Abstract
Neddylation is a type of posttranslational protein modification that has been observed to be overactivated in various cancers. UBC12 is one of two key E2 enzymes in the neddylation pathway. Reports indicate that UBC12 deficiency may suppress lung cancer cells, such that UBC12 could play an important role in tumor progression. However, systematic studies regarding the expression profile of UBC12 in cancers and its relationship to cancer prognosis are lacking. In this study, we comprehensively analyzed UBC12 expression in diverse cancer types and found that UBC12 is markedly overexpressed in most cancers (17/21), a symptom that negatively correlates with the survival rates of cancer patients, including gastric cancer. These results demonstrate the suitability of UBC12 as a potential target for cancer treatment. Currently, no effective inhibitor targeting UBC12 has been discovered. We screened a natural product library and found, for the first time, that arctigenin has been shown to significantly inhibit UBC12 enzyme activity and cullin neddylation. The inhibition of UBC12 enzyme activity was newly found to contribute to the effects of arctigenin on suppressing the malignant phenotypes of cancer cells. Furthermore, we performed proteomics analysis and found that arctigenin intervened with cullin downstream signaling pathways and substrates, such as the tumor suppressor PDCD4. In summary, these results demonstrate the importance of UBC12 as a potential therapeutic target for cancer treatment, and, for the first time, the suitability of arctigenin as a potential compound targeting UBC12 enzyme activity. Thus, these findings provide a new strategy for inhibiting neddylation-overactivated cancers.
Similar content being viewed by others
Introduction
Neddylation is a type of posttranslational protein modification that is mediated by the ubiquitin-like protein NEDD8, and it plays an important role in regulating protein stability [1]. The overactivated neddylation pathway has been observed in various types of cancer and indicates poor patient prognosis [2,3,4,5,6]. Two specific E2 conjugating enzymes have been identified to participate in the neddylation pathway: UBC12 and UBE2F. UBC12 preferentially regulates the neddylation of cullin 1, cullin 2, cullin 3, and cullin 4 [7]. Published results indicate that knocking down UBC12 could inhibit the development of lung cancer and esophageal squamous cell carcinoma [8, 9]. It remains unclear whether UBC12 is involved in the progression of other cancers and whether the expression of UBC12 specifically confers the prognosis of certain cancer types. In this study, we systematically analyzed UBC12 expression in diverse cancers. We found that in most cancers (17/21), UBC12 overexpression was associated with poor survival rates, indicating that UBC12 could be a potential target for cancer treatment.
Targeting neddylation is a promising approach for cancer therapy. MLN4924 is the first-in-class E1 enzyme inhibitor of neddylation and exerts significant antitumor effects [10,11,12,13,14,15]. Despite its remarkable effects and considerable success in clinical trials, MLN4924 has some unavoidable limitations. First, due to its broad inhibition of the entire neddylation pathway, MLN4924 may induce unexpected toxicities. Second, some preclinical studies reported that mutations in NAEβ and UBA3 could induce resistance to MLN4924 [16, 17]. Preclinical studies have designed several inhibitors targeting the interaction between the E2 enzyme UBC12 and the E3 enzyme DCN1; however, the effects of these inhibitors against tumors have not been evaluated systematically, in vitro or in vivo [18,19,20]. Notably, no inhibitors that directly target UBC12 have been designed to restrain neddylation and tumor progression. These results indicate the need to develop more selective inhibitors of the neddylation pathway.
Considering the potential role of UBC12 in tumor progression and prognosis, we screened a natural product library to identify potential compounds targeting UBC12 enzyme activity. We found, for the first time, that arctigenin showed a remarkable effect in inhibiting the E2 enzyme activity of UBC12, leading to a decrease in neddylated cullins. As a result, tumor-related downstream signals and substrates of cullin, such as the cell cycle signal, P53 pathway, and tumor suppressor PDCD4, are changed. We found that the function of arctigenin in inhibiting tumor cells is associated with UBC12 enzyme level. Collectively, our studies validate the potential and attractive prospect of UBC12 as an effective anticancer target and identify arctigenin as a new approach to target the overactivated neddylation pathway for cancer treatment.
Materials and methods
Cell lines and culture
The PC9 and H1299 human lung cancer cell lines, the HGC27 and MGC803 human gastric cancer cell lines, the 5637 human bladder cancer cell line, and the U2OS human osteosarcoma cell line were purchased from the Shanghai Institute of Biochemistry and Cell Biology (Shanghai, China). Human embryonic kidney HEK-293T and 293FT cells were purchased from Invitrogen (Grand Island, NY, USA). The PC9, H1299, HGC27, MGC803, 5637, and U2OS cells were maintained in RPMI-1640 medium (HyClone). The HEK-293T cells were maintained in Dulbecco’s Modified Eagle’s Medium (DMEM, Gibco). The 293FT cells were maintained in DMEM (Gibco) with 1 mM L-glutamine (Invitrogen) and 1% MEM-nonessential amino acids. All culture media were supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin.
Plasmids, reagents, and antibodies
Human UBC12 was amplified from HEK-293T cDNA and then subcloned into the pCDNA3.0 plasmid. The sequences of shUBC12 #1 and shUBC12 #2 were synthesized and subcloned into the PLKO.1 plasmid. A primary antibody against UBC12 was purchased from Santa Cruz Biotechnology. Primary antibodies against cullin 1 and cullin 3 were purchased from Cell Signaling Technology. Primary antibodies against NEDD8 and PDCD4 were purchased from Abcam. A primary antibody against GAPDH was purchased from Diagbio. Arctigenin was purchased from FEIYUBIO (Nantong, China).
Cell proliferation assay
Cell proliferation was evaluated by a sulforhodamine B (SRB) colorimetric assay. To assess drug effects, cells were seeded in 96-well plates, cultured for 24 h, and then treated with DMSO or a dilution series of arctigenin for 72 h. To assess clone formation ability, cells were seeded in six-well plates, cultured for 24 h, and then exposed to arctigenin or DMSO for 10–12 days. At the end of the experiment, the cells were washed with PBS five times and then fixed with 10% trichloroacetic acid (4 °C). Following fixation, the cells were incubated in SRB for 30 min at room temperature. Then, 1% acetic acid was used to wash residual SRB. After drying, the quantity of clones was measured using ImageJ. A 10 mM Tris solution was used to dissolve the SRB dye in 96-well plates, and the optical density was analyzed at 510 nm.
Cell apoptosis analysis
Cells were stained with PI/Annexin V for 15 min and then collected. Flow cytometry was used to detect PI-/Annexin V+ and PI+/Annexin V+ cells for cell apoptosis analysis.
Western blotting
To assess PDCD4 and GAPDH levels, cells were lysed in 4% SDS buffer (4% SDS, 150 mM NaCl, 50 mM triethylamine). To determine the extent of neddylation of cullin 1 and cullin 3, cells were lysed in 1% NP40 buffer (50 mM Tris, 150 mM NaCl, 1% NP40, pH=7.4) containing protease inhibitors (5 mg/mL leupeptin, 0.1 mM Na3VO4, 1 mM PMSF). A BCA reagent kit was used to quantify the protein level of all samples. Proteins were separated by 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS‒PAGE) and transferred to PVDF membranes. To detect NEDD8-conjugated UBC12, cells were lysed in MES buffer (50 mM MES, 150 mM NaCl, 1% NP40, pH = 4.5) containing protease inhibitors (5 mg/ml leupeptin, 0.1 mM Na3VO4, 1 mM PMSF). Proteins were separated using 12% Bis-Tris gels and transferred to PVDF membranes. The membranes were blocked with 5% skim milk, and then incubated with primary antibodies overnight at 4 °C, subsequently incubated with secondary antibodies at room temperature for 1 h and finally, reacted with ECL detection reagent and detected via chemiluminescence.
Lentivirus transduction
Five micrograms of PLKO.1-shUBC12 (sequence #1 or sequence #2), 5 μg of pCMV-P8.9, and 1 μg of pMDG-VSVG were mixed with jetPRIME transfection reagent, added to 293FT and incubated for 16 h. The supernatant was discarded. The cells were then incubated with fresh medium containing sodium pyruvate overnight. The supernatant was filtered with 0.45 μm filters and concentrated. For lentivirus transduction, cells were cultured in six-well plates and infected with lentiviral suspensions in the presence of 6 mg/mL polybrene for 16 h. The supernatant was replaced with fresh medium, and the cells were cultured until the cells were harvested to examine protein expression.
Quantitative proteomics analysis
HGC27 cells were treated with 40 μM arctigenin for 36 h and 72 h and then harvested. Label-free quantitative proteomics was analyzed by PTM BIO.
Expression and survival analysis of online datasets
The expression profile of UBC12 in different types of cancer was analyzed using the Tumor Immune Estimation Resource (TIMER, http://timer.cistrome.org/) [21]. The gene expression level is shown with log2 TPM. The differential expression between tumor and adjacent normal tissues was determined by Wilcoxon test. Kaplan‒Meier Plotter (KM plotter, http://kmplot.com) was used to analyze the correlation between UBC12 expression and overall survival [22]. Samples were assigned to two cohorts based on the best cutoff of UBC12 expression.
Statistical analysis
Data are presented as the mean values ± SD and were analyzed using Student’s t test. IC50 values were determined using the statistical analysis software platform SPSS. Protein expression was measured using ImageJ. We used Pearson’s coefficient correlation to evaluate the correlation of IC50 values with the neddylation level of UBC12. P values less than 0.05 were considered statistically significant (n.s., P > 0.05; *P < 0.05; **P < 0.01; ***P < 0.001).
Results
The overexpression of UBC12 is closely associated with poor prognosis in a subset of human cancers
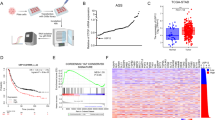
We used the TIMER database to systematically investigate the expression profile of UBC12 in different cancer types. Comprehensive analysis of 21 cancer types from The Cancer Genome Atlas (TCGA) showed that UBC12 was overexpressed in 80.95% of cancer types (17/21), compared to adjacent normal samples. These cancer types include bladder urothelial carcinoma (BLCA), breast invasive carcinoma (BRCA), cervical squamous cell carcinoma and endocervical adenocarcinoma (CESC), cholangiocarcinoma (CHOL), colon adenocarcinoma (COAD), esophageal carcinoma (ECSA), head and neck squamous cell carcinoma (HNSC), kidney chromophobe (KICH), kidney renal clear cell carcinoma (KIRC), liver hepatocellular carcinoma (LIHC), lung adenocarcinoma (LUAD), lung squamous cell carcinoma (LUSC), pheochromocytoma and paraganglioma (PCPG), prostate adenocarcinoma (PRAD), stomach adenocarcinoma (STAD), thyroid carcinoma (THCA), and uterine corpus endometrial carcinoma (UCEC) (Fig. 1a). We further examined whether UBC12 expression was associated with patients’ prognosis. Analyzing the Kaplan‒Meier Plotter database, we found that patients with higher UBC12 expression had lower overall survival rates in five cancer types including STAD, LUAD, KIRC, LIHC, and ESCA (Fig. 1b). Instead, the expression of UBC12 was downregulated in glioblastoma and irrelevant to patient prognosis. These results indicate that, in a subset of human cancers, UBC12 overexpression is closely associated with poor prognosis. Considering the expression level and clinical features, we focused on gastric cancers and lung cancers, which showed significant UBC12 overexpression and worse prognoses, to further explore the fundamental role of UBC12 in cancers.
a UBC12 expression level in human cancers and adjacent normal tissues was determined by TIMER based on TCGA database (*P < 0.05; **P < 0.01; ***P < 0.001. Red, higher in cancer; blue, lower in cancer; black, no significant between cancers and normal tissues). b The relationships between cancer patients’ survival rate and UBC12 expression were analyzed by the Kaplan-Meier plotter database.
Depletion of UBC12 can inhibit malignant phenotypes of cancer cells
After demonstrating that the abnormal activation of UBC12 was closely related to poor prognoses of gastric cancer and lung cancer, we further evaluated the effect of UBC12 downregulation on the malignant phenotypes of these two cancer cell lines. We knocked down UBC12 and evaluated the proliferation abilities of the gastric cancer cell line HGC27 and the lung cancer cell line PC9 (Fig. 2a). The results showed that the downregulation of UBC12 significantly inhibited the proliferation of these two cell lines (Fig. 2b, P < 0.05). Similarly, UBC12 knockdown markedly decreased the clone formation number of HGC27 and PC9 cells (Fig. 2c), providing further evidence of the UBC12 knockdown effect. The relative clone formation rates of shUBC12 #1 in HGC27 cells and PC9 cells were 40% and 9%, respectively. More significant inhibition was observed in the shUBC12 #2 group; and the relative clone formation rates in HGC27 cells and PC9 cells were 27% and 0.4%, respectively. Furthermore, inhibiting UBC12 also induced the apoptosis of HGC27 cells (Fig. 2d). In conclusion, UBC12 knockdown significantly inhibited the malignant phenotypes of gastric cancer cells and lung cancer cells.
a HGC27 and PC9 cells were transfected with lentivirus-shUBC12 or shCtrl (control shRNA) for 72 h and the effect of UBC12 knockdown was examined. b Cell proliferation of HGC27 and PC9 cells transfected with lentivirus-shUBC12 was determined by SRB assay. c Clone formation ability of HGC27 and PC9 cells transduced with lentivirus-shUBC12 was determined by SRB assay and the clone numbers were counted using Image J. d Apoptosis rate of HGC27 and PC9 cells transduced with lentivirus-shUBC12 was determined by flow cytometry. The effect of UBC12 knockdown was examined by Western blotting. *P < 0.05, **P < 0.01, ***P < 0.001.
Arctigenin can effectively restrain the E2 enzyme activity of UBC12
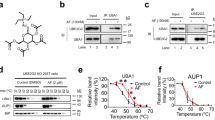
To identify potential compounds capable of targeting UBC12 enzyme activity, we screened a natural compound library consisting of 32 compounds (Supplementary Table 1). As the specific NEDD8-conjugated E2 enzyme, the vital function of UBC12 is to transfer NEDD8 to target proteins, which catalyzes the NEDD8 cascade [7]. Therefore, we examined the protein expression of NEDD8-conjugated UBC12 to evaluate the inhibitory effects of the compound candidates on the E2 enzyme activity of UBC12. MLN4924 was used as a positive control. We found that compound #24 (arctigenin) reduced the level of NEDD8-conjugated UBC12 to the greatest extent (Fig. 3a, b). Arctigenin also inhibited the E2 enzyme activity of UBC12 in a dose-dependent manner (Fig. 3c). Given that UBC12 is known to catalyze the second step of the neddylation pathway, inhibiting UBC12 may decrease the modification levels of cullins. We evaluated the neddylation levels of cullins in HEK-293T cells treated with arctigenin. Arctigenin significantly decreased the neddylation levels of cullin 1, cullin 3, and cullin 4, and modestly decreased the neddylation of cullin 2 (Fig. 3d). Taken together, these results provide new evidence that arctigenin can significantly inhibit UBC12 enzyme activity, leading decreased cullin neddylation.
a UBC12-overexpressing HEK-293T cells were treated with MLN4924 or 32 natural compounds for 24 h. The level of NEDD8-conjugated UBC12 was used to evaluate the enzyme activity of UBC12. b Quantification of inhibitory effects of the 32 natural compounds on E2 enzyme activity of UBC12. c HEK-293T cells were treated with different doses of arctigenin for 24 h and NEDD8-conjugated UBC12 was detected. d UBC12-overexpressing HEK-293T cells were treated with 20 μM arctigenin for 24 h. Protein levels of neddylated cullins were examined.
The inhibition of UBC12 enzyme activity contributes to the abrogation of tumor progression by arctigenin
Considering the inhibitory effect of arctigenin on UBC12 E2 enzyme activity, we investigated the correlation between UBC12 enzyme activity and the antitumor effect of arctigenin. We first evaluated the ability of arctigenin to suppress the proliferation of cancer cells. According to our database analysis, high expression of UBC12 was correlated with poor prognosis in STAD and LUAD. Consequently, we chose the lung cancer cell lines PC9 and H1299 and the gastric cancer cell lines HGC27 and MGC803 to assess the growth inhibitory effect of arctigenin. We also explored this effect on two other cancer types, the 5637 bladder cancer cells and U2OS osteosarcoma cells, in which UBC12 levels were not higher than those determined in the corresponding adjacent normal tissues. Arctigenin treatment dose-dependently inhibited the proliferation of PC9, H1299, and HGC27 cells (with IC50 values of 9.7 μM, 17.2 μM, and 9.5 μM, respectively). In contrast, at similar concentrations ranging from 10 to 20 μM, no cytotoxic effect was observed in MGC803, 5637, and U2OS cells (with IC50 values of 100.3 μM, 140.1 μM, and 117.2 μM, respectively) (Fig. 4a). Considering that UBC12 mainly functions as the E2 enzyme of neddylation and that UBC12 expression is not always associated with its enzyme activity, we evaluated the protein level of NEDD8-conjugated UBC12 in the above cancer cells to clarify whether the sensitivity of cancer cells to arctigenin is associated with the E2 enzyme activity of UBC12. The results showed that the protein levels of NEDD8-conjugated UBC12 in PC9, H1299, and HGC27 cells were significantly higher than those in MGC803, 5637, and U2OS cells (Fig. 4b). Analysis of Pearson’s coefficient correlation confirmed a significant negative correlation between the protein levels of NEDD8-conjugated UBC12 and the IC50 values of arctigenin (r = −0.960, P < 0.05) (Fig. 4c).
a PC9, H1299, HGC27, MGC803, 5637, and U2OS cell lines were treated with arctigenin for 72 h. The cell proliferation was measured by SRB assay (*P < 0.05; **P < 0.01; ***P < 0.001). b Protein levels of NEDD8-conjugated UBC12 expression in different cancer cells were evaluated by Western blotting. c The correlation of ratio of NEDD8-conjugated UBC12 to overall UBC12 with the IC50 values of arctigenin was analyzed using Pearson Correlation Coefficient. HGC27 (d) and PC9 (e) cells were treated with arctigenin for 24 h and protein levels of NEDD8-conjugated UBC12 were determined. HGC27 (f) and PC9 (g) cells were seeded in 6 wells plates with different concentrations of arctigenin treatment for 10 days. *P < 0.05, **P < 0.01, ***P < 0.001.
We further investigated whether arctigenin can inhibit the E2 enzyme activity of UBC12 in gastric and lung cancer cells. Our results showed that arctigenin significantly reduced the protein levels of NEDD8-conjugated UBC12 in a dose-dependent manner in HGC27 and PC9 cells (Fig. 4d, e). The clone formation inhibitory effect of arctigenin was also confirmed (Fig. 4f, g). Collectively, these results demonstrate that the inhibition of UBC12 enzyme activity contributes to the function of arctigenin suppressing the malignant phenotypes of gastric and lung cancer cells.
Arctigenin can increase the PDCD4 protein level by blocking cullin neddylation
Next, we explored the mechanism by which arctigenin inhibits cancer cells. As expected, arctigenin treatment decreased the neddylation levels of cullin 1 and cullin 3 in a time- and concentration-dependent manner in gastric cells (Fig. 5a, b). Inhibition of cullin neddylation results in the accumulation of downstream substrates. Label- free quantitative proteomics was performed to explore the potential mechanism by which arctigenin induces cell death. A total of 4898 quantifiable proteins were identified. The expression levels of a total of 261 and 396 proteins changed significantly in HGC27 cells treated with arctigenin for 36 h and 72 h, respectively. Among these changed proteins, 37 proteins were upregulated and 142 proteins were downregulated in both the arctigenin 36 h group and the arctigenin 72 h group (Fig. 5c). The differentially expressed proteins of the arctigenin 72 h group were classified according to data from the gene ontology database. Arctigenin mainly affected the proteins associated with DNA damage, cell cycle, the p53 class mediator, and nuclear division, which could explain the ability of arctigenin to induce cancer cell death (Fig. 5d). Next, we examined whether any tumor suppressors accumulated among the changed neddylation substrates. Among the upregulated proteins in both the arctigenin 36 h group and 72 h group, we found Programmed Cell Death 4 (PDCD4), a tumor suppressor protein that has been demonstrated to be a substrate of cullin 1 (Fig. 5e) [23]. We then further confirmed the effect of arctigenin on PDCD4 expression in HGC27 cells. We treated HGC27 cells with arctigenin and found that PDCD4 protein accumulated in a concentration- (20 μM and 40 μM) and time-dependent manner (36 h and 72 h) (Fig. 5f, g). In addition, UBC12 overexpression restored the increase in PDCD4 protein levels in HGC27 cells treated with arctigenin (Fig. 5h). Taken together, these findings indicate that arctigenin can inhibit cullin neddylation in gastric cancer cells, resulting in the accumulation of the tumor suppressive protein PDCD4 and changes in key signaling pathways that induce cell death. The inhibition of UBC12 and cullin neddylation contributes to the regulation of protein stability by arctigenin.
Western blotting was performed to detect neddylation level of cullin 1 and cullin 3 in HGC27 cells treated with 40 μM arctigenin for 36 h and 72 h (a) and HGC27 cells treated with 20 and 40 μM arctigenin for 72 h (b). c HGC27 cells were treated with arctigenin for 36 h and 72 h, respectively. Quantitative proteomics was used to identify the proteins regulated by arctigenin. d The Gene ontology (GO) classification was used to analyze the biological functions of the 133 upregulated proteins and 263 downregulated proteins in arctigenin group (72 h). e Heatmap of 37 upregulated proteins overlapped in arctigenin 36 h group and 72 h group. Western blotting analysis was performed to detect the level of PDCD4 in HGC27 cells treated with 40 μM arctigenin for 36 h and 72 h (f) and HGC27 cells treated with 20 and 40 μM arctigenin for 72 h (g). h UBC12-overexpressing HGC27 cells were treated with 20 μM arctigenin for 72 h. Western blotting was performed to detect the level of PDCD4 and UBC12. i A graphical diagram illustrated the entire work.
Discussion
The neddylation pathway is overactivated in various types of tumors [1, 3, 4, 8, 24]. Because UBC12 is the specific E2 conjugating enzyme of the neddylation pathway, inhibition of UBC12 is a promising strategy targeting the overactivated neddylation for cancer treatment. We comprehensively analyzed the expression patterns and clinical features of UBC12 in diverse cancers and propose that UBC12 is a prospective therapeutic target. In addition, UBC12 knockdown resulted in remarkable suppression of malignant tumor characteristics. Furthermore, we are the first to have identified a compound named arctigenin that can inhibit the E2 enzyme activity of UBC12. We determined that the underlying mechanism of the antitumor effect of arctigenin is related to the neddylation pathway, like cullin substrate PDCD4. These findings highlight the crucial role of UBC12 in tumor progression and provide sound evidence for developing arctigenin as a compound that intervenes in the E2 enzyme activity of UBC12 and impairs the overactivation of neddylation in cancers.
A previous study reported that UBC12 is overexpressed in lung cancers [8]. In our systematic analysis of TCGA datasets, we found abnormally increased levels of UBC12 in multiple cancers, and this increase was associated with poor prognosis in some of these cancers. We further validated that knocking down UBC12 can significantly inhibit the progression of gastric cancer. These findings indicate that UBC12 plays an important role in cancer development and is an attractive therapeutic target for cancer treatment.
Although MLN4924 has demonstrated considerable anticancer effects in clinical trials, it still faces challenges that include potential high toxicity and resistance due to unexpected mutations. Here, we provide an innovative UBC12 inhibitor, arctigenin, that markedly inhibits UBC12 enzyme activity and blocks cullin neddylation, especially the neddylation of cullin 1, cullin 3, and cullin 4. This selective inhibition appears to be different from the MLN4924-mediated blockage of the entire neddylation pathway, which could possibly indicate lower toxicity [10]. Overall, our study identified arctigenin as a novel E2 enzyme inhibitor that selectively targets cullin neddylation and, therefore, could be a promising strategy for neddylation-overactivated cancers.
Arctigenin has been reported to possess antitumor activity by regulating the C/EBPalpha, PPARalpha, ROS/p38 MAPK, and Wnt/beta-Catenin pathways [25,26,27,28,29,30]. However, its role in the regulation of the neddylation pathway is not clear. In our studies, arctigenin selectively blocked the neddylation pathway and markedly suppressed the malignant phenotypes of cancer cells. Arctigenin also altered several crucial biological processes, including DNA damage, the cell cycle, the p53 class mediator, and nuclear division, which are crucial to cancer cell death. We also found a high accumulation of PDCD4, which is a crucial tumor suppressor and a substrate of cullin 1 and cullin 3, after arctigenin treatment [23, 31]. Previous studies have established the essential role of PDCD4 in suppressing cancer cell growth, invasion, and neoplastic transformation and in inducing apoptosis [32,33,34,35]. Moreover, PDCD4 is involved in the regulation of protein translation by inhibiting the helicase activity of eIF4A [23, 36, 37]. These results strongly indicate how arctigenin induces the suppression of the malignant tumor phenotypes by regulating the neddylation pathway. Of note, with the exception of PDCD4, the accumulation of a subset of other proteins was also observed in the proteome. Further studies are required to determine how these proteins contribute to the anticancer effect of arctigenin.
Overall, our results elucidate that UBC12 is a prospective and attractive antitumor target. We are the first to report that arctigenin exerts a significant antitumor effect by directly inhibiting the E2 enzyme activity of UBC12, suggesting a new strategy targeting the neddylation pathway in tumor treatment.
References
Watson IR, Irwin MS, Ohh M. NEDD8 pathways in cancer, Sine Quibus Non. Cancer Cell. 2011;19:168–76.
Chen P, Hu T, Liang YP, Li P, Chen XY, Zhang JY, et al. Neddylation inhibition activates the extrinsic apoptosis pathway through ATF4-CHOP-DR5 axis in human esophageal cancer cells. Clin Cancer Res. 2016;22:4145–57.
Li LH, Wang MS, Yu GY, Chen P, Li H, Wei DP, et al. Overactivated neddylation pathway as a therapeutic target in lung cancer. J Natl Cancer I. 2014;106:dju083.
Xie P, Yang JP, Cao Y, Peng LX, Zheng LS, Sun R, et al. Promoting tumorigenesis in nasopharyngeal carcinoma, NEDD8 serves as a potential theranostic target. Cell Death Dis. 2017;8:e2834.
Barbier-Torres L, Delgado TC, Garcia-Rodriguez JL, Zubiete-Franco I, Fernandez-Ramos D, Buque X, et al. Stabilization of LKB1 and Akt by neddylation regulates energy metabolism in liver cancer. Oncotarget. 2015;6:2509–23.
Hua W, Li CJ, Yang ZX, Li LH, Jiang YA, Yu GY, et al. Suppression of glioblastoma by targeting the overactivated protein neddylation pathway. Neuro-Oncol. 2015;17:1333–43.
Huang DT, Ayrault O, Hunt HW, Taherbhoy AM, Duda DM, Scott DC, et al. E2-RING expansion of the NEDD8 cascade confers specificity to cullin modification. Mol Cell. 2009;33:483–95.
Li LH, Kang JH, Zhang WJ, Cai LL, Wang SW, Liang YP, et al. Validation of NEDD8-conjugating enzyme UBC12 as a new therapeutic target in lung cancer. Ebiomedicine. 2019;45:81–91.
Wang SW, Xian JR, Li LH, Jiang YY, Liu Y, Cai LL, et al. NEDD8-conjugating enzyme UBC12 as a novel therapeutic target in esophageal squamous cell carcinoma. Signal Transduct Tar. 2020;5:123.
Soucy TA, Smith PG, Milhollen MA, Berger AJ, Gavin JM, Adhikari S, et al. An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature. 2009;458:732–U67.
Swords RT, Coutre S, Maris MB, Zeidner JF, Foran JM, Cruz J, et al. Pevonedistat, a first-in-class NEDD8-activating enzyme inhibitor, combined with azacitidine in patients with AML. Blood. 2018;131:1415–24.
Sarantopoulos J, Shapiro GI, Cohen RB, Clark JW, Kauh JS, Weiss GJ, et al. Phase I study of the investigational NEDD8-activating enzyme inhibitor pevonedistat (TAK-924/MLN4924) in patients with advanced solid tumors. Clin Cancer Res. 2016;22:847–57.
Shah JJ, Jakubowiak AJ, O’Connor OA, Orlowski RZ, Harvey RD, Smith MR, et al. Phase I study of the novel investigational NEDD8-activating enzyme inhibitor pevonedistat (MLN4924) in patients with relapsed/refractory multiple myeloma or lymphoma. Clin Cancer Res. 2016;22:34–43.
Lockhart AC, Bauer TM, Aggarwal C, Lee CB, Harvey RD, Cohen RB, et al. Phase Ib study of pevonedistat, a NEDD8-activating enzyme inhibitor, in combination with docetaxel, carboplatin and paclitaxel, or gemcitabine, in patients with advanced solid tumors. Invest New Drugs. 2019;37:87–97.
Faessel HM, Mould DR, Zhou X, Faller DV, Sedarati F, Venkatakrishnan K. Population pharmacokinetics of pevonedistat alone or in combination with standard of care in patients with solid tumours or haematological malignancies. Br J Clin Pharmacol. 2019;85:2568–79.
Milhollen MA, Thomas MP, Narayanan U, Traore T, Riceberg J, Amidon BS, et al. Treatment-emergent mutations in NAEbeta confer resistance to the NEDD8-activating enzyme inhibitor MLN4924. Cancer Cell. 2012;21:388–401.
Xu GW, Toth JI, da Silva SR, Paiva SL, Lukkarila JL, Hurren R, et al. Mutations in UBA3 confer resistance to the NEDD8-activating enzyme inhibitor MLN4924 in human leukemic cells. PLoS One. 2014;9:e112004.
Zhou HB, Lu JF, Liu L, Bernard D, Yang CY, Fernandez-Salas E, et al. A potent small-molecule inhibitor of the DCN1-UBC12 interaction that selectively blocks cullin 3 neddylation. Nat Commun. 2017;8:1150.
Kim HS, Hammill JT, Scott DC, Chen YZ, Min J, Rector J, et al. Discovery of novel pyrazolo-pyridone DCN1 inhibitors controlling cullin neddylation. J Med Chem. 2019;62:8429–42.
Zhou W, Ma L, Ding L, Guo Q, He Z, Yang J, et al. Potent 5-Cyano-6-phenyl-pyrimidin-based derivatives targeting DCN1-UBE2M interaction. J Med Chem. 2019;62:5382–403.
Li T, Fu J, Zeng Z, Cohen D, Li J, Chen Q, et al. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 2020;48:W509–W14.
Lanczky A, Gyorffy B. Web-based survival analysis tool tailored for medical research (KMplot): development and implementation. J Med Internet Res. 2021;23:e27633.
Dorrello NV, Peschiaroli A, Guardavaccaro D, Colburn NH, Sherman NE, Pagano M. S6K1- and beta TRCP-mediated degradation of PDCD4 promotes protein translation and cell growth. Science. 2006;314:467–71.
Xie P, Zhang M, He S, Lu K, Chen Y, Xing G, et al. The covalent modifier Nedd8 is critical for the activation of Smurf1 ubiquitin ligase in tumorigenesis. Nat Commun. 2014;5:3733.
Lu Z, Chang L, Zhou H, Liu X, Li Y, Mi T, et al. Arctigenin attenuates tumor metastasis through inhibiting epithelial-mesenchymal transition in hepatocellular carcinoma via suppressing GSK3beta-dependent Wnt/beta-catenin signaling pathway in vivo and in vitro. Front Pharmacol. 2019;10:937.
Lee KS, Lee MG, Kwon YS, Nam KS. Arctigenin enhances the cytotoxic effect of doxorubicin in MDA-MB-231 breast cancer cells. Int J Mol Sci. 2020;21:2997.
Sun Y, Tan YJ, Lu ZZ, Li BB, Sun CH, Li T, et al. Arctigenin inhibits liver cancer tumorigenesis by inhibiting gankyrin expression via C/EBPalpha and PPARalpha. Front Pharmacol. 2018;9:268.
Lu Z, Cao S, Zhou H, Hua L, Zhang S, Cao J. Mechanism of arctigenin-induced specific cytotoxicity against human hepatocellular carcinoma cell lines: Hep G2 and SMMC7721. PLoS One. 2015;10:e0125727.
Hsieh CJ, Kuo PL, Hsu YC, Huang YF, Tsai EM, Hsu YL. Arctigenin, a dietary phytoestrogen, induces apoptosis of estrogen receptor-negative breast cancer cells through the ROS/p38 MAPK pathway and epigenetic regulation. Free Radic Biol Med. 2014;67:159–70.
Wang P, Phan T, Gordon D, Chung S, Henning SM, Vadgama JV. Arctigenin in combination with quercetin synergistically enhances the antiproliferative effect in prostate cancer cells. Mol Nutr Food Res. 2015;59:250–61.
Fiume G, Scialdone A, Rizzo F, De Filippo MR, Laudanna C, Albano F, et al. IBTK Differently modulates gene expression and RNA splicing in HeLa and K562 cells. Int J Mol Sci. 2016;17:1848.
Palamarchuk A, Efanov A, Maximov V, Aqeilan RI, Croce CM, Pekarsky Y. Akt phosphorylates and regulates Pdcd4 tumor suppressor protein. Cancer Res. 2005;65:11282–6.
Loh PG, Yang HS, Walsh MA, Wang Q, Wang X, Cheng Z, et al. Structural basis for translational inhibition by the tumour suppressor Pdcd4. EMBO J. 2009;28:274–85.
Yang HS, Matthews CP, Clair T, Wang Q, Baker AR, Li CC, et al. Tumorigenesis suppressor Pdcd4 down-regulates mitogen-activated protein kinase kinase kinase kinase 1 expression to suppress colon carcinoma cell invasion. Mol Cell Biol. 2006;26:1297–306.
Wei NA, Liu SS, Leung TH, Tam KF, Liao XY, Cheung AN, et al. Loss of programmed cell death 4 (Pdcd4) associates with the progression of ovarian cancer. Mol Cancer. 2009;8:70.
Suzuki C, Garces RG, Edmonds KA, Hiller S, Hyberts SG, Marintchev A, et al. PDCD4 inhibits translation initiation by binding to eIF4A using both its MA3 domains. Proc Natl Acad Sci USA. 2008;105:3274–9.
Yang HS, Jansen AP, Komar AA, Zheng X, Merrick WC, Costes S, et al. The transformation suppressor Pdcd4 is a novel eukaryotic translation initiation factor 4A binding protein that inhibits translation. Mol Cell Biol. 2003;23:26–37.
Acknowledgements
This work was supported by the Zhejiang Provincial Natural Science Foundation of China (No. LD21H310001), the Fundamental Research Funds for the Central Universities (No. 2021XZZX037), the Zhejiang Science and Technology Program of Traditional Chinese Medicine (No. 2022ZQ064), Westlake Laboratory (Westlake Laboratory of Life Sciences and Biomedicine) and Leading Talent of “Ten Thousand Plan” - National High-Level Talents Special Support Plan.
Author information
Authors and Affiliations
Contributions
MDY, BY, QJH and YFC conceived the study and analyzed data; YFC, RZL, and WWY wrote the manuscript; YFC and RZL performed natural products screening assay; YFC, RZL, WWY, and YNY performed cell proliferation and clone formation assay, as well as Western blotting procedure; RZL, SFX, and XJS performed cell apoptosis assay; YQZ and JC provided experimental materials and helped organize the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Supplementary information
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Chen, Yf., Liu, Rz., Ying, Ww. et al. Arctigenin impairs UBC12 enzyme activity and cullin neddylation to attenuate cancer cells. Acta Pharmacol Sin 44, 661–669 (2023). https://doi.org/10.1038/s41401-022-00992-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41401-022-00992-6
Keywords
This article is cited by
-
Protein neddylation and its role in health and diseases
Signal Transduction and Targeted Therapy (2024)
-
The cross talk of ubiquitination and chemotherapy tolerance in colorectal cancer
Journal of Cancer Research and Clinical Oncology (2024)
-
Discovery of neddylation E2s inhibitors with therapeutic activity
Oncogenesis (2023)