Abstract
Acute myeloid leukemia (AML) with FLT3-ITD mutations (FLT3-ITDmut) remains a therapeutic challenge, with a still high relapse rate, despite targeted treatment with tyrosine kinase inhibitors. In this disease, the CD34/CD123/CD25/CD99+ leukemic precursor cells (LPCs) phenotype predicts for FLT3-ITD-positivity. The aim of this study was to characterize the distribution of FLT3-ITD mutation in different progenitor cell subsets to shed light on the subclonal architecture of FLT3-ITDmut AML. Using high-speed cell sorting, we sequentially purified LPCs and CD34+ progenitors in samples from patients with FLT3-ITDmut AML (n = 12). A higher FLT3-ITDmut load was observed within CD34/CD123/CD25/CD99+ LPCs, as compared to CD34+ progenitors (CD123+/−,CD25−,CD99low/−) (p = 0.0005) and mononuclear cells (MNCs) (p < 0.0001). This was associated with significantly increased CD99 mean fluorescence intensity in LPCs. Significantly higher FLT3-ITDmut burden was also observed in LPCs of AML patients with a small FLT3-ITDmut clones at diagnosis. On the contrary, the mutation burden of other myeloid genes was similar in MNCs, highly purified LPCs and/or CD34+ progenitors. Treatment with an anti-CD99 mAb was cytotoxic on LPCs in two patients, whereas there was no effect on CD34+ cells from healthy donors. Our study shows that FLT3-ITD mutations occur early in LPCs, which represent the leukemic reservoir. CD99 may represent a new therapeutic target in FLT3-ITDmut AML.
Similar content being viewed by others
Introduction
Acute myeloid leukemia (AML) is characterized by heterogeneous genetic abnormalities, which significantly influence clinical outcome. Therefore, the definition of molecular profiles is essential at the time of initial diagnosis, to identify specific biomarkers and to design specific treatments1. Although available treatments induce complete remission (CR) in about 80% of AML patients2, 30–40% of them will eventually relapse, due to the emergence of resistant clones.
Multiple genetic abnormalities have shown to account for the clinical heterogeneity of AML and, according to the WHO 2016 classification and the 2017 revised European Leukemia Net (ELN) guidelines3, in addition to karyotype assessment, mutational analyses are required for proper AML classification. Internal tandem duplications of the fms-like tyrosine kinase 3 (FLT3) gene (FLT3-ITDmut) characterize ~25% of AML and represent an independent predictor of poor prognosis, being associated with an increased risk of relapse3,4. The FLT3-ITDmut allelic ratio (AR), reflecting the FLT3-ITDmut leukemic burden, also plays a significant role, especially in the context of NPM1 mutations, where AR < 0.5 or ≥0.5, define favorable or intermediate prognostic categories3.
FLT3-ITD is an unstable marker during AML evolution, with FLT3-ITDmut patients at diagnosis becoming negative or acquiring the mutation at relapse in 25% and 10% of cases, respectively5,6. These data indicate that FLT3-ITDmut testing must be confirmed at the time of disease recurrence to eventually add FLT3-targeted drugs3,7.
We identified minor FLT3-ITDmut subclones at the time of initial AML diagnosis, expressing the CD34/CD123/CD25/CD99+ immunophenotype, in 83% of cases with a FLT3-ITDmut relapse8,9. These leukemic precursor cells (LPC) FLT3-ITDmut subclones may expand at disease recurrence, and their identification and characterization represent an important challenge. Indeed, FLT3-ITD mutations have been shown to occur in primitive AML cells10, representing early drivers of leukemogenesis mainly in patients with a normal karyotype, while additional cooperating mutations may occur later, contributing to disease progression11,12.
In this line, to shed light on the subclonal architecture of FLT3-ITDmut AML and to track FLT3-ITDmut and other clones, we characterized the molecular profile of CD34/CD123/CD25/CD99+ enriched LPCs.
Patients and methods
Patient characteristics
We studied 38 patients with a FLT3-ITDmut AML, aged 21–82 years, diagnosed at the Hematology Unit of Policlinico Tor Vergata in Rome, during the years 2016–2019. AML patients aged <75 years received intensive chemotherapy according to the European Organization of Research and Treatment of Cancer (EORTC)/Gruppo Italiano Malattie EMatologiche dell’Adulto (GIMEMA) protocols, while patients aged >75 years received supportive care or hypomethylating treatment. Treatment included the addition of tyrosine kinase inhibitors (TKIs) in four patients (three received Midostaurin and one Gilterinib), relapsed at a median of 10.5 months from initial diagnosis (range 5–14 months). According to the declaration of Helsinki, all patients gave informed consent for the study, which was approved by the Institutional Review Board of the Policlinico Tor Vergata.
Genetic characterization
Morphologic, immunophenotypic, and molecular routine analysis were carried out on all 38 AML cases at diagnosis and eventually at relapse. For genetic analysis, total RNA was extracted from Ficoll-Hypaque isolated bone marrow (BM) mononuclear cells (MNCs), which contained over 80% of the blast population, and reverse-transcribed using standard procedures13. DNA was extracted from BM-MNCs collected at diagnosis, and for selected cases during follow-up, using a column-based protocol (Qiagen, Hilden, Germany).
Mutations of NPM1 and FLT3 genes were investigated at the DNA and RNA level on MNCs isolated from AML samples and on purified cell populations8,14. As reported elsewhere3, the cut-off for FLT3-ITDmut positivity was AR ≥ 0.05. The validity of this method for quantification of the mutation burden was confirmed by the linear relationship between the value of the AR to the proportion of mutated versus total FLT3 (ITD + WT) allele (n = 99 samples, R = 0.6, p < 0.0001, data not shown). NPM1 molecular analysis was carried out on all AML samples included in this study15 and the NPM1 mutation load was calculated using the same criteria adopted for FLT3-ITDmut AR determination. Amplification products for FLT3-ITDmut and NPM1 genes were analysed on the Applied Biosystems 3130 Genetic Analyzer (Thermo Fisher Scientific), using the GeneMapper™ Software 5.
Patients’ mutational profile was studied in 12 FLT3-ITDmut AML cases at diagnosis, on DNA extracted from BM-MNCs, using NGS and the targeted Oncomine Myeloid-Research Assay (Thermo Fisher Scientific) (Table 1). The assay includes the entire coding region of 17 genes (ASXL1, BCOR, CALR, CEBPA, ETV6, EZH2, IKZF1, NF1, PHF6, PRPF8, RB1, RUNX1, SH2B3, STAG2, TET2, TP53, ZRSR2, and the “hotspot” region of 23 genes (ABL1, BRAF, CBL, CSF3R, DNMT3A, FLT3, GATA2, HRAS, IDH1, IDH2, JAK2, KIT, KRAS, MPL, MYD88, NPM1, NRAS, PTPN11, SETBP1, SF3B1, SRSF2, U2AF1, and WT1). Panel yields 526 amplicons with an average length of 400 bp. Following the manufacturer’s instructions and run in the Ion 530™ chip on the Ion Torrent S5 instrument (Thermo Fisher Scientific), sequence analysis were performed using the Ion Torrent Suite Software v.5.8.0 and the Ion Reporter software v.5.10.3.0 and v.5.10.5.0. Analysis of the raw sequencing data and alignment of sequencing reads to the reference genome (Human genome build 19 of Hg 19) was performed by the Ion Reporter software for Ion S5. Identification of sequence variants was carried out by Ion Torrent Variant Caller Plugin software v4.4-r76860, and coverage of each amplicon was determined by the Coverage Analysis Plugin software v4.4-r77897. The minimum coverage depth was set at 400X for somatic DNA variants. The sensitivity of this NGS assay was 5% variant allele fraction (VAF). 5′ and 3′ untranslated regions (UTRs), intronic donor splice-site variants, and polymorphisms were filtered out.
To assess the mutational profile of AML patients who relapsed after TKIs treatment, we compared the genetic asset of four patients at diagnosis and at relapse (Table 2), studying 30 genes known to be frequently mutated in myeloid malignancies, using NGS and the Myeloid Solution by SOPHiA GENETICS (Saint-Sulpice, Switzerland). The resulting captured libraries were further processed on a MiniSeq sequencing platform (Illumina, San Diego, CA, USA). FASTQ sequencing files were then uploaded on the SOPHiA DDM platform for analysis by the specific technology. Only mutations identified as highly or potentially pathogenic were considered for analysis. The sensitivity of this NGS method was ≥1%. Further details on the NGS analysis have been previously reported16.
Multiparametric flow cytometry (MPFC) studies and high-speed cell sorting
Immunophenotypic studies were performed at diagnosis using standard techniques, as detailed elsewhere17. After completion of diagnostic tests, further sorting, and molecular analysis were performed in particular on 12 AML patients with available cells at diagnosis. The percentage of CD123/CD25/CD99+ cells within the CD34+ cell fraction was studied as previously reported by our group9. Immunophenotype studies were carried out using a CytoFLEX flow cytometer (Beckman Coulter), and data were analysed using the FlowJo software (TreeStar Inc. Ashland, USA). MNCs were isolated by Ficoll Hypaque and were stained with the following anti-human antibodies: CD45 APC-vio770, CD99R PE (purchased from Miltenyi Biotec), CD33 Fitc (e-Bioscience), CD117 PE-Cy7, CD14 PE-Cy5.5 (purchased from Beckman Coulter), CD38 PE-CF594, CD3 Brilliant Violet 785, CD25 Brilliant Violet 421 and CD123 Brilliant Violet 605 (purchased from Becton Dickinson). Progenitor cell populations were sorted by high-speed cell sorting, in the 6-way sorting MoFlo Astrios (Beckman Coulter), using a sequential gating strategy to separate CD34/CD123/CD25/CD99+ LPCs, CD38+ and CD38− cells, CD34+ progenitors (CD123+/−,CD25−,CD99 low/−) and T-lymphocytes (CD45high/CD3+/CD14−/CD33−/CD34). For analysis of CD99 analysis, measurements included the evaluation of the number of cells positively stained and the mean fluorescence intensity (MFI) on leukemic blasts (adjusted for background fluorescence using negative internal controls).
In vitro anti-CD99 mAb treatment assays
To determine whether CD99 may be a relevant therapeutic target, we investigated the ability of a monoclonal antibody (mAb) against CD99 to induce direct cytotoxicity on enriched LPCs. The anti-CD99 mAb clone 3.2–3 (Diatheva, Cartoceto, PU, Italy) is a fully human recombinant IgG4 monoclonal antibody, specific for the extracellular domain of CD99. CD34/CD123/CD99+/CD25− enriched-LPCs sorted from 2 AML patients who relapsed after Midostaurin/chemotherapy treatment and CD34+ cells from 2 healthy donors were seeded at 1 × 105 cells/well in 200 µl of Stemspan SFEM II medium supplemented with CD34+ Expansion Supplement (10×) (Stem Cell Technologies, Carlsbad, California), in a flat-bottom 96-well plate. The anti-CD99 mAb diluted in sterile PBS was then added at a concentration of 25 μg/ml, and cells were incubated for 72 h at 37 °C, in 5% CO2. At the end of the incubation time, cells were stained with a LIVE/DEAD fixable dead cell stain (Life Technologies,Carlsbad,California), according to the manufacturer’s instructions, and analyzed by flow cytometry.
Statistical analysis
Statistical analysis was performed using the GraphPad Prism version 5.0 (La Jolla, CA, USA). P values < 0.05 were considered statistically significant. Differences between categorical variables were evaluated by the Pearson’s chi-squared test, while differences between continuous variables were analysed by unpaired t–test with 95% confidence intervals.
Results
Heterogeneity of FLT3-ITDmut AML at diagnosis and relapse
We studied the subclonal architecture of FLT3-ITDmut AML, using samples from 38 patients with a FLT3-ITDmut AML. The median AR for FLT3-ITDmut in BM-MNCs was 0.34 (range 0.03–1.11) (data not shown). For the purpose of this study, we included three cases with FLT3-ITDmut AR below the <0.05 threshold, which we classified as FLT3-ITDmut after confirmation of the ITD size by cDNA analysis. NPM1 mutations were identified by capillary electrophoresis in 19 AML patients (50%) (median AR:0.75, range 0.44–0.93) (Fig. 1a). The comparison of ARs in 19 FLT3-ITDmut/NPM1 double-mutant AML, showed that FLT3-ITDmut allelic burden was heterogeneous and significantly lower than that of NPM1 (median FLT3-ITDmut AR: 0.36 vs NPM1 AR: 0.75, p = 0.001, Fig. 1a), with no correlations between the mutation loads of the two genes in individual patients (Spearman r = 0.07, p = 0.74). The ITD size was highly variable, with a median of 42 bp (range 16–186 bp). These data confirm that FLT3-ITDmut is present at heterogeneous levels at AML diagnosis, different from NPM1 mutations that are typically heterozygous18.
a FLT3-ITDmut and NPM1mut AR analysis on MNCs in double-mutant AML patients at diagnosis (n = 19). The FLT3-ITDmut and NPM1mut AR were defined as the ratio of the area under the curve of FLT3-ITDmut and FLT3 wild-type alleles. b Targeted NGS analyses performed in primary blasts from FLT3-ITDmut AML cases at diagnosis (n = 12). c Analysis of FLT3-ITDmut AR on MNCs from mutated AML patients (n = 12), including three cases who remained in CCR at 17 months (range 14–20 months), six who relapsed after a median of 7 months (range 3–8 months) from diagnosis and three who death in induction therapy. AR allelic ratio, MNCs mononuclear cells, CCR continuous complete remission.
Looking for differences predictive of outcome, we then analysed the mutation profiles of 12 of these patients at diagnosis, using the Oncomine™ Myeloid-Research Assay panel. Clinical and molecular characteristics of these patients are shown in Table 1. We identified a median of three additional mutations per FLT3-ITDmut AML (range, 1–7), with N-RAS (5 of 12, 41%), RUNX1 (4 of 12, 33%) and DNMT3A (3 of 12, 25%) as the most frequent alterations. Concomitant mutations (number or type) were not predictive for the presence of a FLT3-ITDmut subclone (n = 3, AR < 0.05) or of outcome (Fig. 1b). Also, the number of concomitant mutations did not impact on the probability of relapse (median cumulative incidence of relapse (CIR) on 4.5 months in patients with ≤2 mutations vs 5 months in patients with ≥3 mutations, p = 0.794).
BM-MNC follow-up samples were available for nine patients, including three patients who remained in continuous complete remission (CCR) at 17 months (range 14–20 months), and 6 who relapsed after a median of 7 months (range 3–8 months) from diagnosis. As shown in Fig. 1c, FLT3-ITDmut were not detectable by capillary electrophoresis in CR samples, both in patients who remained in CCR and in those who later relapsed, where the FLT3-ITDmut burden increased in five of six cases. On patient lost the FLT3-ITDmut clone at relapse.
Immunophenotype profiles and FLT3-ITD allele burden in highly purified LPCs at AML diagnosis
We performed additional molecular and immunophenotypic studies in samples sorted from the 12 FLT3-ITDmut patients.
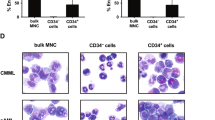
The number of CD34/CD123/CD25/CD99+ LPCs, which corresponds to the LAIP of FLT3-ITDmut AML, was assessed using cytofluorimetry, and accounted for 0.15% of BM-MNCs only (median, range 0.001–12.4%), while CD34+ progenitors represented 0.07% of BM-MNCs (median, range 0.02–75.2%), p = 0.0005 (Fig. 2a). Representative plots of two patients, one with a clear LAIP and one with a minor subclone, are shown in Supplementary Fig. 1.
a Proportion of blasts, LPCs and CD34+ progenitors, defined as the percentage of total MNCs, obtained from AML patients by high-speed sorting at diagnosis (n = 12). b Analysis of FLT3-ITDmut AR in the specific BM subpopulations in AML cases (n = 12). c Enrichment of FLT3-ITDmut load in LPCs purified from AML patients characterized by subclonal FLT3-ITDmut mutations, as compared to MNCs (n = 5). d Analysis of FLT3-ITDmut AR in LPCs separated according to CD38 expression in AML patients at diagnosis (n = 5). e Genescan electropherograms of FLT3-ITDmut PCR fragments amplified from MNCs and LPCs DNA from UPN 6 at AML diagnosis. We observed two different FLT3-ITDmut clones (duplication of 37 and 186 bp) in BM-MNCs cells, while LPCs displayed only the large FLT3-ITDmut clone in a homozygous state. The PCR amplicon size of FLT3 wild-type allele corresponds to 330 bp. f Mean fluorescence intensity (MFI) of CD99 antigen on LPCs, total CD34+ cells and primary blasts purified from AML patients by high-speed sorting at diagnosis (n = 12). CD99 MFI was also evaluated on CD34+ cells obtained from BM samples of 3 HDs. The antibody combination used for cell sorting is indicated. P values were calculated by a paired Student’s t-test. LPCs leukemic precursor cells, AR allelic ratio, MNCs mononuclear cells, HDs healthy donors.
LPCs were characterized by a significantly higher FLT3-ITDmut load, as compared to MNCs (median AR: 0.84, range 0.42–19.3, vs 0.17, range 0.035–0.6 p = 0.0005, n = 12), and CD34+ progenitors (CD123+/−, CD25-, CD99low/-) (median AR = 0.047, range 0–0.8) (p = 0.0005) (Fig. 2b). Lymphocytes sorted according to the CD45high/CD3+/CD14-/CD33-/CD34- immunophenotype, were FLT3-ITD negative, as expected. Of note, a small FLT3-ITDmut clone, observed in the MNCs of six patients at a median AR of 0.04 (range 0.03–0.06), was highly enriched in the corresponding sorted LPCs (median AR = 0.54, range 0.4–0.87, p = 0.003, Fig. 2c). Further sorting of LPCs for CD38 did not enrich for FLT3-ITDmut clones (median AR: 0.87, range 0.15–1, vs 0.88, range 0.05–2, p = 0.62, Fig. 2d).
Using the LPCs data, we could classify as FLT3-ITDmut 3 AML patients, otherwise classified as FLT3-ITD wild-type. Two of these patients had a full-blown FLT3-ITDmut relapse.
Interestingly, in one patient (UPN 6), characterized by two different FLT3-ITDmut clones in the BM-MNCs at diagnosis (duplication of 37 and 186 bp), the LPCs displayed only the larger FLT3-ITD clone in a homozygous state, probably deriving from the copy-neutral loss of heterozygosity (CN-LOH13q) (Fig. 2e).
Since, as previously shown, CD99 expression is significantly associated with FLT3-ITD mutations19, we studied CD99 expression levels in the different BM-subpopulations, expressed as mean fluorescence intensity (MFI) (n = 9 AML cases). CD99-MFI level was significantly higher in LPCs (median: 15647, range 8419–38958), as compared to CD34+ cells (median CD99-MFI 13305, range 2352–28762) (p = 0.005), blasts (median CD99-MFI 4349, range 1605–12283) (p = 0.005) and CD34+ cells purified from three healthy donors (HDs) (median CD99-MFI 2.7, range 1.68–3.9) (p = 0.004) (Fig. 2f).
In this line, the FLT3-ITDmut and NPM1mut burden was different in the cell compartments, with NPM1 mutations homogeneously expressed at high levels both in blasts, LPCs and CD34+ progenitors, and highly variable FLT3-ITDmut loads (p = 0.006) (Supplementary Fig. 2). This reflects the nature of NPM1 mutations, which are considered initiating events in AML.
We were then interested whether the mutation status of sub-populations would be informative of subsequent disease course. We prospectively collected and sorted LPCs and CD34+ progenitors in one of the patients (in UPN1), at diagnosis and at different treatment time-points, after decitabine, and after the following 3 + 7 salvage chemotherapy (Fig. 3). A progressive increase of FLT3-ITDmut burden was observed in LPCs as compared to CD34+ progenitors and blast population. In particular, a high FLT3-ITDmut allele burden was detectable in enriched LPCs at the time of complete morphologic remission (at 6 months), preceding the first hematologic relapse by 2 months (Fig. 3a). Furthermore, comparing the FLT3-ITDmut load in cell subsets, counted as proportion of total MNCs during the disease course, we observed progressive expansion of the FLT3-ITD load, leading to AR > 1, probably due to CN-LOH13q, while the absolute number of LPCs did not expand (Fig. 3b). These data indicate that FLT3-ITDmut clones may persist in LPCs at the time of complete remission, and represent the treatment-resistant FLT3-ITD reservoir, later driving disease relapse. In particular, at the time of the third relapse, an additional FLT3-ITDmut clone (ITD of 27 bp) was detected both in blasts and in LPCs compartments, indicating clonal evolution (Fig. 3c).
a Evaluation of the course of bulk leukemic blasts, LPCs-enriched fraction, and CD34+ progenitors subpopulations, purified from AML patient (UPN 1) who achieved CR after 6 cycles of decitabine, relapse and was treated with the “3 + 7” cytarabine and daunorubicin chemotherapy regimen. Although the median proportion of BM-LPCs was 1.2% (range 1.13–2.38%), the FLT3-ITDmut load persisted at high levels in these cells at the time of CR, preceding relapse by two months. The relative proportion of the different cell subpopulation and the antibody combinations used for cell sorting are indicated. CR complete remission, LPCs leukemic precursor cells. b Genescan electropherograms of FLT3-ITDmut PCR fragments amplified from MNCs at first (at 8 months) and second (at 13 months) hematological relapse. One FLT3-ITDmut clone (duplication of 74 bp) was present at first relapse, and an additional FLT3-ITDmut clone (duplication of 27 bp) emerged at second relapse. P values were calculated by a paired Student’s t-test. MNCs mononuclear cells.
Additional mutations in leukemic progenitor cells at diagnosis
To shed light on the molecular heterogeneity of AML blasts, we performed targeted NGS on the cell sub-populations of 6 FLT3-ITDmut patients, using the Oncomine™-Myeloid-Research-Assay-panel (Fig. 4). The mutational profile of MNCs was compared to that of LPCs and/or to the CD34+ progenitors. The same mutation pattern was present in both blasts and LPCs in four cases (Fig. 4a). In two cases, with the NGS detection limits of 5%, mutations in BCOR and NRAS were identified in MNCs only, probably indicating the acquisition of additional events or expansion of the mutated clones during differentiation. Furthermore, by comparing the VAF of mutations by targeted NGS to the FLT3mut AR in the different cell populations, we confirmed that while the FLT3-ITD burden was again significantly higher in LPCs compared to MNCs, the load of other mutations was similar in the two cell compartments (Fig. 4b).
a Mutational profiling of MNCs and high-speed sorted subpopulations of AML patients at diagnosis (n = 6). Patient samples are displayed as columns, D diagnosis, MNCs mononuclear cells. b Comparison of variant allele frequencies of mutations detected by NGS and FLT3-ITDmut AR studied by capillary electrophoresis, in six diagnostic samples. LPCs leukemic precursor cells, AR allelic ratio, MNCs mononuclear cells. P values were calculated by a paired Student’s t-test.
Mutation profile in AML relapsing after treatment with tyrosin-kynase inhibitors (TKI) and treatment with CD99
TKIs have been recently approved for the treatment of FLT3mut AML. We then studied whether TKI addition to standard chemotherapy (CHT) would modify the mutation profile of AML at relapse. Using the Myeloid Solution panel (SOPHiA GENETICS), we compared the mutation profile at diagnosis and at relapse in 4 FLT3-ITDmut samples from patients who relapsed after 3 + 7/midostaurin or quizartinib in combination with CHT, at a median of 10.5 months (range 6–14) from diagnosis (Table 2 and Fig. 5). In two patients characterized by two different FLT3-ITDmut clones at diagnosis (UPN14/UPN15) only one clone was retained at relapse after midostaurin, while the third patient became FLT3-ITD negative (UPN13). The same FLT3-ITDmut pattern was maintained both at diagnosis and at relapse in one patient treated with quizartinib (UPN16). Minor changes in the mutation type and load occurred for other genes (Fig. 5). These data are in agreement with the specific activity of TK, and with the occurrence of FLT3-ITDmut clonal selection at relapse.
Looking at LPCs and CD34 + progenitors, we confirmed enrichment of FLT3-ITD mutations in sorted LPCs at diagnosis in UPN 11 (FLT3-ITD AR = 0.42), with loss of the mutation at relapse, both in blasts and in CD34/CD123/CD99+ purified cells (supplementary Fig. 3). A second patient (UPN15) was FLT3-ITDmut both at diagnosis and at relapse, with expansion of the FLT3-ITDmut clone to the homozygous state in LPCs (Supplementary Fig. 3).
Since recent studies demonstrated that CD99 may be a potential therapeutic target in AML20, as proof of principle, we tested the activity of the anti-CD99 mAb clone 3.2–3 on LPCs sorted from UPN 13 and UPN 14 at the time of relapse after midostaurin. BM CD34+/CD99- cells isolated from two healthy donors served as controls. As shown in Fig. 6, the anti-CD99 mAb was cytotoxic on enriched LPCs (40.7% and 37.7% dead cells in UPN 13 and UPN 14, respectively, Fig. 6a, b) as compared to untreated cells (UT), while there was no effect on normal CD34+ cells from a healthy donor (Fig. 6c).
The cytotoxic activity of the anti-CD99 mAb clone 3.2–3 was evaluated on CD34/CD123/CD99+ LSCs sorted from (a) UPN 13 and (b) UPN 14 at the time of relapse after midostaurin and on BM CD34+/CD99- cells isolated from 1 HD (c). The anti CD99 mAb cytotoxic effect was assessed by the Live/Dead assay after 72 h of incubation with the CD99 mAb (25 µg/ml). The proportion of dead cells was calculated by subtracting the value obtained in the absence of treatment (UT) from that of treated cells incubated with the anti-CD99 antibody. HD healthy donor.
Discussion
We previously showed that the specific CD34/CD123/CD25/CD99 + LPCs phenotype predicts for FLT3-ITDmut in AML9. Here, we show that the FLT3-ITDmut load is significantly enriched in LPCs purified by high-speed cell sorting, as compared to the bulk blast cell population or to CD34+ progenitors isolated at AML diagnosis. Our data confirm that the onset of FLT3-ITDmut may be an early event occurring at the stem cell level in AML. This is of clinical relevance, since using the LPCs data we could classify as FLT3-ITDmut 3 patients who were under the threshold limits of FLT3-positivity in MNC. According to ELN 2017, the FLT3-ITDmut AR identifies different prognostic subgroups in AML, and high FLT3-ITDmut burden (>0.5) predicts for unfavorable outcome in NPM1mut AML21,22,23,24. The mechanisms behind the variation in AR remains poorly understood, but recent reports suggest that CN-LOH13q may induce high FLT3-ITDmut AR25,26. Clonal evolution is unfortunately common in FLT3-ITDmut AML, with loss of the FLT3 wild-type allele associated with an aggressive phenotype27. In this line, in one of our patients, carrier of two small FLT3-ITDmut clones at diagnosis, the LPCs displayed homozygosity for the larger FLT3-ITDmut clone at relapse, likely originating from CN-LOH13q. Also, FLT3-ITD burden was significantly higher in LPCs as compared to blasts and CD34+ progenitors, while the load for other mutations was similar in the different cell populations.
The role of FLT3-ITDmut as a minimal residual disease marker is debated, due to the instability at relapse and the technical issues related to the patient-specificity of the ITD sequence28. In one of our patients, FLT3-ITDmut LPCs were present at the time of complete remission, while BM-MNC were FLT3-ITDmut negative, preceding morphological relapse by two months. These data indicate that LPCs may represent the leukemic reservoir driving disease relapse. Our data highlight the relevance of characterizing the mutational and immunophenotypic profiles of LPCs, also to eventually add targeted drugs. In this setting, TKIs are a successful therapeutic strategy in AML, and in most recent years several FLT3-targeted agents have been approved by FDA and EMA29,30. The first new molecule approved in nearly two decades has been midostaurin, a multikinase inhibitor active against both FLT3-ITD and TKD mutations. Although midostaurin demonstrated high efficacy when combined with conventional chemotherapy in AML, the relapse rate remains high, at about 50% at a median of two years of follow-up5. In this setting, the results from our study demonstrate that TKIs treatment may impact on clonal evolution of FLT3-ITDmut AML, downregulating specific clones. These data, underscore the importance of repeated mutation testing for FLT3-ITDmut to distinguish patients where TKI may induce long-lasting remission from those where relapse may originate from subclones, which may be FLT3-wild type.
One of the mechanisms of treatment failure may be the persistence of FLT3-ITDmut LPC, as a potential source of relapse. These cells are characterized by overexpression of CD99, as shown by significantly increased MFI compared to CD34+ progenitor cells. To overcome midostaurin resistance and try to target disease-initiating leukemic stem cells, as proof of principle, we tested a CD99-specific antibody on primary patient samples collected at the time of relapse following midostaurin. This treatment was cytotoxic on CD34/CD99+FLT3-ITDmut-LPCs, resistant to midostaurin, while there was no effect on healthy CD34+ cells. One of our patients became FLT3-ITDmut negative at relapse. Loss of FLT3-ITDmut at disease recurrence has been reported in 25% of patients, and this figure may increase after treatment with TK inhibitors. The CD99 antibody may represent an attractive treatment option in these cases21.
In conclusion, we showed that FLT3-ITDmut are present at LPC level and may represent a primary event in leukemogenesis. According to these data, we suggest that the identification of FLT3-ITDmut LPCs driving therapy resistance and/or disease progression in AML may allow to design novel disease-eradicating therapies and eventually improve disease outcome.
References
Papaemmanuil, E. et al. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N. Engl. J. Med. 368, 2209–2221 (2013).
Döhner, H. et al. Diagnosis and management of acute myeloid leukemia in adults: recommendations from an international expert panel, on behalf of the European LeukemiaNet. Blood 115, 453–474 (2010).
Dohner, H. et al. Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood 129, 424–447 (2017).
Thol, F., Schlenk, R. F., Heuser, M. & Ganser, A. How I treat refractory and early relapsed acute myeloid leukemia. Blood 126, 319–327 (2015).
Krönke, J. et al. Clonal evolution in relapsed NPM1-mutated acute myeloid leukemia. Blood 122, 100–108 (2013).
Cloos, J. et al. Stability and prognostic influence of FLT3 mutations in paired initial and relapsed AML samples. Leukemia 20, 1217–1220 (2006).
O’Donnell, M. R. et al. Acute Myeloid Leukemia, Version 3.2017, NCCN Clinical Practice Guidelines in Oncology. J. Natl Compr. Cancer Netw. 15, 926–957 (2017).
Ottone, T. et al. Identification of emerging FLT3 ITD-positive clones during clinical remission and kinetics of disease relapse in acute myeloid leukaemia with mutated nucleophosmin. Br. J. Haematol. 161, 533–540 (2013).
Angelini, D. F. et al. A leukemia-associated CD34/CD123/CD25/CD99+immunophenotype identifies FLT3-mutated clones in acute myeloid Leukemia. Clin. Cancer Res. https://doi.org/10.1158/1078-0432.CCR-14-3186 (2015).
Al-Mawali, A., Gillis, D. & Lewis, I. Immunoprofiling of leukemic stem cells CD34+/CD38-/CD123+ delineate FLT3/ITD-positive clones. J. Hematol. Oncol. 9, 61 (2016).
Papaemmanuil, E. et al. Genomic classification and prognosis in acute myeloid leukemia. N. Engl. J. Med. 374, 2209–2221 (2016).
Welch, J. S. et al. The origin and evolution of mutations in acute myeloid leukemia. Cell 150, 264–278 (2012).
Chomczynski, P. & Sacchi, N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162, 156–159 (1987).
Thiede, C. et al. Analysis of FLT3-activating mutations in 979 patients with acute myelogenous leukemia: association with FAB subtypes and identification of subgroups with poor prognosis. Blood 99, 4326–4335 (2002).
Lin, L.-I. et al. A novel fluorescence-based multiplex PCR assay for rapid simultaneous detection of CEBPA mutations and NPM mutations in patients with acute myeloid leukemias. Leukemia 20, 1899–1903 (2006).
Fabiani, E. et al. Somatic mutations as markers of outcome after azacitidine and allogeneic stem cell transplantation in higher-risk myelodysplastic syndromes. Leukemia 185, 785–790 (2019).
Venditti, A. et al. GIMEMA AML1310 trial of risk-adapted, MRD-directed therapy for young adults with newly diagnosed acute myeloid leukemia. Blood 134, 935–945 (2019).
Falini, B. et al. Cytoplasmic nucleophosmin in acute myelogenous leukemia with a normal karyotype. N. Engl. J. Med. 352, 254–266 (2005).
Vaikari, V. P. et al. Clinical and preclinical characterization of CD99 isoforms in acute myeloid leukemia. Haematologica 105, 999–1012 (2020).
Chung, S. S. et al. CD99 is a therapeutic target on disease stem cells in myeloid malignancies. Sci. Transl. Med. 17, 1910–1921 (2017).
Kottaridis, P. D. et al. Studies of FLT3 mutations in paired presentation and relapse samples from patients with acute myeloid leukemia: implications for the role of FLT3 mutations in leukemogenesis, minimal residual disease detection, and possible therapy with FLT3 inhibitors. Blood 100, 2393–2398 (2002).
Gale, R. E. et al. The impact of FLT3 internal tandem duplication mutant level, number, size, and interaction with NPM1 mutations in a large cohort of young adult patients with acute myeloid leukemia. Blood 111, 2776–2784 (2008).
Terwijn M. et al. Leukemic stem cell frequency: A strong biomarker for clinical outcome in acute myeloid leukemia. PLoS ONE https://doi.org/10.1371/journal.pone.0107587 (2014).
Bacher, U. et al. Quantification of rare NPM1 mutation subtypes by digital PCR. Br. J. Haematol. 167, 710–714 (2014).
Raghavan, M. et al. Genome-wide single nucleotide polymorphism analysis reveals frequent partial uniparental disomy due to somatic recombination in acute myeloid leukemias. Cancer Res. 65, 375–378 (2005).
Dunbar, A. J. et al. 250K single nucleotide polymorphism array karyotyping identifies acquired uniparental disomy and homozygous mutations, including novel missense substitutions of c-Cbl, in myeloid malignancies. Cancer Res. 68, 10349–10357 (2008).
Li, L., Bailey, E., Greenblatt, S., Huso, D. & Small, D. Loss of the wild-type allele contributes to myeloid expansion and disease aggressiveness in FLT3/ITD knockin mice. Blood 118, 4935–4945 (2011).
Cocciardi, S. et al. Clonal evolution patterns in acute myeloid leukemia with NPM1 mutation. Nat. Commun. 10, 2031 (2019).
Stone, R. M. et al. Midostaurin plus chemotherapy for acute myeloid leukemia with a FLT3 mutation. N. Engl. J. Med. 377, 454–464 (2017).
Zhao, J., Song, Y. & Liu, D. Gilteritinib: a novel FLT3 inhibitor for acute myeloid leukemia. Biomark Res. 7, 19 (2019).
Acknowledgements
This work was supported by AIRC 5×1000 call “Metastatic disease: the key unmet need in oncology” to MYNERVA project, #21267 (MYeloid NEoplasms Research Venture AIRC. A detailed description of the MYNERVA project is available at http://www.progettoagimm.it.), by GR-2018-12365529-Santa Lucia to TO, PRIN grant N. 2017WXR7ZT to MTV, and the Innovative Medicines Initiative (IMI) 2 project “HARMONY”, no. 116026 to MTV.
Author information
Authors and Affiliations
Contributions
S.T., T.O., D.F.A., L.B., F.L.C., and M.T.V. made substantial contributions to conception, design of the study, and analysis and interpretation of the data; V.A., G.G., S.L., M.D., A.M.N., M.I.C., E.F., and M.D.B. performed the experiments, B.N., F.F., F.M., G.P., R.C., E.B., M.I.D.P., A.V., and W.A. acquired and analyzed the data; all authors revised and approved the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Travaglini, S., Angelini, D.F., Alfonso, V. et al. Characterization of FLT3-ITDmut acute myeloid leukemia: molecular profiling of leukemic precursor cells. Blood Cancer J. 10, 85 (2020). https://doi.org/10.1038/s41408-020-00352-9
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41408-020-00352-9