mimic the composition, macrostructure or microstructure of native tissues has been developed. However, from laboratory to clinical practice, the gap is still vast. One of the main challenges limiting the clinical translation of tissue engineered constructs is the lack of quality control of the end construct. Indeed, if the chemical, physical and mechanical properties of a biomaterial are reproducible, then the cellular component of such constructs is still subject to interindividual variations or cell manipulation that could hinder the cell-scaffold interaction, and therefore, the overall properties and potentially the efficiency of the construct can be affected. Another major lacking aspect regarding cell viability and cell maturation is the possibility of monitoring 3D constructs during in vitro culture. A real-time, noninvasive monitoring system would be of paramount importance to assess key biological parameters and possibly control cell differentiation processes, matrix production, cell-scaffold interactions, and overall development of the construct.
To date, there is no single and dogmatic assay to study the quality of 2D and 3D constructs. However, depending on need and accessibility, one technique can be chosen over another. The major limitation of the techniques routinely used in tissue engineering/regenerative medicine is that they often involve sample destruction. Due to the invasive character of these techniques, the samples cannot be used for further analyses; therefore, multiple constructs are needed to collect all required information.
In contrast, methods defined as noninvasive, such as confocal or microCT, allow preservation of the sample during the analysis, but they might subject the sample to strong stress, which could then invalidate the final quality. In recent years, further methods, e.g., Raman spectroscopy, have been developed and translated to biological and medical analysis. Though the current lack of RS standardization is the largest challenge for its translation in the clinic, no particular sample preparation is required, and Raman measurements can be carried out both in vitro and in vivo (i.e., intraoperative) conditions.
There is a clear need for the development or translation of noninvasive methodologies to the field of regenerative medicine and tissue engineering to bring the field one step closer to its clinical application. Currently, conservative methods such as minimally invasive sensors for musculoskeletal tissue engineering are completely unexplored but are envisaged to be determinant for the translation of solutions from the preclinical stage. Sensors also have great potential to facilitate the initial optimization of constructs and could be integrated into a more complex workflow that comprises the analysis of different parameters using multiple techniques and possibly allows the monitoring of the same sample over time (at least with some combinations). This “multidimensional” system for the quality control of constructs is highly advisable to exploit the advantages and overcome the limitations of single techniques. For example, above all the advantages of sensors, their main drawback is that they do not provide morphological data, which can in turn be retrieved by combination with other techniques.
As discussed in this review, recent technologies show promising advancements. The implementation of different sets of monitoring technologies will nevertheless remain the only solution for quite a while since the complexity of tissue engineering aspects imposes a plethora of physiologically relevant functions that cannot be assessed using a single technology at hand today. Thus, the combination of different advanced techniques and progress in laboratory applications/representations will improve the quality in the field of tissue engineering toward complete customization of the constructs and increased effectiveness of personalized medicine strategies. In the future, the choice of noninvasive strategies will furthermore outmatch invasive handling due to the limited availability of patient-derived cells as well as its potential to monitor tissue development repeatedly over time, providing insights into crucial morphological development.
Change history
28 December 2021
A Correction to this paper has been published: https://doi.org/10.1038/s41413-021-00174-w
References
Dominici, M. et al. Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy 8, 315–317 (2006).
Khademhosseini, A. & Langer, R. A decade of progress in tissue engineering. Nat. Protoc. 11, 1775–1781 (2016).
Hoffman, T., Khademhosseini, A. & Langer, R. Chasing the paradigm: clinical translation of 25 years of tissue engineering. Tissue Eng. Part A 25, 679–687 (2019).
Salih, V. Standardisation in cell and tissue engineering: methods and protocols. (Elsevier, 2013).
Loebel, C. & Burdick, J. A. Engineering stem and stromal cell therapies for musculoskeletal tissue repair. Cell Stem Cell 22, 325–339 (2018).
Salgado, A. J., Oliveira, J. T., Pedro, A. J. & Reis, R. L. Adult stem cells in bone and cartilage tissue engineering. Curr. Stem Cell Res. Ther. 1, 345–364 (2006).
Im, G. I. Clinical use of stem cells in orthopaedics. Eur. Cells Mater. 33, 183–196 (2017).
Maillacheruvu, P. F., Engel, L. M., Crum, I. T., Agrawal, D. K. & Peeples, E. S. From cord to caudate: characterizing umbilical cord blood stem cells and their paracrine interactions with the injured brain. Pediatr. Res. 83, 205–213 (2018).
Sensebe, L., Gadelorge, M. & Fleury-Cappellesso, S. Production of mesenchymal stromal/stem cells according to good manufacturing practices: a review. Stem Cell. Res. Ther. 4, 66 (2013).
Wuchter, P. et al. Standardization of good manufacturing practice-compliant production of bone marrow-derived human mesenchymal stromal cells for immunotherapeutic applications. Cytotherapy 17, 128–139 (2015).
Harris, D. T. Banking of adipose- and cord tissue-derived stem cells: technical and regulatory issues. Adv. Exp. Med. Biol. 951, 147–154 (2016).
Arutyunyan, I., Fatkhudinov, T. & Sukhikh, G. Umbilical cord tissue cryopreservation: a short review. Stem Cell. Res. Ther. 9, 236 (2018).
Takahashi, K. & Yamanaka, S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–676 (2006).
Andriolo, L., Merli, G., Filardo, G., Marcacci, M. & Kon, E. Failure of autologous chondrocyte implantation. Sports Med. Arthrosc. Rev. 25, 10–18 (2017).
Minas, T. & Bryant, T. The role of autologous chondrocyte implantation in the patellofemoral joint. Clin. Orthop. Relat. Res. 436, 30–39 (2005).
Giannoni, P. et al. Autologous chondrocyte implantation (ACI) for aged patients: development of the proper cell expansion conditions for possible therapeutic applications. Osteoarthr. Cartil. 13, 589–600 (2005).
Malicev, E., Kregar-Velikonja, N., Barlic, A., Alibegovic, A. & Drobnic, M. Comparison of articular and auricular cartilage as a cell source for the autologous chondrocyte implantation. J. Orthop. Res. 27, 943–948 (2009).
Mumme, M. et al. Nasal chondrocyte-based engineered autologous cartilage tissue for repair of articular cartilage defects: an observational first-in-human trial. Lancet 388, 1985–1994 (2016).
Garcia-Gareta, E., Coathup, M. J. & Blunn, G. W. Osteoinduction of bone grafting materials for bone repair and regeneration. Bone 81, 112–121 (2015).
Azi, M. L. et al. Autologous bone graft in the treatment of post-traumatic bone defects: a systematic review and meta-analysis. BMC Musculoskelet. Disord. 17, 465 (2016).
Bourin, P. et al. Stromal cells from the adipose tissue-derived stromal vascular fraction and culture expanded adipose tissue-derived stromal/stem cells: a joint statement of the International Federation for Adipose Therapeutics and Science (IFATS) and the International Society for Cellular Therapy (ISCT). Cytotherapy 15, 641–648 (2013).
Confalonieri, D., Schwab, A., Walles, H. & Ehlicke, F. Advanced therapy medicinal products: a guide for bone marrow-derived MSC application in bone and cartilage tissue engineering. Tissue Eng. Part B Rev. 24, 155–169 (2018).
Izeta, A. et al. Cell-based product classification procedure: what can be done differently to improve decisions on borderline products? Cytotherapy 18, 809–815 (2016).
Giancola, R., Bonfini, T. & Iacone, A. Cell therapy: cGMP facilities and manufacturing. Muscles Ligaments Tendons J. 2, 243–247 (2012).
Patel, K. & Chotai, N. Documentation and records: harmonized GMP requirements. J. Yong. Pharm. 3, 138–150 (2011).
Galvez-Martin, P. et al. Mesenchymal stem cells as therapeutics agents: quality and environmental regulatory aspects. Stem Cells Int. 2016, 1–14 (2016).
Minor, P. International reference preparations for standardization of biological medicinal products. Bundesgesundheitsblatt Gesundheitsforschung Gesundheitsschutz 57, 1145–1151 (2014).
Manghani, K. Quality assurance: Importance of systems and standard operating procedures. Perspect. Clin. Res. 2, 34–37 (2011).
Klein, T. J. et al. Strategies for zonal cartilage repair using hydrogels. Macromol. Biosci. 9, 1049–1058 (2009).
Little, C. B. & Ghosh, P. Variation in proteoglycan metabolism by articular chondrocytes in different joint regions is determined by post-natal mechanical loading. Osteoarthr. Cartil. 5, 49–62 (1997).
Klika, V., Gaffney, E. A., Chen, Y. C. & Brown, C. P. An overview of multiphase cartilage mechanical modelling and its role in understanding function and pathology. J. Mech. Behav. Biomed. Mater. 62, 139–157 (2016).
Responte, D. J., Natoli, R. M. & Athanasiou, K. A. Collagens of articular cartilage: structure, function, and importance in tissue engineering. Crit. Rev. Biomed. Eng. 35, 363–411 (2007).
Moroni, L. et al. Biofabrication: a guide to technology and terminology. Trends Biotechnol. 36, 384–402 (2018).
Murphy, S. V. & Atala, A. 3D bioprinting of tissues and organs. Nat. Biotechnol. 32, 773–785 (2014).
Cui, H., Nowicki, M., Fisher, J. P. & Zhang, L. G. 3D Bioprinting for Organ Regeneration. Adv. Healthc. Mater. 6, https://doi.org/10.1002/adhm.201601118 (2017).
Pare, A. et al. Tailored three-dimensionally printed triply periodic calcium phosphate implants: a preclinical study for craniofacial bone repair. Acs Biomater. Sci. Eng. 6, 553–563 (2020).
Charbonnier, B. et al. Custom-made macroporous bioceramic implants based on triply-periodic minimal surfaces for bone defects in load-bearing sites. Acta Biomater. 109, 254–266 (2020).
Inzana, J. A. et al. 3D printing of composite calcium phosphate and collagen scaffolds for bone regeneration. Biomaterials 35, 4026–4034 (2014).
Korn, P. et al. 3D printing of bone grafts for cleft alveolar osteoplasty - in vivo evaluation in a preclinical model. Front. Bioeng. Biotechnol. 8, 217 (2020).
Cidonio, G. et al. Nanoclay-based 3D printed scaffolds promote vascular ingrowth ex vivo and generate bone mineral tissue in vitro and in vivo. Biofabrication 12, 035010 (2020).
Yang, Y. et al. 3D bioprinted integrated osteochondral scaffold-mediated repair of articular cartilage defects in the rabbit knee. J. Med. Biol. Eng. 40, 71–81 (2019).
Shim, J. H. et al. Three-dimensional bioprinting of multilayered constructs containing human mesenchymal stromal cells for osteochondral tissue regeneration in the rabbit knee joint. Biofabrication 8, 014102 (2016).
Wen, H. et al. Hierarchical patterning via dynamic sacrificial printing of stimuli-responsive hydrogels. Biofabrication 12, 035007 (2020).
Kosik-Koziol, A. et al. 3D bioprinted hydrogel model incorporating beta-tricalcium phosphate for calcified cartilage tissue engineering. Biofabrication 11, 035016 (2019).
Levingstone, T. J. et al. Multi-layered collagen-based scaffolds for osteochondral defect repair in rabbits. Acta Biomater. 32, 149–160 (2016).
Zhang, S. et al. Bi-layer collagen/microporous electrospun nanofiber scaffold improves the osteochondral regeneration. Acta Biomater. 9, 7236–7247 (2013).
Zhao, Y. et al. Role of the calcified cartilage layer of an integrated trilayered silk fibroin scaffold used to regenerate osteochondral defects in rabbit knees. Acs Biomater. Sci. Eng. 6, 1208–1216 (2019).
Kesti, M., Fisch, P., Pensalfini, M., Mazza, E. & Zenobi-Wong, M. Guidelines for standardization of bioprinting: a systematic study of process parameters and their effect on bioprinted structures. BioNanoMaterials 17, 193 (2016).
Ning, L. et al. Process-induced cell damage: pneumatic versus screw-driven bioprinting. Biofabrication 12, 025011 (2020).
Emmermacher, J. et al. Engineering considerations on extrusion-based bioprinting: interactions of material behavior, mechanical forces and cells in the printing needle. Biofabrication 12, 025022 (2020).
Mouser, V. H. M. et al. Bio-ink development for three-dimensional bioprinting of hetero-cellular cartilage constructs. Connect. tissue Res. 61, 137–151 (2020).
Muller, M., Ozturk, E., Arlov, O., Gatenholm, P. & Zenobi-Wong, M. Alginate sulfate-nanocellulose bioinks for cartilage bioprinting applications. Ann. Biomed. Eng. 45, 210–223 (2017).
Vazquez, O. R., Avila, I. O. & Hernandez, E. An overview of mechanical tests for polymeric biomaterial scaffolds used in tissue engineering. J. Res. Updates Polym. Sci. 4, 168–178 (2015).
Trachtenberg, J. E., Vo, T. N. & Mikos, A. G. Pre-clinical characterization of tissue engineering constructs for bone and cartilage regeneration. Ann. Biomed. Eng. 43, 681–696 (2015).
Patel, J. M., Wise, B. C., Bonnevie, E. D. & Mauck, R. L. A systematic review and guide to mechanical testing for articular cartilage tissue engineering. Tissue Eng. Part C: Methods 25, 593–608 (2019).
Marchiori, G., Berni, M., Boi, M. & Filardo, G. Cartilage mechanical tests: Evolution of current standards for cartilage repair and tissue engineering. A literature review. Clin. Biomech. 68, 58–72 (2019).
Food, U. & Administration, D. Guidance for industry: preparation of IDEs and INDs for products intended to repair or replace knee cartilage. Washington, DC: US Food and Drug Administration (2011).
Honigmann, P. et al. Patient-specific surgical implants made of 3D printed peek: material, technology, and scope of surgical application. Biomed. Res. Int. 2018, 4520636 (2018).
Chamo, D. et al. Accuracy assessment of molded, patient-specific polymethylmethacrylate craniofacial implants compared to their 3D printed originals. J. Clin. Med. 9, 832 (2020).
Msallem, B. et al. Evaluation of the dimensional accuracy of 3d-printed anatomical mandibular models using FFF, SLA, SLS, MJ, and BJ printing technology. J. Clin. Med. 9, 817 (2020).
Martorelli, M. et al. Design and analysis of 3D customized models of a human mandible. Procedia CIRP 49, 199–202 (2016).
Winder, J., Cooke, R. S., Gray, J., Fannin, T. & Fegan, T. Medical rapid prototyping and 3D CT in the manufacture of custom made cranial titanium plates. J. Med. Eng. Technol. 23, 26–28 (1999).
Werz, S. M., Zeichner, S. J., Berg, B. I., Zeilhofer, H. F. & Thieringer, F. 3D printed surgical simulation Models as educational tool by maxillofacial surgeons. Eur. J. Dent. Educ. 22, e500–e505 (2018).
Murphy, S. V., De Coppi, P. & Atala, A. Opportunities and challenges of translational 3D bioprinting. Nat. Biomed. Eng. 4, 370–380 (2020).
Stoddart, M. J. (ed) Mammalian cell viability: methods and protocols. New York: Humana Press (2011).
Camarillo, C., Swerdel, M. & Hart, R. P. Comparison of microarray and quantitative real-time PCR methods for measuring MicroRNA levels in MSC cultures. Methods Mol. Biol. 698, 419–429 (2011).
Rao, M. S. et al. Comparison of RNA-Seq and microarray gene expression platforms for the toxicogenomic evaluation of liver from short-term rat toxicity studies. Front. Genet. 9, 636 (2018).
van Dijk, E. L., Auger, H., Jaszczyszyn, Y. & Thermes, C. Ten years of next-generation sequencing technology. Trends Genet. 30, 418–426 (2014).
Goodwin, S., McPherson, J. D. & McCombie, W. R. Coming of age: ten years of next-generation sequencing technologies. Nat. Rev. Genet. 17, 333–351 (2016).
Wile, B. M., Ban, K., Yoon, Y. S. & Bao, G. Molecular beacon-enabled purification of living cells by targeting cell type-specific mRNAs. Nat. Protoc. 9, 2411–2424 (2014).
Mao, S., Ying, Y., Wu, X. & Chen, A. K. Delivering molecular beacons via an electroporation-based approach enables live-cell imaging of single RNA transcripts and genomic loci. Methods Mol. Biol. 2106, 241–252 (2020).
Nitin, N., Santangelo, P. J., Kim, G., Nie, S. & Bao, G. Peptide-linked molecular beacons for efficient delivery and rapid mRNA detection in living cells. Nucleic Acids Res. 32, e58 (2004).
Alexander, J. C., Browne, S., Pandit, A. & Rochev, Y. Biomaterial constructs for delivery of multiple therapeutic genes: a spatiotemporal evaluation of efficacy using molecular beacons. PLoS One 8, e65749 (2013).
Chen, F. H., Rousche, K. T. & Tuan, R. S. Technology insight: adult stem cells in cartilage regeneration and tissue engineering. Nat. Clin. Pr. Rheumatol. 2, 373–382 (2006).
Loh, Q. L. & Choong, C. Three-dimensional scaffolds for tissue engineering applications: role of porosity and pore size. Tissue Eng. Part B Rev. 19, 485–502 (2013).
Rosenberg, L. Chemical basis for the histological use of safranin O in the study of articular cartilage. J Bone Joint Surg. Am. 53, 69–82 (1971).
Ponce, M. C., Zorzi, A. R., Miranda, J. B. d. & Amstalden, E. M. I. Proposal for a New Histological Scoring System for Cartilage Repair. Clinics (Sao Paulo) 73, e562 (2018).
Terry, D., Chopra, R., Ovenden, J. & Anastassiades, T. Differential use of Alcian blue and toluidine blue dyes for the quantification and isolation of anionic glycoconjugates from cell cultures: application to proteoglycans and a high-molecular-weight glycoprotein synthesized by articular chondrocytes. Anal. Biochem. 285, 211–219 (2000).
Koch, M. et al. Tissue engineering of large full-size meniscus defects by a polyurethane scaffold: accelerated regeneration by mesenchymal stromal cells. Stem Cells Int. 2018, 8207071 (2018).
van Gaalen, S. M. et al. Use of fluorochrome labels in in vivo bone tissue engineering research. Tissue Eng. Part B Rev. 16, 209–217 (2010).
Querido, W., Farina, M. & Balduino, A. Giemsa as a fluorescent dye for mineralizing bone-like nodules in vitro. Biomed. Mater. 7, 011001 (2012).
Sim, A. M. et al. A novel fluorescein-bisphosphonate based diagnostic tool for the detection of hydroxyapatite in both cell and tissue models. Sci. Rep. 8, 17360 (2018).
Knabe, C. et al. A method for immunohistochemical detection of osteogenic markers in undecalcified bone sections. Biotech. Histochem. 81, 31–39 (2006).
Yang, R., Davies, C. M., Archer, C. W. & Richards, R. G. Immunohistochemistry of matrix markers in Technovit 9100 New-embedded undecalcified bone sections. Eur. Cell Mater. 6, 57–71 (2003). discussion 71.
James, R., Jenkins, L., Ellis, S. E. & Burg, K. J. L. Histological processing of hydrogel scaffolds for tissue-engineering applications. J. Histotechnol. 27, 133–139 (2004).
Shi, S. R., Shi, Y. & Taylor, C. R. Antigen retrieval immunohistochemistry: review and future prospects in research and diagnosis over two decades. J. Histochem. Cytochem. 59, 13–32 (2011).
Panchal, V. et al. Confocal laser scanning microscopy for rapid optical characterization of graphene. Commun. Phys. 1, 1–7 (2018).
Nguyen, D. H. et al. Biomimetic model to reconstitute angiogenic sprouting morphogenesis in vitro. Proc. Natl. Acad. Sci. U. S. A. 110, 6712–6717 (2013).
Miller, D. S. et al. Xenobiotic transport across isolated brain microvessels studied by confocal microscopy. Mol. Pharm. 58, 1357–1367 (2000).
Evers, J. F., Schmitt, S., Sibila, M. & Duch, C. Progress in functional neuroanatomy: precise automatic geometric reconstruction of neuronal morphology from confocal image stacks. J. Neurophysiol. 93, 2331–2342 (2005).
Smith, L. E., Smallwood, R. & Macneil, S. A comparison of imaging methodologies for 3D tissue engineering. Miscosc. Res. Tech. 73, 1123–1133 (2010).
Muller, M. Introduction to confocal fluorescence microscopy. Vol. 69 (SPIE press, 2006).
Song, W. et al. Three-dimensional morphometric comparison of normal and apoptotic endothelial cells based on laser scanning confocal microscopy observation. Microsc. Res. Tech. 76, 1154–1162 (2013).
Mottet, D. et al. Regulation of hypoxia-inducible factor-1alpha protein level during hypoxic conditions by the phosphatidylinositol 3-kinase/Akt/glycogen synthase kinase 3beta pathway in HepG2 cells. J. Biol. Chem. 278, 31277–31285 (2003).
Zinchuk, V., Zinchuk, O. & Okada, T. Quantitative colocalization analysis of multicolor confocal immunofluorescence microscopy images: pushing pixels to explore biological phenomena. Acta. Histochem. Cytochem. 40, 101–111 (2007).
Basoli, V. et al. Physical stimulation by REAC and BMP4/WNT-1 inhibitor synergistically enhance cardiogenic commitment in iPSCs. PLoS One 14, e0211188 (2019).
Aoki, M., Yasutake, M. & Murohara, T. Derivation of functional endothelial progenitor cells from human umbilical cord blood mononuclear cells isolated by a novel cell filtration device. Stem Cells 22, 994–1002 (2004).
Wangler, S. et al. CD146/MCAM distinguishes stem cell subpopulations with distinct migration and regenerative potential in degenerative intervertebral discs. Osteoarthr. Cartil. 27, 1094–1105 (2019).
Chazotte, B. Labeling mitochondria with fluorescent dyes for imaging. Cold Spring Harb. Protoc. 2009, pdb prot4948 (2009).
Qing, Y. et al. Advanced black phosphorus nanomaterials for bone regeneration. Int. J. Nanomed. 15, 2045–2058 (2020).
Papadopoulos, N. G. et al. An improved fluorescence assay for the determination of lymphocyte-mediated cytotoxicity using flow cytometry. J. Immunol. Methods 177, 101–111 (1994).
Lim, Y. C., Zou, Y.-X., Lee, J. W. & Chan, S.-C. Time-interleaved analog-to-digital-converter compensation using multichannel filters. IEEE Trans. Circuits Syst. I: Regul. Pap. 56, 2234–2247 (2009).
Hillman, E. M. C., Voleti, V., Li, W. & Yu, H. Light-sheet microscopy in neuroscience. Annu. Rev. Neurosci. 42, 295–313 (2019).
Reynaud, E. G., Peychl, J., Huisken, J. & Tomancak, P. Guide to light-sheet microscopy for adventurous biologists. Nat. Methods 12, 30–34 (2015).
Glaser, A. K. et al. Light-sheet microscopy for slide-free non-destructive pathology of large clinical specimens. Nat. Biomed. Eng. 1, 0084 (2017).
Zanacchi, F. C. et al. Live-cell 3D super-resolution imaging in thick biological samples. Nat. methods 8, 1047–1049 (2011).
Santi, P. A. Light sheet fluorescence microscopy: a review. J. Histochem. Cytochem. 59, 129–138 (2011).
de Siqueira, A. F., Cabrera, F. C., Pagamisse, A. & Job, A. E. Segmentation of scanning electron microscopy images from natural rubber samples with gold nanoparticles using starlet wavelets. Microsc. Res. Tech. 77, 71–78 (2014).
Polyakov, B. et al. Manipulation of nanoparticles of different shapes inside a scanning electron microscope. Beilstein J. Nanotechnol. 5, 133–140 (2014).
Thiberge, S. et al. Scanning electron microscopy of cells and tissues under fully hydrated conditions. Proc. Natl. Acad. Sci. U. S. A. 101, 3346–3351 (2004).
Haddad, A. et al. Immunohistochemistry in association with scanning electron microscopy for the morphological characterization and location of astrocytes of the rabbit retina. J. Neurosci. Methods 106, 131–137 (2001).
Goldberg, M. W. Immunolabeling for scanning electron microscopy (SEM) and field emission SEM. Methods Cell Biol. 88, 109–130 (2008).
Marshall, D., Helfrich, M. H. & Aspden, R. M. Scanning electron microscopy of bone. Methods Mol. Med. 80, 311–320 (2003).
Boyde, A. Scanning electron microscopy of bone. Methods Mol. Biol. 816, 365–400 (2012).
Jones, S. J., Boyde, A., Ali, N. N. & Maconnachie, E. Variation in the sizes of resorption lacunae made in vitro. Scan. Electron Microsc. 1571–1580 (1986).
Kubosch, E. J. et al. Clinical trial and in-vitro study comparing the efficacy of treating bony lesions with allografts versus synthetic or highly-processed xenogeneic bone grafts. BMC Musculoskelet. Disord. 17, 77 (2016).
Clark, J. M. & Rudd, E. Cell patterns in the surface of rabbit articular cartilage revealed by the backscatter mode of scanning electron microscopy. J. Orthop. Res. 9, 275–283 (1991).
Richards, R. G. & Kaab, M. J. Microwave-enhanced fixation of rabbit articular cartilage. J. Microsc. 181, 269–276 (1996).
Suso, S., Segur, J. M., Carbonell, J. A., Manero, J. M. & Planell, J. A. Cartilage appearance using an environmental scanning electron microscope. Cell Preservation Technol. 2, 51–54 (2004).
Eberle, A. L. & Zeidler, D. Multi-beam scanning electron microscopy for high-throughput imaging in connectomics research. Front. Neuroanat. 12, 112–112 (2018).
Titze, B. & Genoud, C. Volume scanning electron microscopy for imaging biological ultrastructure. Biol. Cell 108, 307–323 (2016).
Marx, V. (Nature Publishing Group, 2013).
Narayan, K. & Subramaniam, S. Focused ion beams in biology. Nat. Methods 12, 1021–1031 (2015).
Hasegawa, T. et al. Biological application of focus ion beam-scanning electron microscopy (FIB-SEM) to the imaging of cartilaginous fibrils and osteoblastic cytoplasmic processes. J. Oral. Biosci. 59, 55–62 (2017).
Vahabi, S., Nazemi Salman, B. & Javanmard, A. Atomic force microscopy application in biological research: a review study. Iran. J. Med Sci. 38, 76–83 (2013).
Viji Babu, P. K. & Radmacher, M. Mechanics of brain tissues studied by atomic force microscopy: a perspective. Front. Neurosci. 13, 600 (2019).
Maver, U., Velnar, T., Gaberšček, M., Planinšek, O. & Finšgar, M. Recent progressive use of atomic force microscopy in biomedical applications. TrAC Trends Anal. Chem. 80, 96–111 (2016).
Edwardson, J. M. & Henderson, R. M. Atomic force microscopy and drug discovery. Drug Disco. Today 9, 64–71 (2004).
Azeloglu, E. U. & Costa, K. D. in Atomic Force Microscopy in Biomedical Research: Methods and Protocols (eds Braga, P. C. & Ricci, D.) 303-329 (Humana Press, 2011).
Haase, K. & Pelling, A. E. Investigating cell mechanics with atomic force microscopy. J. R. Soc., Interface 12, 20140970 (2015).
Maloney, J. M. et al. Mesenchymal stem cell mechanics from the attached to the suspended state. Biophys. J. 99, 2479–2487 (2010).
Szydlak, R., Majka, M., Lekka, M., Kot, M. & Laidler, P. AFM-based Analysis of Wharton’s Jelly Mesenchymal Stem Cells. Int. J. Mol. Sci. 20, 4351 (2019).
Streppa, L. et al. Prestressed cells are prone to cytoskeleton failures under localized shear strain: an experimental demonstration on muscle precursor cells. Sci. Rep. 8, 8602 (2018).
Klapetek, P. Quantitative Data Processing in Scanning Probe Microscopy: SPM Applications for Nanometrology: Second Edition. (2018).
Butler, H. J. et al. Using Raman spectroscopy to characterize biological materials. Nat. Protoc. 11, 664–687 (2016).
Pappas, D., Smith, B. W. & Winefordner, J. D. Raman spectroscopy in bioanalysis. Talanta 51, 131–144 (2000).
Rippon, W. B., Koenig, J. L. & Walton, A. G. Raman spectroscopy of proline oligomers and poly-L-proline. J. Am. Chem. Soc. 92, 7455–7459 (1970).
Deveney, M. J., Walton, A. G. & Koenig, J. L. Raman spectra of imino acids and poly-L-hydroxyproline. Biopolymers 10, 615–630 (1971).
Lord, R. C. & Yu, N. T. Laser-excited Raman spectroscopy of biomolecules. I. Native lysozyme and its constituent amino acids. J. Mol. Biol. 50, 509–524 (1970).
Puppels, G. J. et al. Studying single living cells and chromosomes by confocal Raman microspectroscopy. Nature 347, 301–303 (1990).
Notingher, I. et al. Application of FTIR and Raman spectroscopy to characterisation of bioactive materials and living cells. Spectroscopy 17, 275–288 (2003).
Verrier, S., Notingher, I., Polak, J. M. & Hench, L. L. In situ monitoring of cell death using Raman microspectroscopy. Biopolymers 74, 157–162 (2004).
Verrier, S., Zoladek, A. & Notingher, I. Raman micro-spectroscopy as a non-invasive cell viability test. Methods Mol. Biol. 740, 179–189 (2011).
Germond, A. et al. Cell type discrimination based on image features of molecular component distribution. Sci. Rep. 8, 11726 (2018).
Kunstar, A. et al. Label-free Raman monitoring of extracellular matrix formation in three-dimensional polymeric scaffolds. J. R. Soc. Interface 10, 20130464 (2013).
Rice, W. L., Kaplan, D. L. & Georgakoudi, I. Quantitative biomarkers of stem cell differentiation based on intrinsic two-photon excited fluorescence. J. Biomed. Opt. 12, 060504 (2007).
Rice, W. L., Kaplan, D. L. & Georgakoudi, I. Two-photon microscopy for non-invasive, quantitative monitoring of stem cell differentiation. PLoS One 5, e10075 (2010).
Mahadevan-Jansen, A., Mitchell, M. F., Ramanujam, N., Utzinger, U. & Richards-Kortum, R. Development of a fiber optic probe to measure NIR Raman spectra of cervical tissue in vivo. Photochem. Photobiol. 68, 427–431 (1998).
Jermyn, M. et al. Intraoperative brain cancer detection with Raman spectroscopy in humans. Sci. Transl. Med. 7, 274–219 (2015).
Wang, J., Bergholt, M. S., Zheng, W. & Huang, Z. Development of a beveled fiber-optic confocal Raman probe for enhancing in vivo epithelial tissue Raman measurements at endoscopy. Opt. Lett. 38, 2321–2323 (2013).
Wood, J. J. et al. Evaluation of a confocal Raman probe for pathological diagnosis during colonoscopy. Colorectal Dis. 16, 732–738 (2014).
Stone, N., Baker, R., Rogers, K., Parker, A. W. & Matousek, P. Subsurface probing of calcifications with spatially offset Raman spectroscopy (SORS): future possibilities for the diagnosis of breast cancer. Analyst 132, 899–905 (2007).
Keller, M. D., Majumder, S. K. & Mahadevan-Jansen, A. Spatially offset Raman spectroscopy of layered soft tissues. Opt. Lett. 34, 926–928 (2009).
Schulmerich, M. V. et al. Noninvasive Raman tomographic imaging of canine bone tissue. J. Biomed. Opt. 13, 020506 (2008).
Alfonso-Garcia, A., Mittal, R., Lee, E. S. & Potma, E. O. Biological imaging with coherent Raman scattering microscopy: a tutorial. J. Biomed. Opt. 19, 71407 (2014).
Galvis, L., Dunlop, J. W., Duda, G., Fratzl, P. & Masic, A. Polarized Raman anisotropic response of collagen in tendon: towards 3D orientation mapping of collagen in tissues. PLoS One 8, e63518 (2013).
Dooley, M. et al. Spatially-offset Raman spectroscopy for monitoring mineralization of bone tissue engineering scaffolds: feasibility study based on phantom samples. Biomed. Opt. Express 10, 1678–1690 (2019).
Liao, Z. et al. Feasibility of spatially offset Raman spectroscopy for in vitro and in vivo monitoring mineralization of bone tissue engineering scaffolds. Anal. Chem. 89, 847–853 (2017).
Talari, A. C. S., Movasaghi, Z., Rehman, S. & Rehman, I. U. Raman spectroscopy of biological tissues. Appl. Spectrosc. Rev. 50, 46–111 (2014).
Guo, S. et al. Comparability of Raman spectroscopic configurations: A large scale cross-laboratory study. Anal. Chem. 92, 15745–15756 (2020).
Power, L. et al. in Imaging, Manipulation, and Analysis of Biomolecules, Cells, and Tissues XVII (2019).
Guldberg, R. E. et al. Analyzing bone, blood vessels, and biomaterials with microcomputed tomography. IEEE Eng. Med. Biol. Mag. 22, 77–83 (2003).
Hedberg, E. L. et al. Methods: a comparative analysis of radiography, microcomputed tomography, and histology for bone tissue engineering. Tissue Eng. 11, 1356–1367 (2005).
Frese, J. et al. Nondestructive monitoring of tissue-engineered constructs. Biomed. Tech. (Berl.) 59, 165–175 (2014).
Ho, S. T. & Hutmacher, D. W. A comparison of micro CT with other techniques used in the characterization of scaffolds. Biomaterials 27, 1362–1376 (2006).
Hilldore, A., Wojtowicz, A. & Johnson, A. W. Micro-CT based quantification of non-mineralized tissue on cultured hydroxyapatite scaffolds. J. Biomed. Mater. Res. A 82, 1012–1021 (2007).
van Lenthe, G. H. et al. Nondestructive micro-computed tomography for biological imaging and quantification of scaffold-bone interaction in vivo. Biomaterials 28, 2479–2490 (2007).
Panzavolta, S. et al. Strontium-substituted hydroxyapatite-gelatin biomimetic scaffolds modulate bone cell response. Macromol. Biosci. 18, e1800096 (2018).
Amadori, S. et al. Highly porous gelatin reinforced 3D scaffolds for articular cartilage regeneration. Macromol. Biosci. 15, 941–952 (2015).
Buzarovska, A., Gualandi, C., Parrilli, A. & Scandola, M. Effect of TiO2 nanoparticle loading on Poly(l-lactic acid) porous scaffolds fabricated by TIPS. Compos. Part B: Eng. 81, 189–195 (2015).
Melchels, F. P. et al. Effects of the architecture of tissue engineering scaffolds on cell seeding and culturing. Acta biomaterialia 6, 4208–4217 (2010).
Appel, A. A., Anastasio, M. A., Larson, J. C. & Brey, E. M. Imaging challenges in biomaterials and tissue engineering. Biomaterials 34, 6615–6630 (2013).
Jones, A. C. et al. Assessment of bone ingrowth into porous biomaterials using MICRO-CT. Biomaterials 28, 2491–2504 (2007).
Jones, A. C. et al. Analysis of 3D bone ingrowth into polymer scaffolds via micro-computed tomography imaging. Biomaterials 25, 4947–4954 (2004).
Thimm, B. W., Wechsler, O., Bohner, M., Muller, R. & Hofmann, S. In vitro ceramic scaffold mineralization: comparison between histological and micro-computed tomographical analysis. Ann. Biomed. Eng. 41, 2666–2675 (2013).
Westhauser, F. et al. Micro-Computed-Tomography-Guided Analysis of In Vitro Structural Modifications in Two Types of 45S5 Bioactive Glass Based Scaffolds. Materials (Basel) 10, 1341 (2017).
Porter, B. D., Lin, A. S., Peister, A., Hutmacher, D. & Guldberg, R. E. Noninvasive image analysis of 3D construct mineralization in a perfusion bioreactor. Biomaterials 28, 2525–2533 (2007).
Russo, A. et al. Magnetic forces and magnetized biomaterials provide dynamic flux information during bone regeneration. J. Mater. Sci. Mater. Med. 27, 51 (2016).
Ciocca, L. et al. Customized hybrid biomimetic hydroxyapatite scaffold for bone tissue regeneration. J. Biomed. Mater. Res B Appl. Biomater. 105, 723–734 (2017).
Bothe, F. et al. Treatment of Focal Cartilage Defects in Minipigs with Zonal Chondrocyte/Mesenchymal Progenitor Cell Constructs. Int. J. Mol. Sci. 20, 1–17 (2019).
Della Bella, E. et al. Osteoinductivity of nanostructured hydroxyapatite-functionalized gelatin modulated by human and endogenous mesenchymal stromal cells. J. Biomed. Mater. Res A 106, 914–923 (2018).
Roshan-Ghias, A., Vogel, A., Rakotomanana, L. & Pioletti, D. P. Prediction of spatio-temporal bone formation in scaffold by diffusion equation. Biomaterials 32, 7006–7012 (2011).
Rousselle, S. D., Ramot, Y., Nyska, A. & Jackson, N. D. Pathology of bioabsorbable implants in preclinical studies. Toxicol. Pathol. 47, 358–378 (2019).
Fricia, M. et al. Osteointegration in custom-made porous hydroxyapatite cranial implants: from reconstructive surgery to regenerative medicine. World Neurosurg. 84, 591 e511–596 (2015).
Zeller-Plumhoff, B. et al. Quantitative characterization of degradation processes in situ by means of a bioreactor coupled flow chamber under physiological conditions using time-lapse SRµCT. Mater. Corros. 69, 298–306 (2018).
Askari, E. et al. Micro-CT based finite element modelling and experimental characterization of the compressive mechanical properties of 3-D zirconia scaffolds for bone tissue engineering. J. Mech. Behav. Biomed. Mater. 102, 103516 (2020).
Yue, S. et al. Synchrotron X-ray microtomography for assessment of bone tissue scaffolds. J. Mater. Sci. Mater. Med. 21, 847–853 (2010).
Dhillon, A. et al. Analysis of sintered polymer scaffolds using concomitant synchrotron computed tomography and in situ mechanical testing. J. Mater. Sci. Mater. Med. 22, 2599–2605 (2011).
Pena Fernandez, M., Barber, A. H., Blunn, G. W. & Tozzi, G. Optimization of digital volume correlation computation in SR-microCT images of trabecular bone and bone-biomaterial systems. J. Microsc. 272, 213–228 (2018).
Dorsey, S. M., Lin-Gibson, S. & Simon, C. G. Jr. X-ray microcomputed tomography for the measurement of cell adhesionand proliferation in polymer scaffolds. Biomaterials 30, 2967–2974 (2009).
Parrilli, A. et al. Three-dimensional cellular distribution in polymeric scaffolds for bone regeneration: a microCT analysis compared to SEM, CLSM and DNA content. J. Microsc. 255, 20–29 (2014).
Faraj, K. A. et al. Micro-computed tomographical imaging of soft biological materials using contrast techniques. Tissue Eng. Part C. Methods 15, 493–499 (2009).
Papantoniou, I. et al. Three-dimensional characterization of tissue-engineered constructs by contrast-enhanced nanofocus computed tomography. Tissue Eng. Part C. Methods 20, 177–187 (2014).
Wang, Y. et al. The synergistic effects of 3-D porous silk fibroin matrix scaffold properties and hydrodynamic environment in cartilage tissue regeneration. Biomaterials 31, 4672–4681 (2010).
Appel, A., Anastasio, M. A. & Brey, E. M. Potential for imaging engineered tissues with X-ray phase contrast. Tissue Eng. Part B Rev. 17, 321–330 (2011).
Nam, S. Y., Ricles, L. M., Suggs, L. J. & Emelianov, S. Y. Imaging strategies for tissue engineering applications. Tissue Eng. Part B Rev. 21, 88–102 (2015).
Appel, A. A. et al. Imaging of poly(alpha-hydroxy-ester) scaffolds with X-ray phase-contrast microcomputed tomography. Tissue Eng. Part C. Methods 18, 859–865 (2012).
Brey, E. M. et al. X-ray imaging of poly(ethylene glycol) hydrogels without contrast agents. Tissue Eng. Part C. Methods 16, 1597–1600 (2010).
Bawolin, N. K., Dolovich, A. T., Chen, D. X. B. & Zhang, C. W. J. Characterization of mechanical properties of tissue scaffolds by phase contrast imaging and finite element modeling. J Biomech. Eng. 137, 081004 (2015).
Giuliani, A. et al. Polyglycolic acid-polylactic acid scaffold response to different progenitor cell in vitro cultures: a demonstrative and comparative X-ray synchrotron radiation phase-contrast microtomography study. Tissue Eng. Part C. Methods 20, 308–316 (2014).
Gil, C. J. et al. In vivo tracking of tissue engineered constructs. Micromachines (Basel) 10, 1–23 (2019).
Guldberg, R. E. et al. 3D imaging of tissue integration with porous biomaterials. Biomaterials 29, 3757–3761 (2008).
Cengiz, I. F., Oliveira, J. M. & Reis, R. L. Micro-computed tomography characterization of tissue engineering scaffolds: effects of pixel size and rotation step. J. Mater. Sci. Mater. Med. 28, 129 (2017).
McDougald, W. A., Collins, R., Green, M. & Tavares, A. A. S. High dose MicroCT does not contribute toward improved MicroPET/CT image quantitative accuracy and can limit longitudinal scanning of small animals. Front. Phys. 5, 1–11 (2017).
Fragogeorgi, E. A. et al. In vivo imaging techniques for bone tissue engineering. J. Tissue Eng. 10, 2041731419854586 (2019).
Geng, H. et al. A correlative imaging based methodology for accurate quantitative assessment of bone formation in additive manufactured implants. J. Mater. Sci. Mater. Med. 27, 112 (2016).
Palmquist, A., Shah, F. A., Emanuelsson, L., Omar, O. & Suska, F. A technique for evaluating bone ingrowth into 3D printed, porous Ti6Al4V implants accurately using X-ray micro-computed tomography and histomorphometry. Micron 94, 1–8 (2017).
Maronpot, R. R. et al. Regulatory forum opinion piece*: imaging applications in toxicologic pathology-recommendations for use in regulated nonclinical toxicity studies. Toxicol. Pathol. 45, 444–471 (2017).
Beaudette, K. et al. Optical coherence tomography for the identification of musculoskeletal structures of the spine: a pilot study. Biomed. Opt. Expr. 3, 533–542 (2012).
Shubhakaran, K. The application of optical coherence tomography in neurologic diseases. Neurol. Clin. Pr. 6, 9–10 (2016).
Yang, Y. et al. Investigation of optical coherence tomography as an imaging modality in tissue engineering. Phys. Med. Biol. 51, 1649–1659 (2006).
Wang, L., Xu, M., Zhang, L., Zhou, Q. & Luo, L. Automated quantitative assessment of three-dimensional bioprinted hydrogel scaffolds using optical coherence tomography. Biomed. Opt. Expr. 7, 894–910 (2016).
Wang, L., Xu, M. E., Luo, L., Zhou, Y. & Si, P. Iterative feedback bio-printing-derived cell-laden hydrogel scaffolds with optimal geometrical fidelity and cellular controllability. Sci. Rep. 8, 2802 (2018).
Larin, K. V. & Sampson, D. D. Optical coherence elastography—OCT at work in tissue biomechanics [Invited]. Biomed. Opt. Expr. 8, 1172–1202 (2017).
Ghosn, M. G., Tuchin, V. V. & Larin, K. V. Depth-resolved monitoring of glucose diffusion in tissues by using optical coherence tomography. Opt. Lett. 31, 2314–2316 (2006).
Justino, C. I. L., Duarte, A. C. & Rocha-Santos, T. A. P. Recent progress in biosensors for environmental monitoring: a review. Sensors (Basel) 17, 2918 (2017).
Rocchitta, G. et al. Enzyme biosensors for biomedical applications: strategies for safeguarding analytical performances in biological fluids. Sensors (Basel) 16, 780 (2016).
Farina, D. et al. Development of a biosensor telemetry system for monitoring fermentation in craft breweries. Food Chem. 218, 479–486 (2017).
Barberis, A. et al. Real-time monitoring of glucose and phenols intestinal absorption through an integrated Caco-2TC7cells/biosensors telemetric device: hypoglycemic effect of fruit phytochemicals. Biosens. Bioelectron. 88, 159–166 (2017).
Rocchitta, G. et al. Simultaneous telemetric monitoring of brain glucose and lactate and motion in freely moving rats. Anal. Chem. 85, 10282–10288 (2013).
Hasan, A. et al. Recent advances in application of biosensors in tissue engineering. Biomed. Res. Int. 2014, 307519 (2014).
Ilinoiu, E. C., Manea, F., Serra, P. A. & Pode, R. Simultaneous/selective detection of dopamine and ascorbic acid at synthetic zeolite-modified/graphite-epoxy composite macro/quasi-microelectrodes. Sensors 13, 7296–7307 (2013).
Migheli, R. et al. Novel integrated microdialysis-amperometric system for in vitro detection of dopamine secreted from PC12 cells: design, construction, and validation. Anal. Biochem. 380, 323–330 (2008).
Shi, B.-X., Wang, Y., Zhang, K., Lam, T.-L. & Chan, H. L.-W. Monitoring of dopamine release in single cell using ultrasensitive ITO microsensors modified with carbon nanotubes. Biosens. Bioelectron. 26, 2917–2921 (2011).
Calia, G. et al. Biotelemetric monitoring of brain neurochemistry in conscious rats using microsensors and biosensors. Sensors 9, 2511–2523 (2009).
Kumar, A. et al. Amperometric microsensor based on nanoporous gold for ascorbic acid detection in highly acidic biological extracts. Anal. Chim. Acta 1095, 61–70 (2020).
Sanna, D. et al. Synthesis of nitric oxide donors derived from Piloty’s acid and study of their effects on dopamine secretion from PC12 cells. Pharmaceuticals 10, 74 (2017).
Bazzu, G. et al. Real-time monitoring of brain tissue oxygen using a miniaturized biotelemetric device implanted in freely moving rats. Anal. Chem. 81, 2235–2241 (2009).
Serra, P. A. et al. Real-time telemetry monitoring of oxygen in the central complex of freely-walking Gromphadorhina portentosa. PLoS One 14, e0224932 (2019).
Rocchitta, G. et al. Development and characterization of an implantable biosensor for telemetric monitoring of ethanol in the brain of freely moving rats. Anal. Chem. 84, 7072–7079 (2012).
Rocchitta, G. et al. Propylene glycol stabilizes the linear response of glutamate biosensor: potential implications for in-vivo neurochemical monitoring. Chemosensors 6, 58 (2018).
Clark Jr, L. C., Wolf, R., Granger, D. & Taylor, Z. Continuous recording of blood oxygen tensions by polarography. J. Appl. Physiol. 6, 189–193 (1953).
Marvin, J. S. et al. An optimized fluorescent probe for visualizing glutamate neurotransmission. Nat. Methods 10, 162–170 (2013).
Ganesana, M., Trikantzopoulos, E., Maniar, Y., Lee, S. T. & Venton, B. J. Development of a novel micro biosensor for in vivo monitoring of glutamate release in the brain. Biosens. Bioelectron. 130, 103–109 (2019).
Kucherenko, D. Y. et al. A highly selective amperometric biosensor array for the simultaneous determination of glutamate, glucose, choline, acetylcholine, lactate and pyruvate. Bioelectrochemistry 128, 100–108 (2019).
Díaz‐García, C. M. & Yellen, G. Neurons rely on glucose rather than astrocytic lactate during stimulation. J. Neurosci. Res. 97, 883–889 (2019).
Banica, F.-G. Chemical sensors and biosensors: fundamentals and applications. (John Wiley & Sons, 2012).
McMahon, C. P. et al. Control of the oxygen dependence of an implantable polymer/enzyme composite biosensor for glutamate. Anal. Chem. 78, 2352–2359 (2006).
Kieninger, J. et al. Sensor access to the cellular microenvironment using the sensing cell culture flask. Biosensors 8, 44 (2018).
Kieninger, J., Weltin, A., Flamm, H. & Urban, G. A. Microsensor systems for cell metabolism–from 2D culture to organ-on-chip. Lab Chip 18, 1274–1291 (2018).
Laschi, S. & Mascini, M. Planar electrochemical sensors for biomedical applications. Med. Eng. Phys. 28, 934–943 (2006).
Lauks, I. R. Microfabricated biosensors and microanalytical systems for blood analysis. Acc. Chem. Res. 31, 317–324 (1998).
Abouzeid, J., Darwish, G. & Karam, P. Biosensors for optimal tissue engineering: recent developments and shaping the future. Tissue Eng. Artif. Organs: Regenerative Med., Smart Diagnostics Personalized Med. 1, 143–167 (2017).
Weltin, A. et al. Accessing 3D microtissue metabolism: lactate and oxygen monitoring in hepatocyte spheroids. Biosens. Bioelectron. 87, 941–948 (2017).
Kieninger, J., Aravindalochanan, K., Sandvik, J. A., Pettersen, E. O. & Urban, G. A. Pericellular oxygen monitoring with integrated sensor chips for reproducible cell culture experiments. Cell Prolif. 47, 180–188 (2014).
Modena, M. M., Chawla, K., Misun, P. M. & Hierlemann, A. Smart cell culture systems: Integration of sensors and actuators into microphysiological systems. ACS Chem. Biol. 13, 1767–1784 (2018).
Weltin, A. et al. Cell culture monitoring for drug screening and cancer research: a transparent, microfluidic, multi-sensor microsystem. Lab Chip 14, 138–146 (2014).
Telsnig, D. et al. Characterization of an amperometric biosensor for the determination of biogenic amines in flow injection analysis. Int. J. Electrochem. Sci. 7, 10476–10486 (2012).
von Woedtke, T., Julich, W. D., Hartmann, V., Stieber, M. & Abel, P. U. Sterilization of enzyme glucose sensors: problems and concepts. Biosens. Bioelectron. 17, 373–382 (2002).
Druhmann, D. et al. Utilizing Roche Cedex Bio analyzer for in process monitoring in biotech production. BMC Proc. 5, P106 (2011). Suppl 8.
Ausubel, L. J., Lopez, P. M. & Couture, L. A. GMP scale-up and banking of pluripotent stem cells for cellular therapy applications. Methods Mol. Biol. 767, 147–159 (2011).
Tang, W., Hu, Z., Muallem, H. & Gulley, M. L. Quality assurance of RNA expression profiling in clinical laboratories. J. Mol. Diagn. 14, 1–11 (2012).
Acknowledgements
We would like to acknowledge the SNSF Spark project (CRSK-3_190410, Real-time quality monitoring of engineered tissue for regenerative medicine—RT-ET), AO Foundation (Osteochondral defect collaborative research program), AOSpine International, Joint Approach Eurostar project (co-funded by Eureka member countries and the European Union Horizon 2020 framework program), and Graubünden Innovationsstiftung for their financial support. Unless otherwise stated, figures were prepared via BioRender.com. The authors are grateful to Prof. Andrea J. Vernengo for help in English proof-reading of the manuscript.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflicts of interest.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Zuncheddu, D., Della Bella, E., Schwab, A. et al. Quality control methods in musculoskeletal tissue engineering: from imaging to biosensors. Bone Res 9, 46 (2021). https://doi.org/10.1038/s41413-021-00167-9
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41413-021-00167-9