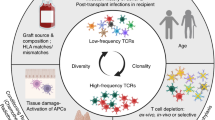
Abstract
Detectable in biopsies and body fluids, the measurement of a single or panels of microRNAs have been reported to be quite sensitive and specific for the prediction, diagnosis, and prognosis of many diseases. Interest in the use of microRNAs as biomarkers and eventual therapeutic targets has increased exponentially in the last decade. However, in the field of graft-versus-host disease (GVHD), the discovery of their involvement in biological processes and their predictive value is only emerging. With 30–75% of patients developing GVHD following allogeneic hematopoietic cell transplant and the absence of routinely used predictive biomarkers, microRNAs are promising for early detection and follow-up of this condition. We aim to summarize the current knowledge on the involvement of these small RNAs in the pathophysiology of this disease. We also review studies investigating the potential of miRNAs as biomarkers for early detection, follow-up, and prognosis of GVHD.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout
Similar content being viewed by others
References
Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–54.
Tetreault N, De Guire V. miRNAs: their discovery, biogenesis and mechanism of action. Clin Biochem. 2013;46:842–5.
De Guire V, Robitaille R, Tetreault N, Guerin R, Menard C, Bambace N, et al. Circulating miRNAs as sensitive and specific biomarkers for the diagnosis and monitoring of human diseases: promises and challenges. Clin Biochem. 2013;46:846–60.
Okamura K, Phillips MD, Tyler DM, Duan H, Chou YT, Lai EC. The regulatory activity of microRNA* species has substantial influence on microRNA and 3′ UTR evolution. Nat Struct Mol Biol. 2008;15:354–63.
Eulalio A, Huntzinger E, Izaurralde E. Getting to the root of miRNA-mediated gene silencing. Cell. 2008;132:9–14.
Flynt AS, Lai EC. Biological principles of microRNA-mediated regulation: shared themes amid diversity. Nat Rev Genet. 2008;9:831–42.
Huang Y, Shen XJ, Zou Q, Wang SP, Tang SM, Zhang GZ. Biological functions of microRNAs: a review. J Physiol Biochem. 2011;67:129–39.
Lawrie CH, Gal S, Dunlop HM, Pushkaran B, Liggins AP, Pulford K, et al. Detection of elevated levels of tumour-associated microRNAs in serum of patients with diffuse large B-cell lymphoma. Br J Haematol. 2008;141:672–5.
Weber JA, Baxter DH, Zhang S, Huang DY, Huang KH, Lee MJ, et al. The microRNA spectrum in 12 body fluids. Clin Chem. 2010;56:1733–41.
Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci USA. 2008;105:10513–8.
Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–9.
Arroyo JD, Chevillet JR, Kroh EM, Ruf IK, Pritchard CC, Gibson DF, et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci USA. 2011;108:5003–8.
Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, et al. Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci USA. 2002;99:15524–9.
Chen X, Hu Z, Wang W, Ba Y, Ma L, Zhang C, et al. Identification of ten serum microRNAs from a genome-wide serum microRNA expression profile as novel noninvasive biomarkers for nonsmall cell lung cancer diagnosis. Int J Cancer. 2012;130:1620–8.
Li LM, Hu ZB, Zhou ZX, Chen X, Liu FY, Zhang JF, et al. Serum microRNA profiles serve as novel biomarkers for HBV infection and diagnosis of HBV-positive hepatocarcinoma. Cancer Res. 2010;70:9798–807.
Heneghan HM, Miller N, Kelly R, Newell J, Kerin MJ. Systemic miRNA-195 differentiates breast cancer from other malignancies and is a potential biomarker for detecting noninvasive and early stage disease. Oncologist. 2010;15:673–82.
Mastroianni J, Stickel N, Andrlova H, Hanke K, Melchinger W, Duquesne S, et al. miR-146a controls immune response in the melanoma microenvironment. Cancer Res. 2019;79:183–95.
Danylesko I, Shimoni A, Nagler A. Allogeneic stem cell transplantation and targeted immunotherapy for multiple myeloma. Clin Lymphoma Myeloma Leuk. 2013;13:S330–48.
Juric MK, Ghimire S, Ogonek J, Weissinger EM, Holler E, van Rood JJ, et al. Milestones of hematopoietic stem cell transplantation—from first human studies to current developments. Front Immunol. 2016;7:470.
Shlomchik WD. Graft-versus-host disease. Nat Rev Immunol. 2007;7:340–52.
Jagasia M, Arora M, Flowers ME, Chao NJ, McCarthy PL, Cutler CS, et al. Risk factors for acute GVHD and survival after hematopoietic cell transplantation. Blood. 2012;119:296–307.
Reddy P, Zeiser R. Editorial: non-coding RNAs and graft versus host disease. Front Immunol. 2018;9:2713.
Peltier D, Reddy P. Non-coding RNA mediated regulation of allogeneic T cell responses after hematopoietic transplantation. Front Immunol. 2018;9:1110.
Xiao B, Wang Y, Li W, Baker M, Guo J, Corbet K, et al. Plasma microRNA signature as a noninvasive biomarker for acute graft-versus-host disease. Blood. 2013;122:3365–75.
Reddy P, Ferrara JL. Immunobiology of acute graft-versus-host disease. Blood Rev. 2003;17:187–94.
Koenecke C, Krueger A. MicroRNA in T-cell development and t-cell mediated acute graft-versus-host disease. Front Immunol. 2018;9:992.
Atarod S, Dickinson AM. MicroRNAs: the missing link in the biology of graft-versus-host disease? Front Immunol. 2013;4:420.
Ranganathan P, Heaphy CE, Costinean S, Stauffer N, Na C, Hamadani M, et al. Regulation of acute graft-versus-host disease by microRNA-155. Blood. 2012;119:4786–97.
Rodriguez A, Vigorito E, Clare S, Warren MV, Couttet P, Soond DR, et al. Requirement of bic/microRNA-155 for normal immune function. Science. 2007;316:608–11.
Thai TH, Calado DP, Casola S, Ansel KM, Xiao C, Xue Y, et al. Regulation of the germinal center response by microRNA-155. Science. 2007;316:604–8.
Zhang R, Wang X, Hong M, Luo T, Zhao M, Shen H, et al. Endothelial microparticles delivering microRNA-155 into T lymphocytes are involved in the initiation of acute graft-versus-host disease following allogeneic hematopoietic stem cell transplantation. Oncotarget. 2017;8:23360–75.
Gam R, Shah P, Crossland RE, Norden J, Dickinson AM, Dressel R. Genetic association of hematopoietic stem cell transplantation outcome beyond histocompatibility genes. Front Immunol. 2017;8:380.
Kohlhaas S, Garden OA, Scudamore C, Turner M, Okkenhaug K, Vigorito E. Cutting edge: the Foxp3 target miR-155 contributes to the development of regulatory T cells. J Immunol. 2009;182:2578–82.
Baumjohann D, Ansel KM. MicroRNA-mediated regulation of T helper cell differentiation and plasticity. Nat Rev Immunol. 2013;13:666–78.
Lu LF, Thai TH, Calado DP, Chaudhry A, Kubo M, Tanaka K, et al. Foxp3-dependent microRNA155 confers competitive fitness to regulatory T cells by targeting SOCS1 protein. Immunity. 2009;30:80–91.
Jeker LT, Zhou X, Gershberg K, de Kouchkovsky D, Morar MM, Stadthagen G, et al. MicroRNA 10a marks regulatory T cells. PLoS One. 2012;7:e36684.
Hippen KL, Loschi M, Nicholls J, MacDonald KPA, Blazar BR. Effects of microRNA on regulatory T cells and implications for adoptive cellular therapy to ameliorate graft-versus-host disease. Front Immunol. 2018;9:57.
Chen S, Smith BA, Iype J, Prestipino A, Pfeifer D, Grundmann S, et al. MicroRNA-155-deficient dendritic cells cause less severe GVHD through reduced migration and defective inflammasome activation. Blood. 2015;126:103–12.
Zitzer NC, Taylor PA, Ngankeu A, Efebera YA, Devine SM, Blazar BR, et al. MiR-155 impacts T cell migration in acute graft-versus-host disease (aGVHD). Blood. 2015;126:3080.
Atarod S, Ahmed MM, Lendrem C, Pearce KF, Cope W, Norden J, et al. miR-146a and miR-155 expression levels in acute graft-versus-host disease incidence. Front Immunol. 2016;7:56.
Stickel N, Prinz G, Pfeifer D, Hasselblatt P, Schmitt-Graeff A, Follo M, et al. MiR-146a regulates the TRAF6/TNF-axis in donor T cells during GVHD. Blood. 2014;124:2586–95.
Stickel N, Hanke K, Köhler M, Prinz G, Pfeifer D, Schmitt-Graeff A, et al. Recipient dendritic cells are regulated by MiR-146a during acute GvHD. Blood. 2015;126:146.
Stickel N, Hanke K, Marschner D, Prinz G, Kohler M, Melchinger W, et al. MicroRNA-146a reduces MHC-II expression via targeting JAK/STAT signaling in dendritic cells after stem cell transplantation. Leukemia. 2017;31:2732–41.
Zeiser R, Burchert A, Lengerke C, Verbeek M, Maas-Bauer K, Metzelder SK, et al. Ruxolitinib in corticosteroid-refractory graft-versus-host disease after allogeneic stem cell transplantation: a multicenter survey. Leukemia. 2015;29:2062–8.
Boldin MP, Taganov KD, Rao DS, Yang L, Zhao JL, Kalwani M, et al. miR-146a is a significant brake on autoimmunity, myeloproliferation, and cancer in mice. J Exp Med. 2011;208:1189–201.
Chen S, Zeiser R. The role of microRNAs in myeloid cells during graft-versus-host disease. Front Immunol. 2018;9:4.
Park H, Huang X, Lu C, Cairo MS, Zhou X. MicroRNA-146a and microRNA-146b regulate human dendritic cell apoptosis and cytokine production by targeting TRAF6 and IRAK1 proteins. J Biol Chem. 2015;290:2831–41.
He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–33.
Ventura A, Young AG, Winslow MM, Lintault L, Meissner A, Erkeland SJ, et al. Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell. 2008;132:875–86.
Olive V, Li Q, He L. mir-17-92: a polycistronic oncomir with pleiotropic functions. Immunol Rev. 2013;253:158–66.
Jiang S, Li C, Olive V, Lykken E, Feng F, Sevilla J, et al. Molecular dissection of the miR-17-92 cluster’s critical dual roles in promoting Th1 responses and preventing inducible Treg differentiation. Blood. 2011;118:5487–97.
Wu Y, Heinrichs J, Bastian D, Fu J, Nguyen H, Schutt S, et al. MicroRNA-17-92 controls T-cell responses in graft-versus-host disease and leukemia relapse in mice. Blood. 2015;126:1314–23.
Wu Y, Schutt S, Paz K, Zhang M, Flynn RP, Bastian D, et al. MicroRNA-17-92 is required for T-cell and B-cell pathogenicity in chronic graft-versus-host disease in mice. Blood. 2018;131:1974–86.
Hennion-Tscheltzoff O, Leboeuf D, Gauthier SD, Dupuis M, Assouline B, Gregoire A, et al. TCR triggering modulates the responsiveness and homeostatic proliferation of CD4+ thymic emigrants to IL-7 therapy. Blood. 2013;121:4684–93.
Kroesen BJ, Teteloshvili N, Smigielska-Czepiel K, Brouwer E, Boots AM, van den Berg A, et al. Immuno-miRs: critical regulators of T-cell development, function and ageing. Immunology. 2015;144:1–10.
Li QJ, Chau J, Ebert PJ, Sylvester G, Min H, Liu G, et al. miR-181a is an intrinsic modulator of T cell sensitivity and selection. Cell. 2007;129:147–61.
Sang W, Zhang C, Zhang D, Wang Y, Sun C, Niu M, et al. MicroRNA-181a, a potential diagnosis marker, alleviates acute graft versus host disease by regulating IFN-gamma production. Am J Hematol. 2015;90:998–1007.
Strimbu K, Tavel JA. What are biomarkers? Curr Opin HIV AIDS. 2010;5:463–6.
Paczesny S. Acute graft-versus-host disease prognosis: are biomarkers ready for preemptive clinical trials? Clin Chem 2017;63:1561–3.
Hartwell MJ, Ozbek U, Holler E, Renteria AS, Major-Monfried H, Reddy P, et al. An early-biomarker algorithm predicts lethal graft-versus-host disease and survival. JCI Insight. 2017;2:e89798.
Presland RB. Biology of chronic graft-vs-host disease: immune mechanisms and progress in biomarker discovery. World J Transplant. 2016;6:608–19.
Xie LN, Zhou F, Liu XM, Fang Y, Yu Z, Song NX, et al. Serum microRNA155 is increased in patients with acute graft-versus-host disease. Clin Transplant. 2014;28:314–23.
Jalapothu D, Boieri M, Crossland RE, Shah P, Butt IA, Norden J, et al. Tissue-specific expression patterns of microRNA during acute graft-versus-host disease in the rat. Front Immunol. 2016;7:361.
Crossland RE, Norden J, Kralj Juric M, Pearce KF, Lendrem C, Bibby LA, et al. Serum and extracellular vesicle microRNAs miR-423, miR-199, and miR-93* as biomarkers for acute graft-versus-host disease. Front Immunol. 2017;8:1446.
Crossland RNJ, Collin M, Dickinson AM. Urinary micrornas miR-377, miR-423, miR-93 and miR-199 as biomarkers for graft versus host disease. Blood. 2014;124:1088.
Crossland RE, Norden J, Juric MK, Green K, Pearce KF, Lendrem C, et al. Expression of serum microRNAs is altered during acute graft-versus-host disease. Front Immunol. 2017;8:308.
Gimondi S, Dugo M, Vendramin A, Bermema A, Biancon G, Cavane A, et al. Circulating miRNA panel for prediction of acute graft-versus-host disease in lymphoma patients undergoing matched unrelated hematopoietic stem cell transplantation. Exp Hematol. 2016;44:624–34 e1.
Zhao XS, Wang YN, Lv M, Kong Y, Luo HX, Ye XY, et al. miR-153-3p, a new bio-target, is involved in the pathogenesis of acute graft-versus-host disease via inhibition of indoleamine- 2,3-dioxygenase. Oncotarget. 2016;7:48321–34.
Cheng HH, Yi HS, Kim Y, Kroh EM, Chien JW, Eaton KD, et al. Plasma processing conditions substantially influence circulating microRNA biomarker levels. PLoS ONE. 2013;8:e64795.
Ranganathan P, Ngankeu A, Zitzer NC, Leoncini P, Yu X, Casadei L, et al. Serum miR-29a Is upregulated in acute graft-versus-host disease and activates dendritic cells through TLR binding. J Immunol. 2017;198:2500–12.
Wang Y, Zhao X, Ye X, Luo H, Zhao T, Diao Y, et al. Plasma microRNA-586 is a new biomarker for acute graft-versus-host disease. Ann Hematol. 2015;94:1505–14.
Funding
This work was supported in part by grants from the Cancer Research Society of Canada (grants no. 22669 to MG), and the Fonds de recherche du Québec-Santé (FRQS).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Newmarch, M., Kostantin, E., Tsongalis, G.J. et al. MicroRNAs in graft-versus-host disease: a review of the latest data. Bone Marrow Transplant 55, 1014–1020 (2020). https://doi.org/10.1038/s41409-019-0764-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41409-019-0764-1