Abstract
Grincamycins (GCNs) are a class of angucycline glycosides isolated from actinomycete Streptomyces strains that have potent antitumor activities, but their antitumor mechanisms remain unknown. In this study, we tried to identify the cellular target of grincamycin B (GCN B), one of most dominant and active secondary metabolites, using a combined strategy. We showed that GCN B-selective-induced apoptosis of human acute promyelocytic leukemia (APL) cell line NB4 through increase of ER stress and intracellular reactive oxygen species (ROS) accumulation. Using a strategy of combining phenotype, transcriptomics and protein microarray approaches, we identified that isocitrate dehydrogenase 1(IDH1) was the putative target of GCN B, and confirmed that GCNs were a subset of selective inhibitors targeting both wild-type and mutant IDH1 in vitro. It is well-known that IDH1 converts isocitrate to 2-oxoglutarate (2-OG), maintaining intracellular 2-OG homeostasis. IDH1 and its mutant as the target of GCN B were validated in NB4 cells and zebrafish model. Knockdown of IDH1 in NB4 cells caused the similar phenotype as GCN B treatment, and supplementation of N-acetylcysteine partially rescued the apoptosis caused by IDH1 interference in NB4 cells. In zebrafish model, GCN B effectively restored myeloid abnormality caused by overexpression of mutant IDH1(R132C). Taken together, we demonstrate that IDH1 is one of the antitumor targets of GCNs, suggesting wild-type IDH1 may be a potential target for hematological malignancies intervention in the future.
Similar content being viewed by others
Introduction
Marine microorganism-derived natural products have been one of the most important sources of clinical and pharmaceutical compounds for many decades [1, 2]. These compounds possess enormous chemical and biological diversity and are evolutionarily optimized as drug-like molecules; therefore, identification of the biological targets and the mode of action of bioactive natural products would greatly benefit new drug development.
The angucycline group of antibiotics, produced by type-II polyketide synthase (PKS), is the largest group of polycyclic aromatic polyketides; these compounds bear an unsymmetrically assembled benz [a] anthracene tetracyclic skeleton with an angularly condensed tetracyclic ring [3]. In the decades following the first discovery, >120 members of them have been identified, most of which are derived from the secondary metabolites of marine microorganisms [3, 4]. These compounds are rich in chemical scaffolds and have diverse biological activities, ranging from cytotoxins to antiviral, antibacterial (especially against drug-resistant bacteria), and anti-cancer activities [4]. Grincamycin group antibiotics, derived from the fermentation extracts of actinomycete Streptomyces strains, are a class of angucyclines linked with several oligosaccharide side chains [5,6,7]. Previous studies have demonstrated that some members of this class of compounds have promising antitumor activities against a panel of tumor cells [5, 6]. The presence of deoxy sugar moieties increases the structural biodiversity of angucyclines and, without these moieties, these compounds exhibit distinct bioactivity. However, the mechanism by which grincamycins (GCNs) inhibit tumor activity is not well delineated.
Isocitrate dehydrogenase (IDH) enzymes catalyze the oxidative decarboxylation of isocitrate to produce 2-oxoglutarate (2-OG) and concomitantly produce NADPH/NADH from NADP+/NAD+. In mammalian cells, there are three IDH isoenzymes: IDH1 (cytosolic, NADP-dependent), IDH2 (mitochondrial, NADP-dependent), and IDH3 (mitochondrial, NAD-dependent). IDH1 maintains cellular redox homeostasis by yielding NADPH and 2-OG in the cytoplasm and peroxisomes. IDH1 has a physiological function in protecting liver cells from oxidative stress by regulating the intracellular NADP + /NADPH ratio [8]. Interestingly, other studies also indicated that IDH1 was capable of converting NADPH, 2-OG, and CO2 to produce isocitrate in the cytosol, which is a reverse reaction of dehydrogenase and a critical step of reductive glutamine metabolism [9].
Thus, we tried to identify the cellular target of GCN B using a combined strategy. We demonstrated that GCNs are a novel class of inhibitors of IDH1. Through targeting IDH1, GCN B disrupts the cellular redox balance and 2-OG homeostasis, triggers ROS accumulation and eventually causes cell death.
Materials and methods
Reagents and antibodies
GCN B and its analogs were supplied by Dr. Jian-hua Ju from the South China Sea Institution of Oceanology (Guangzhou, China). Doxorubicin, thapsigargin, Taxol, dimethylooxalylglycine (DMOG), glucose-6-phosphate (G6P), glucose-6-phosphate dehydrogenase (G6PD) recombinant protein, nicotinamide adenine dinucleotide phosphate (NADP+), NADPH, DL-isocitrate, 2-oxoglutarate (2-OG), 2ʹ,7ʹ-dichlorodihydrofluorescein diacetate (DCF-DA), menadione, phenazine ethosulfate (PES), 1-phenyl-2-thiourea (PTU), flag peptide, streptavidin-linked agarose beads and anti-Flag M2 agarose beads were purchased from Sigma- Aldrich (Merck, Germany). Anti-IDH1, anti-ATF4, anti-GADD34, anti-phospho-eIF2α S51, anti-phospho-PERK, and anti-p53 antibodies were purchased from ABclonal Inc. (Wuhan, China). Anti-GAPDH antibody was purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA). Anti-caspase-3, anti-caspase-9, and anti-PARP antibodies were purchased from Cell Signaling Technology (Danvers, MA, USA).
Quantitative analysis of intracellular 2-OG and 2-HG
The metabolites were extracted from cells by adding 1 mL chilled mixture of methanol/water (v/v, 4/1) into each well of the plates immediately after removing the culture medium and shaking at 1400 × g for 10 min at 4 °C for 15 min. The extraction was then transferred into a clean 1.5 mL Eppendorf tube for centrifugation at 18,000 × g for 10 min at 4 °C. The supernatant (50 μL) was transferred into a 150 μL insert fixed in a 2 mL vial for analysis. The concentrations of 2-HG and 2-OG were measured using an liquid chromatography–mass spectrometry (LC-MS/MS) method, which was validated to assure acceptable accuracy and precision (accuracy >90% with relative standard deviation (RSD) <10%). Specifically, prepared samples were separated on an Agilent 1260 Infinity liquid chromatography instrument (Agilent, CA, USA) equipped with a 4.6 × 50 mm Poroshell 120 EC-C18 2.7 μm column (Agilent, CA, USA) held at 20 °C. The mobile phase consisted of water including 0.3% formic acid (solvent A) and MeOH/water (v/v; 95/5), including 0.3% formic acid (solvent B). The gradient program started at 95% (v/v) of solvent A for 1 min, then decreased to 5% solvent A linearly over 1 min with a 2 min hold, then went back to 95% of solvent A linearly in 0.5 min, and then equilibrated for 2 min until next analysis. The flow rate of the mobile phase was 0.5 mL/min. The injection volume was 5 μL, and the samples were kept at 10 °C in an autosampler. The analytes were monitored by negative mode electrospray ionization (ESI) tandem mass spectrometry in MRM mode on an AB Sciex QTrap 6500 Mass Spectrometer (AB Sciex, MA, USA). The gas flow rate was fixed for curtain gas, ion source gas 1 and ion source gas 2 as 20, 55, 55 L/h, respectively, and the collision gas was set as a medium. ESI was performed with the ionspray voltage set at –4500 V and the source temperature set at 550 °C. By direct infusion of the single compound solution at a concentration of 1 μM, declustering potential (DP), collision energy (CE), and collision cell exit potential (CXP) for each analyte were optimized to achieve the maximum intensity of the precursor and production ions, respectively. DP/CE/CXP parameters were optimized as –29.0/–12.0/–12.0 and –34.0/–12.0/–10.8 for 2-OG and 2-HG, respectively. The corresponding precursor and dominant daughter ions used to set up the transition monitored in the MRM mode were 145.0/100.9 and 146.8/128.9 for 2-OG and 2-HG, respectively. The dwell time was set as 100 ms for each compound.
Gas chromatography–mass spectrometer (GC-MS)
Each sample was loaded into a screw-cap V-type bottom glass vial and dried with a vacuum or nitrogen evaporator. Next, 50 μL of pyridine containing 20 mg/mL of methoxyamine hydrochloride was added, and the sample was incubated at 37 °C for 16–24 h. Derivatization was performed at 70 °C for 30 min by adding 20–40 μL of MTBSTFA, and then 1 μL of derivatized sample was loaded onto an Agilent 6890-5973 (Agilent, CA, USA) for GC-MS analysis as described previously [10].
Cytotoxicity assay
The cytotoxicity assay was performed as described previously [11]. In brief, cells were seeded at a density of 1000–5000/well in a 96-well plate in a final volume of 190 μL/well. Compounds were then diluted and added into the wells. Seventy-two hours later, cells were incubated with 10 μg/mL Alamar blue solution for 4 h, and fluorescence at Ex560/Em590 was determined using an Envision plate reader (PerkinElmer Inc., USA)
Intracellular ROS detection
Intracellular levels of ROS were determined as previously described [12] with minor modifications. Briefly, 1 × 105 NB4 cells were treated with the indicated concentrations of compounds together with DCF-DA at a final concentration of 10 μM for 2 h at 37 °C. Then, the cells were collected, washed, and subjected to fluorescence-activated cell sorting (FACS) Calibur flow cytometry (BD Biosciences, San Jose, CA, USA). The levels of ROS were analyzed by measuring the mean fluorescence intensity (MFI) of DCF-DA.
Protein microarray
Human proteome microarrays, which consist of more than 17,000 human proteins, were used as described previously [13]. In brief, the glass-based chips conjugated with N-terminal GST fusion proteins were first blocked with 0.5% BSA and then separately soaked with 10, 50, or 100 μM GCN B for 30 min. The chips were then washed with phosphate-buffered saline (PBS) three times, dried, and scanned with a GenePix 4300A Microarray System (Molecular Devices) at Ex450/Em600. Spot intensity was analyzed using GenePix Pro 7 software. The candidate target is determined as follows: X equals the scanning signal of each protein, the cutoff is X ≥ means + 3 × Stdev, and n equals the number of proteins on the proteome chip.
Transcription profiling with RNA-seq
NB4 cells (2 × 106 per well) were seeded into 6-well plates and separately treated with dimethyl sulfoxide (DMSO), 0.25 μM GCN B, or 0.5 μM GCN B for 6 h. Total RNA was extracted by TRIzol. The cDNA library was generated using the TruSeq Stranded mRNA Library Prep Kit (Illumina Inc.) following the manufacturer’s instructions. Samples were then fragmented and tagged with adapters prior to being sequenced (200 bp/cluster) by HiSeq2000. Data were processed for gene set enrichment analysis (GSEA) using GSEA 3.0 (http://www.broad.mit.edu/gsea)
Computational modeling
The crystal structure of IDH1 (PDB code 5DE1) was used to construct the models of compounds binding with IDH1. The protein was prepared, and hydrogen atoms were added using the Protein Preparation Wizard module of the Schrodinger software. Among the several binding sites predicted by the Sitemap module, the binding site that was consistent with the ligand within the crystal structure was selected for subsequent docking. Then, different conformations of the compounds were docked into the binding site using the Glide module with the SP precision mode. The other parameters were set as default. A 20-nanosecond molecular dynamics simulation was used for postdocking optimization with the AMBER 11 software package using the FF03 force field. The SHAKE algorithm was used, and the simulation step was set to 2 femtoseconds. After clustering the structures extracted from the trajectory into five clusters, the representative structure from the most principal cluster was used for subsequent analysis.
Streptavidin pull-down assay
One microgram of purified IDH1 proteins was incubated with streptavidin beads (20 μL/tube) and 10 μM free GCN B or DMSO (control) in 50 μL 1% BSA buffer at 4 °C. One hour later, biotinylated GCN B was added into each tube to a final concentration of 1 μM. After rotating for another 1 h at 4 °C, each sample was washed with PBS three times, and after centrifuging, the beads were boiled and subjected to sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and Coomassie brilliant blue R250 staining.
Thermal shift assay
Purified protein was aliquoted into PCR tubes (3 μg/tube) and incubated with 10 μM GCN B in a final volume of 50 μL at different temperatures for 30 s. Samples were then centrifuged at 12,000 r.p.m. for 10 min at 4 °C, and then 30 μL of the supernatant was transferred to a new tube. The supernatant was then mixed with 6 μL of 6× Laemmli sample buffer and boiled at 100 °C for 10 min. Twenty-five microliters of each sample was subjected to SDS-PAGE. The gel was then stained with Coomassie brilliant blue R250, scanned, and analyzed by ImageJ Software.
Measurement of the enzymatic activity of IDH1
The oxidative activity (conversion of isocitrate to 2-OG) of wild-type IDH1 and reductive activity (conversion of 2-OG to 2-HG) of R132C or R132H IDH1 were measured using a fluorescence intensity assay. Assays were conducted in a 100 μL volume in a 96-well microplate. For the oxidative activity assay, each compound was preincubated with recombinant protein in the presence NADP+ (6 nM IDH1, 250 μM NADP+) in 95 μL of 100 mM Tris-HCl buffer (pH 8.0) at 37 °C for 30 min. The reaction was initiated by adding 5 μL of substrate (500 mM DL-isocitrate, 100 mM MgCl2 in Tris-HCl). For the reductive activity assay, each compound was preincubated with reaction mixtures containing 6 nM recombinant R132C or R132H IDH1, 125 μM NADPH and 100 mM Tris-HCl buffer, pH 8.0, at 37 °C for 30 min. The reaction was initiated by adding 5 μL of substrate (500 mM 2-OG, 100 mM MgCl2 in Tris-HCl). The increase or decrease in NADPH fluorescence was monitored (excitation/emission, 340/450 nm) for 30 min at room temperature by an Envision plate reader (PerkinElmer Inc., USA).
Measurement of the intracellular NADPH/NADP+ ratio
NB4 cells were seeded at 1 × 107 per well into a 6-well microplate, treated with each candidate compound for 6 h at 37 °C, and collected by centrifuging at 1500 r.p.m. for 5 min. Pellets were resuspended in 300 μL of extraction buffer (20 mM NAM, 20 mM NaHCO3, 100 mM Na2CO3) and ultrasonicated for 30 s on ice. The supernatants were then collected after centrifugation at 12,000 r.p.m. for 10 min at 4 °C. For NADPH extraction, 150 μL of the supernatant was incubated at 60 °C for 30 min. Then, 160 μL of NADP cycling buffer (100 mM Tris-HCl, pH 8.0; 0.5 mM thiazolyl blue; 2 mM phenazine ethosulfate; 5 mM EDTA) containing 1.3 U of G6PD was added to a 96-well microplate with 20 μL of cell extracts in each well. After incubation in the dark for 1 min at room temperature, absorbance at 570 nm was measured every 30 s for 10 min at 30 °C using a SpectraMax M5 Microplate Reader (Molecular Devices, LLC., San Jose, CA, USA). The concentration of NADP+ was calculated by subtracting the amount of NADPH (heated sample) from the total amount of NADP+ and NADPH (unheated sample).
Zebrafish maintenance, injections, and pharmacological treatments
Zebrafish maintenance and breeding were performed using standard method [14]. Embryos were injected at the one-cell stage with 50 pg control or R132C IDH1 linearized DNA. For the pharmacological treatments, embryos were manually dechorionated and incubated with 2.5 μM GCN B from 12 hpf (hours post fertilization) to the indicated stage. To prevent melanin pigment formation, embryos were incubated in egg water containing 0.045% 1-phenyl-2-thiourea (PTU) after the gastrulation stage.
Zebrafish whole-mount in situ hybridization (WISH)
The pu.1, mpeg1, and mpo probes were transcribed in vitro with T3 or T7 polymerase (Ambion) using the Digoxigenin RNA Labeling Kit (Roche). WISH was performed as previously described using NBT and BCIP (Sigma) as substrates [15, 16]. Images were obtained using an Olympus SZX16 microscope equipped with an Olympus DP80 CCD.
Statistical analysis
All statistical processing was performed using GraphPad Prism 5 software. All experiments were repeated at least two times and performed in triplicate unless stated otherwise. The data are presented as the means ± SEM. Significance was evaluated using Student’s two-tailed t-test for two dataset comparisons. In all cases of more than two experimental conditions, statistical analysis was performed using one-way or two-way ANOVA. Symbols for significant levels were *P < 0.05, **P < 0.01, ***P < 0.001.
Results
GCN family antibiotics selectively kill hematological tumor cells
First, we measured the antitumor activity of the natural product GCN and its six analogs (Fig. 1), all isolated from the deep-sea-derived actinomycete Streptomyces lusitanus SCSIO LR32, in the human acute promyelocytic leukemia (APL) cell line NB4. As shown in Table 1, GCN, GCN-1b, and P-1894B, all of which have a classic angular condensed ring, are relatively more active than GCN B and GCN-2b, which are the tricyclic vinemycinone-B2 type analogs of GCN and GCN-1b, respectively. Interestingly, GCN-2a, which bears two reductive hydroxyl groups instead of the carbonyl groups in GCN B, completely lacks cytotoxicity (IC50 > 10 μM). This SARs (structure–activity relationships) result indicates that the carbonyl groups of the terminal hexasaccharide side chains are required for its antitumor activity.
Since the angular ring easily rearranges to a tricyclic compound under acidic conditions [5, 6], GCN B and GCN-2b are the main products during fermentation. Thus, we used GCN B as a representative compound for further study. Next, we determined the cytotoxicity of GCN B on a panel of tumor cell lines covering both solid and hematological malignancies. All four myeloid-derived cell lines (NB4, BC-3, JSC, DB) were sensitive to GCN B in the nanomolar range, while the other solid tumor cells (BxPC-3, HepG2, HeLa and PC-3) were 5–10 times less sensitive to GCN B with IC50 values in the micromolar range (Table 2). This differential sensitivity against hematological and solid tumor cell lines prompted us to further investigate the biological mechanisms of GCN B cytotoxicity.
GCN B induces endoplasmic reticulum (ER) stress, ROS accumulation, and apoptotic cell death
Inspired by the skeleton similarity between angucycline and the widely used chemotherapeutic anthracycline antibiotics, we first investigated whether GCN B acted through simple DNA intercalation. By determining the binding ability of GCN B to a random DNA chain, we found that GCN B barely bound to DNA compared with doxorubicin (Supplementary Fig. S1a), the most well-known anthracycline. This result indicates that GCN B exerts cytotoxic activity against tumor cells with distinct mechanisms.
Changes in cell viability can be physiologically related to cell cycle arrest or cell death. Therefore, we examined cell cycle progression by performing propidium iodide (PI) staining and flow cytometry. Our data demonstrated that the distribution of cells across the various stages of the cell cycle was similar between GCN B-treated and control (DMSO-treated) cells. Taxol, which significantly blocks the cell cycle at the G2/M phase (88.89%), was used as a positive control (Supplementary Fig. S1c). This result ruled out the possibility that the antitumor activity of GCN B was due to cell cycle arrest.
A number of studies have indicated that natural products cause cell death through oxidative stress; thus, we next evaluated the ER stress response upon GCN B treatment. Fig. 2a shows that GCN B significantly increased the messenger RNA (mRNA) levels of CHOP and GADD34 (ER stress molecular markers) in a dose-dependent manner but did not affect the mRNA level of another stress sensor activating transcription factor 6 (ATF6). Consistent with these results, the p-PERK, p-eIF2α, ATF4, and GADD34 proteins were also induced in a time-dependent manner upon GCN B exposure (Fig. 2b). Moreover, an XBP-1 splicing experiment further confirmed that NB4 cells underwent a typical stress response upon GCN B stimulation (Fig. 2c). Since ROS are produced downstream of and as a consequence of the unfolded protein response (UPR), the levels of ROS production, as analyzed by DCF-DA fluorescence, were further determined. Following exposure to different concentrations of GCN B for 3 h, the intensity of DCF-DA fluorescence increased in a dose-dependent manner, suggesting that ROS accumulated upon GCN B exposure (Fig. 2d).
a mRNA levels of genes related to ER stress (ATF6, ATF4, CHOP, GADD34) in NB4 cells treated with the indicated compounds for 12 h. Values are the mean ± SD (n = 3). *P < 0.05, **P < 0.01, ***P < 0.001. b Protein levels of the indicated ER stress markers in NB4 cells treated with 1.25 μM GCN B for the indicated time. c XBP-1 splicing was determined by RT-PCR. uXbp-1, unspliced forms of Xbp-1 mRNA; sXbp-1, spliced forms of Xbp-1. d Flow cytometry analysis of NB4 cells stained with H2-DCFDA incubated with indicated concentration of GCN B, 0.1% DMSO (as negative control) or 0.5 μM TBHP (as positive control) for 2 h. e Apoptosis of NB4 cells treated with GCN B by FACS. Cells were treated with DMSO, 0.3 µM GCN B, 0.6 µM GCN B, or 0.5 µM TG (as positive control) for 24 h and double stained with PI and Annexin-V-FITC before being loaded onto FACS flow cytometer for apoptosis determination. f Western blot of apoptosis markers in NB4 cells treated with GCN B. Cells were treated with DMSO, 0.3 µM GCN B, or 0.6 µM GCN B for 24 h. Lysates from these cells were analyzed by Western blot for PARP, caspase-9, and caspase-3.
Since excessive ROS accumulation induces apoptosis, we next evaluated the apoptosis of NB4 cells by performing double staining with PI and Annexin-V-FITC. Similar to thapsigargin (TG), GCN B induced the apoptosis of NB4 cells in a dose-dependent manner following 24 h of treatment (Fig. 2e). Interestingly, there was a false-positive shift of the PI channel in the GCN B-treated samples since GCN B is a fluorescent reagent with excitation at 450 nm and emission at 600 nm (Supplementary Fig. S2a). Apoptosis was also confirmed using Western blot detection of the activation of caspase-3, caspase-9, and PARP (Fig. 2f).
Some compounds, such as thapsigargin (TG), cause unfolded protein responses and cell death by blocking the sarco/endoplasmic reticulum (ER) Ca2+-ATPase (SERCA) and disrupting calcium homeostasis. To clarify whether calcium flux is involved in the GCN B-induced ER stress response, we examined the changes in intracellular calcium levels by using the ratiometric Ca2+ indicator Fura 8-AM. As shown in Supplementary Fig. S1b, GCN B fails to cause calcium release from ER stores or subsequent calcium release-activated calcium (CRAC) entry, which is distinct from the effect observed in the TG-treated cells.
As the NB4 cell line is an APL cell line that can be induced to differentiate by drugs, such as ATRA or arsenite, we also checked whether GCN B induces the differentiation of NB4 cells. FACS analysis demonstrated that neither CD11b nor CD11c were significantly upregulated upon GCN B treatment (Supplementary Fig. S1d), suggesting that GCN B has a minimal effect on the differentiation of NB4 cells.
Cellular 2-OG metabolism is disrupted in GCN B-treated NB4 cells
Next, to gain insight into the cellular pathways that are regulated by GCN B, we compared the gene expression profiles of NB4 cells treated with different concentrations of GCN B or DMSO by performing RNA sequencing and GSEA. The GSEA data showed that hypoxia, p53, and mammalian target of rapamycin (mTOR) signaling pathways were the most highly upregulated pathways upon 6 h of GCN B exposure (Fig. 3a, b), which is in line with previous observations of ROS accumulation and alteration in p53 expression (Fig. 2b). In addition, the most significantly perturbed gene sets were closely related to histone methylation status, including H3K27me3, H3K4me3, and EZH2 targets (Fig. 3c, d). Interestingly, both histone trimethylation and hypoxia are directly associated with 2-oxoglutarate-dependent dioxygenase (2-OGDO) enzymes, which includes HIFs, prolyl hydroxylases, asparagine hydroxylase, collagen hydroxylases, Jumonji C domain-containing histone lysine demethylases (KDM2-7) and so on [17]. In addition, the GSEA profile of GCN B treatment is highly similar to that of TOSEDOSTAT, a potent aminopeptidase inhibitor with antiproliferation activity through amino acid deprivation and stress response [18], while 2-OG is an essential metabolite linking the TCA cycle and amino acid metabolism. Putting all the clues together, we suspected that cellular 2-OG homeostasis was disrupted by GCN B treatment. To prove this hypothesis, intracellular 2-OG in NB4 cells upon GCN B treatment was quantified by LC-MS/MS. As shown in Fig. 3e, the 2-OG levels were significantly decreased after 6 h of treatment with GCN B, achieving a 66% reduction at the highest concentration (1.25 μM), while no significant changes were observed in the AGI-5198 (a specific IDH1 mutant inhibitor) control group. DMOG is a cell-permeant 2-OG analog that inhibits 2-OGDO enzymes [19, 20]. Thus, we tested whether DMOG aggravated the perturbation of cellular 2-OG caused by GCN B. The results showed that NB4 cells were more susceptible to the combination of DMOG with GCN B than GCN B alone, with a shift of IC50 from 55.59 to 16.88 nM (Fig. 3f). These results indicate that the mechanism of action of GCN B is highly related to cellular 2-OG homeostasis.
a, c GSEA was performed to identify pathways that were perturbed by GCN B treatment. The top five most down- and upregulated gene sets from the Hallmark gene sets (h.all.v5.1.symbols) or Chemical and Genetic Perturbation sets (c2.cgp.v5.2.symbols) are shown. b, d Enrichment plots for the MTORC1 signaling gene set and the MIKKELSEN_ES_LCP_WITH_H3K4ME3 gene set. Genes annotated as mTOR signaling pathway components were upregulated while genes associated with histone methylation were downregulated after GCN B treatment. e GCN B decreased the intracellular 2-OG levels of NB4 cells. Each treatment condition was performed in sextuplicate (n = 6), and the experiment was repeated twice. f Cytotoxicity of GCN B against NB4 cells in the presence or absence of DMOG.
GCN B binds to IDH1 in vitro
It is well-known that IDHs, which convert isocitrate to 2-OG, are critical regulators of intracellular 2-OG homeostasis. Therefore, we further explored whether IDHs are potential molecular targets of GCN B. First, a protein microarray [21] was applied to identify potential GCN B binding proteins; a library of 16,368 unique full-length human ORFs [22] was screened in a single binding assay by utilization of the spontaneous fluorescence (excitation at 450 nm and emission at 600 nm) of GCN B. Interestingly, IDH1 was one of the 24 positive hits (Supplementary Fig. S2c). Next, this finding was validated by performing in vitro binding assays. As expected, after incubation with GCN B for 1 h, binding of GCN B to the IDH1 recombinant protein was detected, and the amount of fluorescence signal increased in a dose-dependent manner (Fig. 4a, lanes 1–3). In contrast, almost no fluorescence signal was observed for the negative control, G6PD recombinant protein. (Fig. 4a, lanes 5–6). Interestingly, GCN-2a, an analog of GCN B that has a similar fluorescence spectrum (Supplementary Fig. S2b), barely bound to the IDH1 recombinant protein (Fig. 4a, lane 4), which indicates the specificity of GCN B binding. Next, we synthesized a biotin-labeled GCN B probe, confirmed its antitumor activities (Supplementary Fig. S2d), and carried out streptavidin affinity purification and a competition assay. Recombinant IDH1 was pulled down by 1 μM biotinylated probe, and binding could be competed with 10 μM free GCN B (Fig. 4b). Similar pull-down results were acquired by using a HeLa whole cell lysate instead of recombinant protein (Supplementary Fig. S2f). In addition, a thermal denaturation-based method, the thermal shift assay (TSA), was used to validate the GCN B binding capacity to IDH1. When purified IDH1 was incubated with GCN B, a clear shift in melting temperature (Tm) from 48.90 °C to 51.96 °C was observed in the thermal denaturation curve (Fig. 4d), and GCN B also increased the thermal stability of IDH1 in a dose-dependent manner (Fig. 4e).
a Native PAGE of IDH1 incubated with different concentrations of GCN B or GCN-2a. The left picture was scanned with Typhoon to visualize fluorescence signal corresponding to GCN B and GCN-2a, the right picture shows the same gel after silver staining. G6PD was loaded as a negative control. b Pull-down of wild-type IDH1 by the biotinylated probe GCN B-Biotin. Samples treated with GCN B-Biotin in the presence or absence of free GCN B were pulled down with streptavidin beads, separated by SDS-PAGE and stained with silver. The final concentration of DMSO was 1%, Biotin-GCN B was 1 μM and free GCN B was 10 μM. c Docking of GCN B to IDH1 (dimer). d Thermal shift assay of GCN B-treated IDH1 and artificial mutants. Samples incubated with GCN B or DMSO were loaded into 10% PAGE gels and scanned after Coomassie staining. The relative gray intensity values were plotted, and Tm was calculated by Boltzmann sigmoidal fitting. e Thermal stability of wild-type form of IDH1 and N271L, L120A, and S280C mutants incubated with indicated concentration of GCN B at 52 °C.
To gain more precise binding information, we used molecular docking to predict the potential GCN B binding site (s). The virtual simulation results suggested that GCN B binds at the dimer surface of IDH1, where the compound is predicted to form hydrogen bonds with N271 and S280 of one monomer and with L120 of the other (Fig. 4c). To test this hypothesis, three artificial mutant IDH1s, IDHL120A, IDHN271L and IDHS280C, were constructed, and the capacity of each artificial mutant to bind to GCN B was evaluated by thermal shift assay. We found that the IDHS280C mutation eliminated the shift caused by GCN B binding and failed to enhance the thermal stability as wild-type does, which suggests that the S280 residue is critical for GCN B-IDH1 binding (Fig. 4d, e). Accordingly, the S280C mutant exhibits dramatically decreased enzymatic activities, as well as sensitivities to GCN B inhibition (Supplementary Fig. S2g).
Since IDH1 functions as a dimer, we next assessed whether GCN B was able to affect IDH1 dimer formation. Using blue native PAGE (BN-PAGE), we observed that GCN B did not disrupt IDH1 dimer formation even at a concentration of 100 μM (Supplementary Fig. S2e).
GCN B allosterically inhibits the enzymatic activity of IDH1
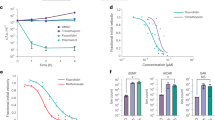
Next, we determined whether GCN B inhibits the activity of IDH1. Flag-tagged IDH1 or IDH2 was overexpressed in HEK293T cells and enriched by affinity purification, and the inhibitory activity of GCN B against each enzyme was determined. Fig. 5a shows that GCN B inhibited IDH1 activity in a dose-dependent manner, with an IC50 of approximately 1 μM in vitro. To our surprise, IDH2, which shares 60% homology with IDH1, was barely inhibited by GCN B, even at a concentration of 5 μM (Fig. 5a). G6PD, a cytosolic NADPH-producing dehydrogenase, was used as a control. This result confirmed that GCN B is a selective inhibitor targeting IDH1. Next, we determined the model of binding of the inhibition by performing kinetics analysis. As shown in Fig. 5b, with the increase in GCN B concentration, the apparent Vm(app) of the isocitrate substrate for IDH1 decreased from ~2.28 μM/min to 0.31 μM/min in a dose-dependent manner, while the apparent Kmax(app) was slightly affected, which indicates that GCN B behaves as a noncompetitive inhibitor for the binding of isocitrate. Additionally, similar noncompetitive inhibition was observed in the binding of NADPH (Fig. 5b).
a The relative enzyme activities of IDH1, IDH2 and G6PD in response to GCN B treatment. Different concentration of GCN B was pretreated with IDH1, IDH2, or G6PD for 30 min on ice, and the enzyme reaction was performed as described in “Materials and Methods”. ***P < 0.001 compared to DMSO. b Lineweaver–Burk plot for Km and Vmax values of IDH1 in the presence of different concentrations of GCN B. Data were obtained as mean values of 1/V (the inverse of the increase in NADPH concentration per min) from three independent tests with different concentrations of isocitrate(left) or NADP+ (right). c Dose response enzyme inhibition curve showing the activity of GCN B against wild-type IDH1 and the R132C and R132H mutants. d Relative amounts of 2-HG produced by IDH R132C in an enzymatic activity assay in the presence of different concentrations of GCN B. GC-MS was used to detect 2-HG amounts. **P < 0.01, ***P < 0.001 compared to DMSO.
Considering that somatic mutations of IDH1 play important roles in glioma and AML tumorigenesis [23], we next evaluated the inhibitory activity of GCN B towards the IDH1 R132C and R132H mutants. Fig. 5c shows that GCN B inhibited wild-type IDH1 (wtIDH1), R132C mutant and R132H mutant forms of IDH1 with IC50 values of 1.67, 0.78, and 3.44 μM, respectively. This inhibitory effect was also confirmed by GC-MS detection of 2-HG content in an in vitro enzymatic assay using recombinant R132C mutant protein purified from E. coli (Fig. 5d). In addition, we measured the inhibitory activity of other GCN class analogs. Table 3 shows the enzyme activity IC50 values for the enzyme activity of each compound against wild-type IDH1 and the R132C mutant form of IDH1. As expected, these compounds also exhibit inhibitory activity against both wild-type and mutant IDH1 with IC50 values in the micromolar range except for GCN-2a, which barely binds to IDH1, as shown in Fig. 4a before.
IDH1 is the biological target of GCN B in cells
Next, we aimed to clarify whether IDH1 is the cellular target of GCN B-induced ROS production and apoptosis in AML cells. First, we examined the correlation between the IC50 of GCN B and the expression level of cellular IDH1. We found that all four hematological cell lines, NB4, BC-3, JSC, and DB, exhibited lower expression levels of IDH1 (Fig. 6a), and the decreased expression of IDH1 in hematological cells was correlated with a decreased IC50 of GCN B (r = 0.7520, P = 0.0314) (Fig. 6b). Since IDH1 is the major producer of cytosolic NADPH, we first evaluated the redox balance upon GCN B exposure. Consistent with our hypothesis, GCN B-treated cells had a significantly lower intracellular NADPH/NADP+ ratio than control cells (Fig. 6c). Next, we added the antioxidant N-acetylcysteine (NAC) to restore the GSH redox balance. When NB4 cells were pretreated with 2.5 mM NAC, they were less sensitive to GCN B treatment, with a shift in IC50 from 150 to 664 nM (Fig. 6d).
a Western blot analysis of the expression of IDH1 in eight cancer cell lines. b The correlation analysis of IDH1 expression and IC50 of the eight cancer cells to GCN B. r = 0.7520, P = 0.0314. c Intracellular NADPH/NADP ratio of NB4 cells with treatment of GCN B. NB4 cells were treated with DMSO, different concentrations of GCN B, or Menadione (as positive control) for 2 h. Samples were loaded into a 96-well plate and mixed with G6PD buffer containing PES and MTT. The absorbance at 560 nm was measured every 30 s for 10 min after the reaction initialized by adding G6P. *P < 0.05, **P < 0.01. d Effect of GCN B on cellular viability of NB4 cells when pretreated with NAC. Cells were pretreated with 2.5 mM NAC for 2 h and then IC50 of GCN B against NB4 cells was determined as described in “Materials and Methods”. e, f Apoptosis of IDH1 knockdown cell lines. e Cells were harvested 36 h after infection and stained with Annexin-V-PE for detection by flow cytometry. f Growth curve of IDH1 knockdown cells. Cell growth was measured every day for a total of 4 days following cells seeding. Fluorescence values were normalized to the values at the day of seeding. ***P < 0.001. g, h Determination of apoptosis in IDH1 knockdown NB4 cells treated with NAC. NB4 cells were treated with 2.5 mM NAC for 24 h after shRNA infection, and then stained with Annexin-V-PE for detection by flow cytometry (g) or subjected to Western blot (h). n = 3 biological replicates. *P < 0.05, **P < 0.01, ***P < 0.001.
To examine the role of IDH1 in NB4 cells, we depleted IDH1 by expressing specific short-hairpin RNAs (shRNAs). We observed that IDH1 interference caused a significant block in cell growth (Fig. 6f) and an induction of apoptosis, as demonstrated by annexin-V staining and cleavage of PARP, caspase-3, and caspase-9 (Fig. 6h and Supplementary Fig. S3a). To further validate the specificity of cell death, we also utilized knockdown-resistant cells. As we expected, the apoptosis caused by IDH1 interference was partially reversed in NB4 cells resistant to IDH1 interference (Supplementary Fig. S4a). Next, to explore the mechanism of apoptosis, we further evaluated ROS levels and ER stress in IDH1 knockdown cells. We found that 24 h after shRNA infection, IDH1 interference caused an increase in ROS levels and oxidative stress (Supplementary Fig. S3b, c). When we supplemented IDH1 KD cells with NAC, we found that apoptosis caused by IDH1 interference was partially reversed by the addition of NAC (Fig. 6g, h). Based on these results, we have shown that GCN B induces the apoptosis of NB4 cells, at least partially, by targeting IDH1.
GCN B inhibits IDH1 activity in a zebrafish model
Encouraged by our finding that GCN B inhibits IDH1 in vitro, we tested whether IDH1 was a biological target in vivo by monitoring the effect of GCN B in a zebrafish model. Since zebrafish idh1 mutants show defects in primitive hematopoiesis [24] and play important roles in leukemia initiation, we evaluated whether GCN B was able to ameliorate the leukemogenicity potential of human IDH1 mutation. Pu.1 is an essential transcriptional factor that governs embryonic myelopoiesis, and mpeg1 and mpo are two specific marker genes indicating the number and maturation of myeloid cells. In the WISH analysis of zebrafish embryos by pu.1, mpeg1, and mpo probes, myeloid cells that developed during primitive myelopoiesis mainly resided on the yolk surface and then migrated into circulation or to various tissues. The statistical analysis of the number of myeloid cells provided evidence that ectopic expression of R132C IDH1 caused expansion of myelopoiesis, which could be normalized to the level of myelopoiesis in embryos by GCN B treatment (Fig. 7). This result suggests that GCN B inhibits the activity of mutant IDH1 during abnormal myeloid differentiation in vivo.
GCN B reversed the phenotype caused by overexpression of mutant IDH1(R132C) in zebrafish. Wild-type zebrafish (Ctrl) were injected with IDH1 R132C plasmid and treated with 3 μM GCN B. The expression levels of pu.1 (a), mpeg1 (b), and mpo (c) mRNA were determined at 32 hpf. Left: representative images of each staining. Right: Statistical analysis of the staining, n = 10 embryos each group, *P < 0.05, **P < 0.01, ***P < 0.001.
Discussion
Identification of cellular targets and the mode of action of natural products remains one of the biggest challenges in chemistry and biology today. In this study, by using a strategy combining phenotype, bioinformation, and protein microarray data, IDH1 was identified as a novel target of the GCN class of natural products. Classical approaches to direct protein target identification usually involve a series of linear steps to narrow down the possible modes of action, which rely on the biological activities of each compound. Here, combined methods including RNA-seq and microarray were used to rapidly identify the targets of small molecules. Notably, a protein microarray, which is convenient and unbiased in a genome-wide way, was utilized in this study. Considering that all human recombinant proteins are equally printed onto the nitrocellulose-coated glass slide, low-abundance proteins and some membrane proteins can be detected more easily in comparison with the classic “pull-down” strategy from cell lysates. In addition, this method is also especially suitable for compounds that exhibit fluorescence because synthesis of the probe structure is not needed as usual.
In this study, we show evidence that GCN B selectively triggers the apoptosis of NB4 APL cells through the inhibition of IDH1 and the elevation of intracellular ROS levels. Increased generation of ROS and an altered redox status have long been observed in most cancer cells, including leukemia cells [25, 26]. This biochemical property of leukemia cells makes them more vulnerable to exogenous oxidative stress and can be exploited for therapeutic benefits as a “ROS-stimulating strategy”. Substantial data have shown that arsenic trioxide (ATO), a drug used worldwide that induces complete clinical remission in 90% of APL patients, acts at least partially through a ROS-mediated mechanism [27]. In fact, in addition to the data shown in the main text, we also found that GCNs caused stress granule accumulation and phosphorylation of eIF2 (data not shown), which is very similar to typical oxidative stress responses upon ATO treatment. Therefore, the selectivity of GCN B in hematological malignancies might be closely related to its oxidative stress status. We postulate that enhancing the ROS-generating capacity using GCNs will effectively target all types of cells with high ROS levels and will not be limited to APL. Hence, the use of GCN B in preclinical applications in other types of AML cells or solid tumor cells with high ROS levels is worth exploring in the future.
Here, we report that IDH1 interference in NB4 cells causes caspase-dependent cell death, which is at least partially attributed to a ROS-mediated mechanism. Calvert et al. [28] reported that overexpression of wild-type IDH1 promotes glioma tumor growth by regulating NADPH-dependent lipid metabolism and decreasing ROS level. Additionally, Idh1-deficient mice have a high basal level of ROS in hepatocytes [8]. These data indirectly support our hypothesis that wild-type IDH1 is required for redox homeostasis and might be a desirable target for therapeutic intervention in AML patients. There are already known wild-type IDH1 small molecule inhibitors undergoing drug development; therefore, it is worth testing whether those inhibitors could selectively kill cells that bare high basal levels of ROS. Moreover, IDH1 mutation is believed to promote leukemogenesis and is regarded as a targetable driver of human AML [29] by causing aberrant accumulation of the oncometabolite 2-HG [30]. Several specific inhibitors targeting mutant IDH1 have been explored in the treatment of glioma and AML in clinical trials [31,32,33,34]. As we have shown the inhibitory activity of GCN B against mutant IDH1, it will be worthwhile to evaluate the efficacy of GCN B in AML cells harboring mutations in IDH1 in the future.
Protein microarray data revealed that there are other potential candidate targets of GCN B in vitro. Since pretreatment with NAC only partially rescued the cytotoxicity caused by IDH1 knockdown, we postulate that there might be other targets accounting for GCN B-induced cytotoxicity. We have confirmed the capacity of GCN B to bind to several candidate targets, such as SLC2A5 (data not shown). SLC2A5 has been reported to be responsible for AML and lung cancer metabolism [35, 36]. It is possible that the cytotoxicity of GCN family compounds is mediated by multiple molecular targets. In the future, we will examine whether there are other possible targets of GCN B in cells.
In conclusion, the evidence indicates that GCN B could be a selective lead compound that induces human NB4 cell death by targeting IDH1 through a ROS-mediated mechanism. We could not test the compound in an animal model because of the limited production of GCN B. In collaboration with chemists to totally synthesize GCN B and its analogs, the therapeutic potential of marine-originated angucycline glycoside antibiotics on human malignant tumors is worthy of further research.
References
Shen B. A new golden age of natural products drug discovery. Cell. 2015;163:1297–300.
Schumacher M, Kelkel M, Dicato M, Diederich M. Gold from the sea: marine compounds as inhibitors of the hallmarks of cancer. Biotechnol Adv. 2011;29:531–47.
Kharel MK, Pahari P, Shepherd MD, Tibrewal N, Nybo SE, Shaaban KA, et al. Angucyclines: biosynthesis, mode-of-action, new natural products, and synthesis. Nat Prod Rep. 2012;29:264–325.
Rohr J, Thiericke R. Angucycline group antibiotics. Nat Prod Rep. 1992;9:103–37.
Huang H, Yang T, Ren X, Liu J, Song Y, Sun A, et al. Cytotoxic angucycline class glycosides from the deep sea actinomycete Streptomyces lusitanus SCSIO LR32. J Nat Products. 2012;75:202–8.
Zhu X, Duan Y, Cui Z, Wang Z, Li Z, Zhang Y, et al. Cytotoxic rearranged angucycline glycosides from deep sea-derived Streptomyces lusitanus SCSIO LR32. J Antibiot (Tokyo). 2017;70:819–22.
Hayakawa Y, Iwakiri T, Imamura K, Seto H, Otake N. Studies on the isotetracenone antibiotics. III. A new isotetracenone antibiotic, grincamycin. J Antibiot (Tokyo). 1987;40:1785–7.
Itsumi M, Inoue S, Elia AJ, Murakami K, Sasaki M, Lind EF, et al. Idh1 protects murine hepatocytes from endotoxin-induced oxidative stress by regulating the intracellular NADP+/NADPH ratio. Cell Death Differ. 2015;22:1837–45.
Metallo CM, Gameiro PA, Bell EL, Mattaini KR, Yang J, Hiller K, et al. Reductive glutamine metabolism by IDH1 mediates lipogenesis under hypoxia. Nature. 2011;481:380–4.
Wang P, Wu J, Ma S, Zhang L, Yao J, Hoadley KA, et al. Oncometabolite D-2-hydroxyglutarate inhibits ALKBH DNA repair enzymes and sensitizes IDH mutant cells to alkylating agents. Cell Rep. 2015;13:2353–61.
Raz B, Iten M, Grether-Buhler Y, Kaminsky R, Brun R. The Alamar Blue assay to determine drug sensitivity of African trypanosomes (T.b. rhodesiense and T.b. gambiense) in vitro. Acta Tropica. 1997;68:139–47.
Chan WH, Chang YJ. Dosage effects of resveratrol on ethanol-induced cell death in the human K562 cell line. Toxicol Lett. 2006;161:1–9.
Fan B, Lu KY, Reymond Sutandy FX, Chen YW, Konan K, Zhu H, et al. A human proteome microarray identifies that the heterogeneous nuclear ribonucleoprotein K (hnRNP K) recognizes the 5’ terminal sequence of the hepatitis C virus RNA. Mol Cell Proteom: MCP. 2014;13:84–92.
Westerfield M. The zebrafish book. A guide for the laboratory use of zebrafish (Danio rerio). 4th ed. Eugene: Univ. of Oregon Press; 2000.
Gao L, Li D, Ma K, Zhang W, Xu T, Fu C, et al. TopBP1 governs hematopoietic stem/progenitor cells survival in zebrafish definitive hematopoiesis. PLoS Genet. 2015;11:e1005346.
Jia XE, Ma K, Xu T, Gao L, Wu S, Fu C, et al. Mutation of kri1l causes definitive hematopoiesis failure via PERK-dependent excessive autophagy induction. Cell Res. 2015;25:946–62.
Markolovic S, Wilkins SE, Schofield CJ. Protein hydroxylation catalyzed by 2-oxoglutarate-dependent oxygenases. J Biol Chem. 2015;290:20712–22.
Krige D, Needham LA, Bawden LJ, Flores N, Farmer H, Miles LE, et al. CHR-2797: an antiproliferative aminopeptidase inhibitor that leads to amino acid deprivation in human leukemic cells. Cancer Res. 2008;68:6669–79.
Elvidge GP, Glenny L, Appelhoff RJ, Ratcliffe PJ, Ragoussis J, Gleadle JM. Concordant regulation of gene expression by hypoxia and 2-oxoglutarate-dependent dioxygenase inhibition-the role of HIF-1 alpha, HIF-2 alpha, and other pathways. J Biol Chem. 2006;281:15215–26.
Hamada S, Kim TD, Suzuki T, Itoh Y, Tsumoto H, Nakagawa H, et al. Synthesis and activity of N-oxalylglycine and its derivatives as Jumonji C-domain-containing histone lysine demethylase inhibitors. Bioorg Med Chem Lett. 2009;19:2852–5.
Zhu H, Bilgin M, Bangham R, Hall D, Casamayor A, Bertone P, et al. Global analysis of protein activities using proteome chips. Science. 2001;293:2101–5.
Jeong JS, Jiang L, Albino E, Marrero J, Rho HS, Hu J, et al. Rapid identification of monospecific monoclonal antibodies using a human proteome microarray. Mol Cell Proteom: MCP. 2012;11:O111 016253.
Yang H, Ye D, Guan KL, Xiong Y. IDH1 and IDH2 mutations in tumorigenesis: mechanistic insights and clinical perspectives. Clin Cancer Res. 2012;18:5562–71.
Shi X, He BL, Ma AC, Guo Y, Chi Y, Man CH, et al. Functions of idh1 and its mutation in the regulation of developmental hematopoiesis in zebrafish. Blood. 2015;125:2974–84.
Zhou F, Shen Q, Claret FX. Novel roles of reactive oxygen species in the pathogenesis of acute myeloid leukemia. J Leukoc Biol. 2013;94:423–9.
Cairns RA, Harris IS, Mak TW. Regulation of cancer cell metabolism. Nat Rev Cancer. 2011;11:85–95.
Miller WH Jr, Schipper HM, Lee JS, Singer J, Waxman S. Mechanisms of action of arsenic trioxide. Cancer Res. 2002;62:3893–903.
Calvert AE, Chalastanis A, Wu Y, Hurley LA, Kouri FM, Bi Y, et al. Cancer-associated IDH1 promotes growth and resistance to targeted therapies in the absence of mutation. Cell Rep. 2017;19:1858–73.
Rakheja D, Konoplev S, Medeiros LJ, Chen W. IDH mutations in acute myeloid leukemia. Hum Pathol. 2012;43:1541–51.
Ward PS, Patel J, Wise DR, Abdel-Wahab O, Bennett BD, Coller HA, et al. The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting alpha-ketoglutarate to 2-hydroxyglutarate. Cancer Cell. 2010;17:225–34.
Nassereddine S, Lap CJ, Haroun F, Tabbara I. The role of mutant IDH1 and IDH2 inhibitors in the treatment of acute myeloid leukemia. Ann Hematol. 2017;96:1983–91.
Rohle D, Popovici-Muller J, Palaskas N, Turcan S, Grommes C, Campos C, et al. An inhibitor of mutant IDH1 delays growth and promotes differentiation of glioma cells. Science. 2013;340:626–30.
Fujii T, Khawaja MR, DiNardo CD, Atkins JT, Janku F. Targeting isocitrate dehydrogenase (IDH) in cancer. Discov Med. 2016;21:373–80.
Okoye-Okafor UC, Bartholdy B, Cartier J, Gao EN, Pietrak B, Rendina AR, et al. New IDH1 mutant inhibitors for treatment of acute myeloid leukemia. Nat Chem Biol. 2015;11:878–86.
Chen WL, Wang YY, Zhao A, Xia L, Xie G, Su M, et al. Enhanced fructose utilization mediated by SLC2A5 is a unique metabolic feature of acute myeloid leukemia with therapeutic potential. Cancer Cell. 2016;30:779–91.
Chen WL, Jin X, Wang MS, Liu D, Luo Q, Tian HC, et al. GLUT5-mediated fructose utilization drives lung cancer growth by stimulating fatty acid synthesis and AMPK/mTORC1 signaling. JCI Insight. 2020;5:e131596. https://doi.org/10.1172/jci.insight.131596.
Acknowledgements
This work was supported by the National Key Research and Development Program of China [2018YFC0310900], the National Natural Science Foundation of China [21877016; 81372768; 21572038; 21672248; 81820108030], the Laboratory for Marine Drugs and Bioproducts of Qingdao National Laboratory for Marine Science and Technology [LMDBKF201704], and the Fund of the State Key Laboratory of Bioorganic and Natural Products Chemistry [SKLBNPC16233].
Author information
Authors and Affiliations
Contributions
ZW performed experiments, analyzed data, and wrote the manuscript; ZXL performed experiments and wrote the manuscript; WCZ performed the zebrafish experiment; HBH prepared all GCNs; JYL and HZ analyzed RNA-seq data and docking. RNW, DZ, and WL synthesized the biotinylated GCN B probe and performed in vitro binding experiments; JQW, ZC, WLX, and WJ performed the in vivo experiment; LSZ performed the GC-MS analysis; JYL performed LC-MS/MS analysis, HZ performed the microarray assay, DY, LY, CL, WJP, JHJ, and YJD designed and directed the study.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Supplementary information
Rights and permissions
About this article
Cite this article
Wang, Z., Li, Zx., Zhao, Wc. et al. Identification and characterization of isocitrate dehydrogenase 1 (IDH1) as a functional target of marine natural product grincamycin B. Acta Pharmacol Sin 42, 801–813 (2021). https://doi.org/10.1038/s41401-020-0491-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41401-020-0491-6
Keywords
This article is cited by
-
Natural product 1,2,3,4,6-penta-O-galloyl-β-D-glucopyranose is a reversible inhibitor of glyceraldehyde 3-phosphate dehydrogenase
Acta Pharmacologica Sinica (2022)