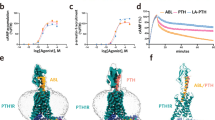
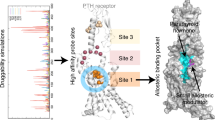
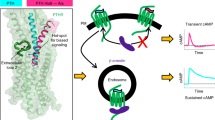
Abstract
Parathyroid hormone (PTH) and PTH-related peptide (PTHrP) are two endogenous hormones recognized by PTH receptor-1 (PTH1R), a member of class B G protein- coupled receptors (GPCRs). Both PTH and PTHrP analogs including teriparatide and abaloparatide are approved drugs for osteoporosis, but they exhibit distinct pharmacology. Here we report two cryo-EM structures of human PTH1R bound to PTH and PTHrP in the G protein-bound state at resolutions of 2.62 Å and 3.25 Å, respectively. Detailed analysis of these structures uncovers both common and unique features for the agonism of PTH and PTHrP. Molecular dynamics (MD) simulation together with site-directed mutagenesis studies reveal the molecular basis of endogenous hormones recognition specificity and selectivity to PTH1R. These results provide a rational template for the clinical use of PTH and PTHrP analogs as an anabolic therapy for osteoporosis and other disorders.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
Cryo-EM maps have been deposited in the Electron Microscopy Data Bank under accession codes: EMD-34585 (PTH-bound PTH1R-Gs complex), EMD-34587 (PTHrP-bound PTH1R-Gs complex) and EMD-34598 (dimer of PTH-bound PTH1R-Gs complex). The atomic coordinates have been deposited in the Protein Data Bank under accession codes: 8HA0 (PTH-bound PTH1R-Gs complex), 8HAF (PTHrP-bound PTH1R-Gs complex) and 8HAO (dimer of PTH-bound PTH1R-Gs complex).
References
Gardella TJ, Vilardaga JP. International Union of Basic and Clinical Pharmacology. XCIII. The parathyroid hormone receptors–family B G protein-coupled receptors. Pharmacol Rev. 2015;67:310–37.
Zhao LH, Ma S, Sutkeviciute I, Shen DD, Zhou XE, de Waal PW, et al. Structure and dynamics of the active human parathyroid hormone receptor-1. Science. 2019;364:148–53.
Nishimura Y, Esaki T, Isshiki Y, Furuta Y, Mizutani A, Kotake T, et al. Lead optimization and avoidance of reactive metabolite leading to PCO371, a potent, selective, and orally available human parathyroid hormone receptor 1 (hPTHR1) agonist. J Med Chem. 2020;63:5089–99.
Kir S, White JP, Kleiner S, Kazak L, Cohen P, Baracos VE, et al. Tumour-derived PTH-related protein triggers adipose tissue browning and cancer cachexia. Nature. 2014;513:100–4.
Wider J, Undyala VVR, Lanske B, Datta NS, Przyklenk K. Parathyroid hormone-related peptide and its analog, abaloparatide, attenuate lethal myocardial ischemia-reperfusion injury. J Clin Med. 2022;11:2273; https://doi.org/10.3390/jcm11092273.
Chew CK, Clarke BL. Abaloparatide: recombinant human PTHrP (1-34) anabolic therapy for osteoporosis. Maturitas. 2017;97:53–60.
Srivastava M, Deal C. Osteoporosis in elderly: prevention and treatment. Clin Geriatr Med. 2002;18:529–55.
Bilezikian JP, Rubin MR, Finkelstein JS. Parathyroid hormone as an anabolic therapy for women and men. J Endocrinol Invest. 2005;28:41–9.
Quattrocchi E, Kourlas H. Teriparatide: a review. Clin Ther. 2004;26:841–54.
Gonnelli S, Caffarelli C. Abaloparatide. Clin Cases Min Bone Metab. 2016;13:106–9.
Pioszak AA, Parker NR, Gardella TJ, Xu HE. Structural basis for parathyroid hormone-related protein binding to the parathyroid hormone receptor and design of conformation-selective peptides. J Biol Chem. 2009;284:28382–91.
Dean T, Vilardaga JP, Potts JT Jr., Gardella TJ. Altered selectivity of parathyroid hormone (PTH) and PTH-related protein (PTHrP) for distinct conformations of the PTH/PTHrP receptor. Mol Endocrinol. 2008;22:156–66.
Hattersley G, Dean T, Corbin BA, Bahar H, Gardella TJ. Binding selectivity of abaloparatide for PTH-Type-1-receptor conformations and effects on downstream signaling. Endocrinology. 2016;157:141–9.
Okazaki M, Ferrandon S, Vilardaga JP, Bouxsein ML, Potts JT Jr., Gardella TJ. Prolonged signaling at the parathyroid hormone receptor by peptide ligands targeted to a specific receptor conformation. Proc Natl Acad Sci USA. 2008;105:16525–30.
Zhao LH, Lin J, Ji SY, Zhou XE, Mao C, Shen DD, et al. Structure insights into selective coupling of G protein subtypes by a class B G protein-coupled receptor. Nat Commun. 2022;13:6670.
Kobayashi K, Kawakami K, Kusakizako T, Miyauchi H, Tomita A, Kobayashi K, et al. Endogenous ligand recognition and structural transition of a human PTH receptor. Mol Cell. 2022;82:3468–83. e5
Maeda S, Koehl A, Matile H, Hu H, Hilger D, Schertler GFX, et al. Development of an antibody fragment that stabilizes GPCR/G-protein complexes. Nat Commun. 2018;9:3712.
Chan P, Gabay M, Wright FA, Kan W, Oner SS, Lanier SM, et al. Purification of heterotrimeric G protein alpha subunits by GST-Ric-8 association: primary characterization of purified G alpha(olf). J Biol Chem. 2011;286:2625–35.
Dixon AS, Schwinn MK, Hall MP, Zimmerman K, Otto P, Lubben TH, et al. NanoLuc complementation reporter optimized for accurate measurement of protein interactions in cells. ACS Chem Biol. 2016;11:400–8.
Ma S, Shen Q, Zhao LH, Mao C, Zhou XE, Shen DD, et al. Molecular basis for hormone recognition and activation of corticotropin-releasing factor receptors. Mol Cell. 2020;77:669–80 e4.
Zivanov J, Nakane T, Scheres SHW. Estimation of high-order aberrations and anisotropic magnification from cryo-EM data sets in RELION-3.1. Iucrj. 2020;7:253–67.
Rohou A, Grigorieff N. CTFFIND4: fast and accurate defocus estimation from electron micrographs. J Struct Biol. 2015;192:216–21.
Punjani A, Rubinstein JL, Fleet DJ, Brubaker MA. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat Methods. 2017;14:290–6.
Sanchez-Garcia R, Gomez-Blanco J, Cuervo A, Carazo JM, Sorzano COS, Vargas J. DeepEMhancer: a deep learning solution for cryo-EM volume post-processing. Commun Biol. 2021;4:874.
Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, et al. UCSF Chimera–a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–12.
Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–32.
Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–21.
Chen VB, Arendall WB, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D. 2010;66:12–21.
Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596:583–9.
Jo S, Cheng X, Lee J, Kim S, Park S-J, Patel DS, et al. CHARMM-GUI 10 years for biomolecular modeling and simulation. J Comput Chem. 2017;38:1114–24.
Lee J, Hitzenberger M, Rieger M, Kern NR, Zacharias M, Im W. CHARMM-GUI supports the Amber force fields. J Chem Phys. 2020;153:35103.
Tian C, Kasavajhala K, Belfon KAA, Raguette L, Huang H, Migues AN, et al. ff19SB: amino-acid-specific protein backbone parameters trained against quantum mechanics energy surfaces in solution. J Chem Theory Comput. 2020;16:528–52.
He X, Man VH, Yang W, Lee TS, Wang J. A fast and high-quality charge model for the next generation general AMBER force field. J Chem Phys. 2020;153:114502.
Roe DR, Cheatham TE III. PTRAJ and CPPTRAJ: software for processing and analysis of molecular dynamics trajectory data. J Chem theory Comput. 2013;9:3084–95.
Harrigan MP, Sultan MM, Hernández CX, Husic BE, Eastman P, Schwantes CR, et al. MSMBuilder: statistical models for biomolecular dynamics. Biophys J. 2017;112:10–5.
Miller BR 3rd, McGee TD Jr., Swails JM, Homeyer N, Gohlke H, Roitberg AE. MMPBSA.py: an efficient program for end-state free energy calculations. J Chem Theory Comput. 2012;8:3314–21.
Pioszak AA, Xu HE. Molecular recognition of parathyroid hormone by its G protein-coupled receptor. Proc Natl Acad Sci USA. 2008;105:5034–9.
Ehrenmann J, Schoppe J, Klenk C, Pluckthun A. New views into class B GPCRs from the crystal structure of PTH1R. FEBS J. 2019;286:4852–60.
Ehrenmann J, Schoppe J, Klenk C, Rappas M, Kummer L, Dore AS, et al. High-resolution crystal structure of parathyroid hormone 1 receptor in complex with a peptide agonist. Nat Struct Mol Biol. 2018;25:1086–92.
Josephs TM, Belousoff MJ, Liang YL, Piper SJ, Cao J, Garama DJ, et al. Structure and dynamics of the CGRP receptor in apo and peptide-bound forms. Science. 2021;372:eabf7258. https://doi.org/10.1126/science.abf7258.
Mannstadt M, Clarke BL, Vokes T, Brandi ML, Ranganath L, Fraser WD, et al. Efficacy and safety of recombinant human parathyroid hormone (1-84) in hypoparathyroidism (REPLACE): a double-blind, placebo-controlled, randomised, phase 3 study. Lancet Diabetes Endocrinol. 2013;1:275–83.
Zhai X, Mao C, Shen Q, Zang S, Shen DD, Zhang H, et al. Molecular insights into the distinct signaling duration for the peptide-induced PTH1R activation. Nat Commun. 2022;13:6276.
Cary BP, Gerrard EJ, Belousoff MJ, Fletcher MM, Jiang Y, Russell IC, et al. Molecular insights into peptide agonist engagement with the PTH1 receptor. Biorxiv. 2022. https://doi.org/10.1101/2022.09.04.506565.
Acknowledgements
The cryo-EM data were collected by Wen Hu and Kai Wu at Advanced Center for Electron Microscopy at Shanghai Institute of Materia Medica, Chinese Academy of Sciences. We are grateful to them for collecting the cryo-EM data. This work was supported by National Natural Science Foundation of China (32071203 to LHZ; 82073904 to MWW and 81973373 to DHY), the National Key R&D Program of China (2019YFA0904200), the Young Innovator Association of CAS (2018325 to LHZ) and SA-SIBS Scholarship Program to LHZ and DHY; Ministry of Science and Technology (China) grants (2018YFA0507002 to HEX and 2018YFA0507000 to MWW), the Shanghai Municipal Science and Technology Major Project (2019SHZDZX02 to HEX; 18ZR1447800 to LHZ and 21JC1401600 to DHY), the CAS Strategic Priority Research Program (XDB08020303 to HEX).
Author information
Authors and Affiliations
Contributions
LHZ designed the expression constructs, purified the complexes, prepared the final samples for negative stain and data collection toward the structures, participated in model building and performed structure and function data analysis, prepared figures and wrote the manuscript; LHZ prepared the cryo-EM grids and QNY collected cryo-EM images and performed map calculations, built and refined the structure models; YWX participated in model refinement; XHH conducted MD simulations; ATD, CWC, CZ and YZ performed signaling experiments under the supervision of DHY and MWW; LHZ and HEX conceived the project, wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhao, Lh., Yuan, Qn., Dai, At. et al. Molecular recognition of two endogenous hormones by the human parathyroid hormone receptor-1. Acta Pharmacol Sin 44, 1227–1237 (2023). https://doi.org/10.1038/s41401-022-01032-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41401-022-01032-z