Abstract
Glioblastoma (GBM) is the most common and lethal primary brain tumor in adults, but there is no effective drug available for GBM. Avasimibe is a potent inhibitor of acyl-coenzyme A: cholesterol acyltransferase-1 (ACAT-1), which was used to treat atherosclerosis. Experimental evidence and bioinformatics have shown that avasimibe has anticancer activity. In this study we investigated the anticancer effects of avasimibe on human glioblastoma cells and the underlying mechanisms. Our results showed that avasimibe dose-dependently inhibited the proliferation of U251 and U87 human glioblastoma cells with IC50 values of 20.29 and 28.27 μM, respectively, at 48 h. Avasimibe (7.5, 15, 30 μM) decreased the DNA synthesis, and inhibited the colony formation of the tumor cells. Treatment of avasimibe also dose-dependently increased the apoptotic rate of tumor cells, decreased the mitochondrial membrane potential, induced the activity of caspase-3/7, and increased the protein expression of cleaved caspase-9, cleaved PARP and Bax in U251 and U87 cells. RNA-sequencing analyses revealed that avasimibe suppressed the expression of CDK2, cyclin E1, CDK4, cyclin D, CDK1, cyclin B1, Aurora A, and PLK1, while induced the expression of p53, p21, p27, and GADD45A, which was validated by Western blot analysis. These results demonstrated that avasimibe induced mitochondria-dependent apoptosis in glioblastoma cells, which was associated with arresting the cell cycle at G0/G1 phase and G2/M phase by regulating the p53/p21 pathway, p53/GADD45A and Aurora A/PLK1 signaling pathways. In U87 xenograft nude mice model, administration of avasimibe (15, 30 mg·kg−1·d−1, ip, for 18 days) dose-dependently inhibit the tumor growth. Taken together, our results demonstrated that avasimibe might be a promising chemotherapy drug in the treatment of GBM.
Similar content being viewed by others
Introduction
Glioma is the most common primary intracranial tumor, accounting for ~81% of all intracranial tumors. In addition, glioblastoma is the most common glioma (accounting for ~45% of all gliomas) and is characterized by a short course of disease and high mortality [1,2,3]. Although the incidence of glioblastoma (4.67–5.73 cases per 100,000 people) is less than that of most other systemic tumors, the tumors grow rapidly and can invade important life-regulating centers around the tumor. Even for patients who receive chemotherapy and radiotherapy, the median survival time is only from 7 to 15 months. The 5-year survival rate is only from 0.05% to 4.7% [4]. Furthermore, the tumors are not sensitive to current treatment programs, and the prognosis for glioblastoma is far worse than that of other systemic tumors. Currently, the widely accepted treatment options include neurosurgery with radiotherapy and temozolomide chemotherapy. Temozolomide is an oral chemotherapy drug with good central nervous system permeability. It was approved to treat glioblastoma by the FDA in 1999. It is still the most effective chemotherapeutic drug for glioblastoma, and it can prolong the survival time of patients and enhance the patient’s quality of life [5,6,7]. Compared with radiotherapy alone, temozolomide administration with radiotherapy increased the patient median survival time from 12.1 months to 14.6 months, and the 2-year survival rate increased from 10.4% to 26.1% [3, 8]. However, temozolomide still has many deficits, such as side effects and drug resistance. Therefore, further development of glioblastoma drugs is necessary.
Avasimibe, a potent small molecule inhibitor of acyl-CoA cholesterol acyltransferase-1 (ACAT-1), is used to remove vascular plaque and treat atherosclerosis [9, 10]. Avasimibe has been tested in a number of preclinical and clinical trials to treat atherosclerosis and has a good human safety record [11, 12]. A study has shown that avasimibe can be used for tumor immunotherapy to promote antitumor effects by promoting T cell signaling and killing [13]. However, the function and underlying mechanism of avasimibe in glioblastomas are still unclear.
Here, we found that treating U251 and U87 glioblastoma cells with avasimibe inhibited cell proliferation and colony formation. Avasimibe also increased the apoptotic rate of these cells, decreased the mitochondrial membrane potential, induced the activity of caspase-3/7, and increased the protein expression of cleaved caspase-9, cleaved PARP and Bax in the U251 and U87 cells. Furthermore, according to RNA sequencing and Western blot analysis results, avasimibe was found to arrest the cell cycle in the G0/G1 and G2/M phases by regulating the p53/p21, p53/GADD45A, and Aurora A/PLK1 signaling pathways. Taken together, our findings suggest that avasimibe might be a promising chemotherapy drug in the treatment of glioblastoma.
Materials and methods
Cell culture
The U251 and U87 human glioblastoma cell lines were purchased from the Cell Bank of the Chinese Academy of Sciences (Beijing, China). The cells were cultured in a humidified incubator with an atmosphere containing 5% CO2 at 37 °C and maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% FBS and antibiotics (0.1 μg/mL penicillin and 0.1 μg/mL streptomycin).
Cell proliferation assay
CCK8 kit (Beyotime, China) was used to determine cell proliferation according to the manufacturer’s instructions. Briefly, the cells were cultured in 96-well plates at a density of 3 × 103 per well at 37 °C with 0 to 240 μM avasimibe for 24 or 48 h. Then, 10 μL of CCK8 was added and incubated for 1 h. The absorbance value of each well was measured at 450 nm. IC50 values were calculated using GraphPad Prism 7 software (GraphPad Software Inc., San Diego, California, USA).
EdU-DNA synthesis assay
An EdU Apollo 567 in vitro imaging kit (RiboBio, Guangzhou, China) was used to assess DNA synthesis according to the manufacturer’s instructions. Briefly, the cells were cultured in 96-well plates at a density of 1 × 104 per well at 37 °C with 0, 7.5, 15, and 30 μM avasimibe for 48 h. Then, EdU was added at a concentration of 50 µM and incubated for 2 h. Then, the cells were fixed in 4% paraformaldehyde for 30 min, permeated with 0.5% Triton X-100 for 10 min, and then stained with 10 µM Apollo 567 for 30 min. The cells were then counterstained with Hoechst 33342 for 30 min and photographed with a high-content imaging system [14].
Soft agar colony formation assay
A colony formation assay was used to determine the tumorigenicity of the U251 and U87 cells. Briefly, cells were seeded in 6-well plates. 2 mL 0.7% agar in complete medium was added as the bottom layer, and 1 mL 0.35% agar in complete medium containing 3000 cells with 7.5, 15, and 30 µM of avasimibe was added as the top layer. After cultured at 37 °C with 5% CO2 for 3–4 weeks, the colonies were stained with 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide (MTT) solution (200 µL/well) and photographed [15].
Hoechst 33342 staining assay
A Hoechst 33342 staining assay was carried out to observe the morphological characteristics of the apoptotic U251 and U87 cells. Briefly, the cells were cultured in 96-well plates at a density of 3 × 103 per well at 37 °C with 0, 7.5, 15, and 30 μM avasimibe for 48 h. Then, the cells were washed with PBS, fixed with 4% paraformaldehyde, permeabilized with 0.1% Triton X-100, stained with 1 μg/mL Hoechst 33342 solution for 10 min and photographed with a high-content imaging system.
Flow cytometric analysis of the apoptosis rate with Annexin V-EGFP/PI staining
To determine the apoptosis rate, an Annexin V-EGFP cell apoptosis detection kit (KeyGen BioTECH, Nanjing, China) was used to analyze apoptotic cells with flow cytometry, according to the manufacturer’s instructions. Briefly, cells were cultured in 6-cm dishes with 0, 7.5, 15, and 30 μM avasimibe for 48 h. After harvested, the cells were incubated with Annexin V-EGFP and PI for 15 min at room temperature. Flow cytometry was then performed to analyze the apoptosis rate. The data were analyzed by using Flow Jo software (Tristar, CA, USA).
Analysis of the mitochondrial membrane potential (MMP, Δψm)
Mitochondrial membrane potential is associated with cell apoptosis. 5,5ʹ,6,6ʹ-tetrachloro-1,1ʹ,3,3ʹ-tetraethylbenzimidazolocarbocyanine iodide (JC-1) staining was used to determine whether the mitochondrial membrane potential of the cells was changed by avasimibe treatment. U251 and U87 cells were cultured in 6-cm dishes with 0, 7.5, 15, and 30 μM avasimibe for 48 h. After treatment of the drug, the medium was removed, and the cells were incubated with JC-1 at a final concentration of 10 μg/mL for 20 min. The fluorescence value of JC-1 monomers was detected at 488 nm excitation and 530 nm emission wavelengths by a multifunction fluorescence microplate reader. The fluorescence of the JC-1 polymers was detected at 529 nm excitation and 590 nm emission wavelengths. The ratio of the monomer fluorescence value to the polymer fluorescence value was calculated [16].
Analysis of caspase-3/7 activity
A caspase-3/7 live cell fluorescence real-time detection kit (KeyGen BioTECH, Nanjing, China) was used to detect caspase-3/7 activity. Briefly, cells were cultured in 6 cm dishes with 0, 7.5, 15, and 30 μM avasimibe for 48 h. After harvested, the cells were incubated with a caspase-3/7 live cell fluorescence real-time detection kit for 1 h. Flow cytometry was performed to analyze the activity levels of caspase-3/7. The data were analyzed by using Flow Jo software (Tristar, CA, USA).
RNA sequencing
U87 cells were cultured in 6 cm dishes with 15 μM avasimibe for 48 h. Then, the cells were dissolved in TRIzol reagent (Life Technologies, USA) and sent to CapitalBio Corporation (Beijing, China) for next-generation sequencing. A Bioanalyzer 2100 was used to qualify the RNA. NEBNext multiplex oligos were used to establish libraries for Illumina sequencing. The libraries were then sequenced using an Illumina HiSeq 2500 sequencing platform (Illumina Inc., San Diego, CA, USA) [14].
Identification of differentially expressed genes
FASTQ data were aligned to the genome (GRh38) using HISAT2. After removing duplicate reads, the remaining SAM files were converted and sorted into BAM files using SAMtools software [17]. The differential expression was analyzed using Cufflinks software [18]. We used the following criteria to screen differentially expressed genes: |log2FC| ≥ 1 and P-value ≤ 0.05 [14].
Gene Ontology and pathway enrichment analyses
The DAVID website (https://david.ncifcrf.gov/home.jsp) was used to perform gene ontology and pathway enrichment analyses. The Gene Ontology terms were determined using a GOA data set. The enriched biological pathways were determined with the KEGG datasets. Cytoscape 3.6.0 was used to visualize the coexpression modules [14].
Western blotting
Cells were cultured in 60-mm dishes with 0, 7.5, 15, and 30 μM avasimibe for 48 h and then harvested and lysed with RIPA (Applygen, Beijing, China) lysis buffer at 4 °C for 30 min to determine the total protein level. The BCA method (Beyotime, Guangzhou, China) was used to measure the protein concentrations. The equivalent amount of each protein was loaded and separated on a 10% SDS-PAGE gel and then transferred to a PVDF membrane (Millipore, Billerica, MA). The membranes were blocked in 5% fat-free milk for 2 h and incubated overnight with primary antibodies at 4 °C. The primary antibodies against cleaved caspase-9, cleaved PARP, p21, p27, CDK2, cyclin E1, CDK4, cyclin D, and PLK1 were purchased from Cell Signaling Technology; antibodies against Bax, p53, Aurora A, CDK1, cyclin B1, and GAPDH were purchased from Proteintech. Then, the membrane was incubated with goat anti-mouse or anti-rabbit HRP-conjugated secondary antibody (CWBio, Beijing, China). Finally, the immunoreactive bands were visualized using an ECL Western blot kit (CWBio, Beijing, China).
In vivo antitumor activity
Six-week-old female BALB/c-nu nude mice (Beijing Vital River Laboratory Animal Technology Co., Ltd., Beijing, China) weighing 15–18 g were used in this study. All animal experiments were conducted according to the guidelines issued by the NIH Guide for the Care and Use of Laboratory Animals and were approved by the Ethics Committee for Laboratory Animal Care and Use at the Institute of Materia Medica, CAMS & PUMC. Briefly, U87 cells (1 × 107 cells per mouse) were subcutaneously implanted into the right flanks of the mice [19]. After 14 days, when the tumor volume was at least increased to 100 mm3, the mice were randomly divided into three groups (n = 6). The mice were then intraperitoneally treated with or without avasimibe (30 or 15 mg/kg) once per day for 18 days. The tumor volume was measured using a Vernier caliper and calculated according to the following modified ellipsoid formula: tumor volume (mm3) = (length) × (width)2 × 0.5. At the end of the experiment, all the mice were sacrificed, and the tumor tissues were harvested, weighed and photographed.
Statistical analysis
The data are presented as the mean ± standard deviation (SD) and represent at least three independent experiments. One-way ANOVA was used to calculate differences between the various study groups. A P-value < 0.05 was considered statistically significant.
Results
Avasimibe inhibited the viability and proliferation of glioblastoma cells
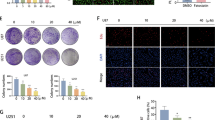
To determine the inhibitory effect of avasimibe (Fig. 1a) on the viability and proliferation of glioblastoma cells, an CCK8 assay was performed, and changes of cell morphology were also recorded. The results showed that avasimibe inhibited the proliferation of glioblastoma cells and that the inhibitory effect was superior in the U251 and U87 cells than it was in the A172 and SF268 cells (Fig. 1b). The IC50 of avasimibe for the U251 and U87 cells was measured by CCK8 assay. The IC50 of avasimibe for the U251 cells at 48 h was 20.29 μM, and the IC50 of avasimibe for the U87 cells at 48 h was 28.27 μM (Fig. 1c). The morphology of the cells was observed with a microscope after 48 h of avasimibe treatment at concentrations of 0, 7.5, 15, and 30 μM. Fig. 1d, e shows that the morphology of the U251 and U87 cells in the drug treatment groups was significantly changed compared with the morphology of the cells in the control groups and that the changes were dose-dependent.
Avasimibe inhibited the proliferation of glioma cells. a The structure of avasimibe. b CCK8 assay showed that avasimibe inhibited cell viability in the A172, SF268, U251, and U87 cells. c IC50 values were determined by CCK8 assay after cell treatment with avasimibe. d, e Morphological features showed that avasimibe changed the morphology of the U251 and U87 cells. Scale bar = 100 μm. f, g EdU staining assays showed that avasimibe inhibited DNA synthesis in the U251 and U87 cells. Scale bar = 50 μm. Error bars, s.d. (n = 3, *P < 0.05)
To confirm the inhibitory effect of avasimibe on the proliferation of the U251 and U87 cells, EdU staining was performed. As shown in Fig. 1f, g, after drug treatment, the DNA synthesis in the cells was significantly inhibited in a dose-dependent manner. Taken together, our results showed that avasimibe could inhibit the viability and proliferation of glioblastoma cells.
Avasimibe is an ACAT-1 inhibitor. To determine the role of ACAT-1 in the growth of glioblastomas, the mRNA and protein levels of ACAT-1 expression in the U87 and U251 cells were knocked down by human ACAT-1 siRNA or avasimibe treatment, and the proliferation of the cells was measured. Our results showed that the expression of ACAT-1 was significantly reduced by ACAT-1 siRNA and avasimibe treatment. In addition, the proliferation of glioblastoma cells was also significantly inhibited (Fig. S1). Altogether, ACAT-1 plays an important role in the growth of glioblastoma cells.
Avasimibe inhibited the colony formation of the glioblastoma cells
To validate the effect of avasimibe on glioblastoma, colony formation assays were performed to examine the effect of avasimibe on the formation of colonies. The results showed that avasimibe inhibited colony formation of U251 and U87 cells in the soft agar (Fig. 2a, b).
Avasimibe-induced apoptosis of the glioblastoma cells
To investigate whether avasimibe induces cell apoptosis, the cells were stained with Hoechst 33342 dye. The results showed that the nuclei of the U251 and U87 cells were concentrated and aggregated after treatment with avasimibe, and granular substances appeared. As the concentration of the drug was increased, more nuclei were lysed, resulting in vacuoles and the formation of apoptotic bodies. These results suggested that avasimibe induced apoptosis of the U251 and U87 cells in a dose-dependent manner (Fig. 3a, b). To validate the effect of avasimibe on the apoptosis of the U251 and U87 cells, Annexin V-EGFP/PI double staining was used to determine whether avasimibe induces cell apoptosis. The results showed that avasimibe induced apoptosis of the U251 and U87 cells in a dose-dependent manner (Fig. 3c, d).
Avasimibe-induced apoptosis in the U251 and U87 cells. a, b Hoechst 33342 staining assays showed that avasimibe induced cell apoptosis in the U251 and U87 cells. Scale bar = 100 μm. c, d Flow cytometric analysis of apoptosis with cells double stained with Annexin V-EGFP/PI, showing that avasimibe increased the apoptosis rate of the U251 and U87 cells (n = 3)
To further explore the mechanism of cell apoptosis induced by avasimibe, the changes in mitochondrial membrane potential (MMP) and the activity of caspase-3/7 were measured. JC-1 staining was used to determine mitochondrial membrane potential. After treatment of avasimibe for 48 h, JC-1 staining was carried out, and the fluorescence values of the JC-1 monomer and JC-1 polymer were measured. As shown in Fig. 4a, b, the ratio of JC-1 monomer/JC-1 polymer was increased as the concentration of the drug increased. These results indicated that avasimibe reduced the mitochondrial membrane potential of the U251 and U87 cells and induced apoptosis in a dose-dependent manner. The activity of caspase-3/7 was also determined by flow cytometry. As shown in Fig. 4c, d, the caspase-3/7 activity gradually increased with increasing avasimibe concentrations. The results suggested that avasimibe increased the caspase-3/7 activity in the U251 and U87 cells and induced apoptosis in a dose-dependent manner.
Avasimibe-induced apoptosis in a mitochondria-dependent manner. a, b JC-1 staining assays showed that avasimibe reduced the mitochondrial membrane potential of the U251 and U87 cells. MMP was calculated as the ratio of the red fluorescence to green fluorescence. c, d Flow cytometric analysis of caspase-3/7 activity showed that avasimibe induced caspase-3/7 activity in the U251 and U87 cells. e, f Western blot analysis showed that avasimibe induced the expression of apoptosis-related proteins in the U251 and U87 cells. Error bars, s.d. (n = 3, *P < 0.05)
The expression of apoptosis-related proteins was determined by Western blotting. In the U251 cells, the expression levels of cleaved PARP, cleaved caspase-9 and Bax were increased with the increase in the concentration of avasimibe (Fig. 4e), which suggested that avasimibe may initiate apoptosis in the mitochondrial pathway by upregulating Bax. In the U87 cells, the expression levels of cleaved caspase-9 and Bax were increased with the increase in the concentrations of avasimibe (Fig. 4f). Taken together, these results suggest that avasimibe inhibits cell proliferation by inducing apoptosis in a mitochondria-dependent manner and that this induction was mediated by activating caspase activity and upregulating Bax.
Differential gene expression and enrichment analyses of the U87 cells treated with avasimibe
To explore the potential mechanisms of avasimibe on glioblastoma cells, RNA sequencing was carried out in the U87 cells treated with 15 μM avasimibe for 48 h and in control cells. Compared with gene expression in the control cells, 442 upregulated genes and 398 downregulated genes were found in the cells treated with avasimibe (|log2FC| ≥ 1 and P-value ≤ 0.05, Fig. 5a). The results from the Gene Ontology enrichment analyses of these differentially expressed genes showed that the genes enriched in biological processes (BP) category involved the cell cycle, cell death, M phase, apoptosis, and mitotic cell cycle (Fig. 5b). In the cell components (CC) category, the differentially expressed genes were involved in intracellular nonmembrane-bounded organelles, nonmembrane-bounded organelles, the nuclear lumen, the nucleoplasm, chromosomes, and so on (Fig. 5c). In the molecular function (MF) category, the differentially expressed genes were involved in DNA binding, nucleotide binding, ribonucleotide binding, ATP binding, and transcription regulator activity (Fig. 5d). The results from the KEGG pathway enrichment analysis showed that the genes were enriched in pathways that involved the cell cycle, DNA replication, pyrimidine metabolism, p53 signaling pathway, and mismatch repair (Fig. 5e, f).
Differential expression and enrichment analysis of the U87 cells treated with avasimibe. a Volcano plot of differential expression results (upregulated genes are in red; downregulated genes are in blue; nonregulated genes are in black (|log2FC| ≥ 1 and P value ≤ 0.05). b–d GO enrichment analysis of differentially expressed genes. e, f KEGG pathway analysis of differentially expressed genes
Avasimibe arrests the cells at the G0/G1 checkpoint by regulating the p53/p21 signaling pathway
Avasimibe may arrest the cell cycle, as was implied by the KEGG pathway enrichment analysis. The expression levels of the proteins associated with G0/G1 cell cycle checkpoints in the U251 and U87 cells treated with avasimibe for 48 h were determined by Western blot analysis. Fig. 6a shows that the expression levels of CDK2, cyclin E1, CDK4, and cyclin D were downregulated, while the expression levels of p53, p21, and p27 proteins were upregulated (Fig. 6a) in the cells treated with avasimibe. Avasimibe also activated downstream proteins p21 and p27 by activating p53, thereby inhibiting the activity of the CDK2/cyclin E1 and CDK4/cyclin D complexes, arresting the cell cycle in the G0/G1 phase. All these results suggest that avasimibe arrested the cell cycle in the G0/G1 phase through a p53-dependent pathway.
Avasimibe induced cell cycle arrest in the U251 and U87 cells. a Western blot analysis showed that avasimibe induced cell cycle arrest in the G0/G1 phase by regulating the p53/p21/p27 pathway. b Western blot analysis showed that avasimibe arrested the cell cycle in the G2/M phase by regulating the p53/GADD45A and Aurora A/PLK1 pathways. c Schematic model for the mechanism of avasimibe in cell cycle arrest
Avasimibe arrests the cells at the G2/M checkpoint by regulating the p53/GADD45A and Aurora A/PLK1 signaling pathways
To explore whether avasimibe arrests the cell cycle at the G2/M phase, the expression levels of proteins associated with G2/M cell cycle checkpoints were determined by Western blotting. The results showed that the expression levels of CDK1 and cyclin B were downregulated by treatment with avasimibe in cells (Fig. 6b). The expression levels of p53 and GADD45A proteins were increased (Fig. 6b). All the results suggested that avasimibe arrested the cell cycle at the G2/M phase through a p53-dependent pathway. GADD45A was upregulated by p53 transcriptional regulation and interacted with the CDK1-cyclin B1 complex, inhibiting the activity of CDK1 kinase, thereby arresting the cell cycle at the G2/M checkpoint. In addition, avasimibe inhibited the expression levels of Aurora A and PLK1, which are critical for the activity of the CDK1-cyclin B1 complex (Fig. 6b), indicating that avasimibe can arrest the cell cycle at the G2/M phase by regulating the Aurora A/PLK1 pathway. Overall, avasimibe arrested the cell cycle at the G2/M checkpoint by regulating the p53/GADD45A and Aurora A/PLK1 signaling pathways.
Avasimibe suppressed tumorigenicity in vivo
To explore whether avasimibe has antitumor activity in vivo, a xenograft model was established by inoculating U87 nude mice with U87 cells and then treating the mice with 30 or 15 mg/kg of avasimibe per mouse every day for 18 days (Fig. 7a). As shown, the intraperitoneal administration of avasimibe significantly reduced tumor volume and tumor weight in a dose-dependent manner (Fig. 7b–e). During the administration of avasimibe, no obvious weight loss or abnormal behavior was observed in the nude mice. No mortality nor obvious metastasis was found in the vital organs, including the liver, kidney, brain, and lung. These results showed that avasimibe had an antitumor effect in vivo in a dose-dependent manner.
Avasimibe suppressed tumor growth in a human glioblastoma xenograft mouse model. a Schematic of xenograft mouse model establishment and avasimibe administration. b Avasimibe inhibited tumor growth as measured by tumor volume. c, d Avasimibe inhibited solid tumor growth. e Avasimibe inhibited tumor growth as measured by tumor weight
Discussion
Drug repurposing, also known as drug repositioning, refers to the reuse of old drugs to treat non-indication diseases. Drug repurposing has been a hot spot in drug development in recent years [20]. One of the earliest examples was mustard gas, which was firstly used as a weapon of war but was later found to possess anticancer properties [21]. Other examples include Viagra and Cialis, changed from use as treatments for erectile dysfunction to treatments for heart failure [22]; thalidomide, changed from use as a sedative to a treatment for erythema nodosum leprosum [23]; and imipramine, changed from use as a tricyclic antidepressant to treatment for small-cell lung cancer [24].
In this study, we firstly demonstrated that avasimibe, which was used in atherosclerosis, has an anti-glioblastoma effect by inducing cell apoptosis and arresting the cell cycle. Avasimibe inhibited cell proliferation, reduced DNA synthesis, increased the apoptotic rate of cells, decreased the mitochondrial membrane potential, and increased the activity of caspase-3/7 and the protein expression of cleaved caspase-9, cleaved PARP and Bax in the U251 and U87 cells. In addition, avasimibe was found to inhibit the expression of CDK1, CDK2, CDK4, cyclin E1, cyclin D, cyclin B1, Aurora A, and PLK1 while increasing the expression of p53, p21, p27, and GADD45A. Our results suggested that avasimibe arrested the cell cycle at the G0/G1 phase and G2/M phase by regulating the p53/p21, p53/GADD45A, and Aurora A/PLK1 signaling pathways.
Apoptosis is a key component of the innate tumor-suppression mechanism that inhibits oncogenesis at multiple stages, ranging from transformation to metastasis [25]. Apoptosis is known to be mediated by the mitochondrial pathway or the death receptor pathway. Bcl-2 family members, such as Bcl-2 and Bax, are crucial mediators of apoptosis in the mitochondrial pathway [26, 27]. In apoptosis, a subgroup of caspase-3, caspase-7, and caspase-9, named executioners, are crucial for the cleavage of PARP, an associated hallmark of apoptosis [28, 29]. A previous study showed that avasimibe-induced apoptosis was caspase-3-dependent and activated by caspase-8 [30]. In our study, we also demonstrated that avasimibe induced apoptosis in a mitochondria-dependent manner. Avasimibe can reduce the mitochondrial membrane potential, which is an indicator of early apoptosis. In addition to caspase-3, avasimibe increased the activity of caspase-7, caspase-9, and PARP and induced the expression of Bax in the U251 and U87 cells. These results suggest that avasimibe may induce cell apoptosis via the mitochondria-dependent pathway, and that the pro-apoptotic protein Bax regulates the pathway.
RNA-seq analysis was used to identify differentially expressed genes in the untreated cells and the cells treated with avasimibe for 48 h and characterize the potential biological pathways targeted by avasimibe. The results showed that, among the top ten enriched pathways, five pathways were associated with the cell cycle, such as the cell cycle, DNA replication, pyrimidine metabolism, mismatch repair, and nucleotide excision repair pathways. Therefore, avasimibe may affect cell proliferation by regulating the cell cycle.
The cell cycle is a set of organized and monitored events responsible for proper DNA replication and cell division. The cell cycle progresses from quiescence (G0 phase) to proliferation (G1, S, G2, and M phases) and returns to quiescence [31, 32]. Three cyclin-dependent kinases, CDKs, and their activating cyclins (B, D, and E) play key roles in mammalian cell cycle regulation [33]. It has been established that CDK4/cyclin D and CDK2/cyclin E promote cell passage through the G1 and S phases, whereas CDK1/cyclin B regulates the cell transition through late G2 phase and mitosis [33, 34]. The activities of the CDK–cyclin complexes depend on the balance between cyclins and cyclin-dependent kinase inhibitors (CKIs), such as p27(kip1) and p21(cip1) [35]. The p53 tumor suppressor protein controls the expression of p21 and p27 [36,37,38,39,40]. The results from our study revealed that treatment with avasimibe significantly downregulated cyclin D and cyclin E1 and upregulated p53, p21, p27, which indicated that avasimibe can arrest the cell cycle at the G0/G1 phase through the combined effects on the positive (cyclin D and cyclin E1) and negative (p21 and p27) regulators of CDK.
We also examined if avasimibe arrests the cell cycle during the G2/M transition by analyzing cyclin B1 production. Cyclin B1 is a key regulator of the cell cycle, crucial for controlling the G2/M transition. Cyclin B1 is involved in regulating the events of mitosis. Its synthesis, which is increased in the early G2 phase, is necessary for the transition from G2 through M [37]. Our results showed that avasimibe arrested the cell cycle at the G2/M phase by reducing the expression of cyclin B1 and CDK1. GADD45A, Aurora A, and PLK1 were found to be involved in the regulatory mechanism.
In normal cells, GADD45A is upregulated by p53 transcriptional regulation and interacts with the CDK1-cyclin B1 complex, inhibiting the activity of CDK1 kinase, thereby arresting the cell cycle at the G2/M checkpoint [41,42,43,44]. Aurora A and PLK1 are also redundant to the activation of CDK1-cyclin B1 during G2/M checkpoint recovery [45,46,47]. Aurora A promotes mitotic entry by controlling the activation of CDK1-cyclin B1 [48, 49]. It also activates PLK1, a key regulator of the cell cycle, in G2 through direct phosphorylation of PLK1 [50,51,52]. PLK1 has a peak of expression in G2/M and acts as a regulator of the cell cycle, which activates the CDK1-cyclin B1 complex that promotes mitosis entrance [53]. In this study, we found that avasimibe increased the expression of GADD45A and decreased the expression of Aurora A and PLK1, indicating that avasimibe arrested the cell cycle at the G2/M phase by regulating the p53/GADD45A and Aurora A/PLK1 signaling pathways.
In a mouse xenograft glioblastoma tumor model, we checked the in vivo antitumor effect of avasimibe treatment. After 18 days of avasimibe treatment, the growth of subcutaneous tumors was significantly inhibited, the tumor volume decreased, and the tumor weight decreased, indicating that avasimibe not only inhibited glioblastoma at the cellular level but also had a good antitumor effect in vivo. Although the therapeutic effect of avasimibe on tumors is not as profound as that of the first-line drug temozolomide in glioblastoma, it can reduce the side effects of traditional tumor drug treatment due to its high safety profile.
Conclusions
This is the first study that avasimibe inhibits the cell cycle in more than one phase through the regulation of p53/p21, p53/GADD45A, and Aurora A/PLK1 signaling pathways. This dual effect highlights the potent antiproliferative effect of avasimibe and suggests that avasimibe might stop the proliferation of glioblastoma cells at any stage in the cell cycle. Avasimibe also exerts antitumor effects on glioblastoma cells by inducing mitochondria-dependent apoptosis. Moreover, avasimibe also has antitumor effects in vivo. Therefore, avasimibe might be used as an anti-glioblastoma drug.
Data availability
The majority of the data generated in this study are included in this publication. The remaining raw data are available from the corresponding author upon reasonable request.
References
Reifenberger G, Wirsching H-G, Knobbe-Thomsen CB, Weller M. Advances in the molecular genetics of gliomas—implications for classification and therapy. Nat Rev Clin Oncol. 2016;14:434–52.
Morgan LL. The epidemiology of glioma in adults: a “state of the science” review. Neuro Oncol. 2015;17:623–4.
Bush NA, Chang SM, Berger MS. Current and future strategies for treatment of glioma. Neurosurg Rev. 2017;40:1–14.
Huang SW, Ali ND, Zhong L, Shi J. MicroRNAs as biomarkers for human glioblastoma: progress and potential. Acta Pharmacol Sin. 2018;39:1405–13.
Saito T, Sugiyama K, Takeshima Y, Amatya VJ, Yamasaki F, Takayasu T, et al. Prognostic implications of the subcellular localization of survivin in glioblastomas treated with radiotherapy plus concomitant and adjuvant temozolomide. J Neurosurg. 2018;128:679–84.
Reardon DA, Lassman AB, van den Bent M, Kumthekar P, Merrell R, Scott AM, et al. Efficacy and safety results of ABT-414 in combination with radiation and temozolomide in newly diagnosed glioblastoma. Neuro Oncol. 2017;19:965–75.
Brada M, Hoang-Xuan K, Rampling R, Dietrich PY, Dirix LY, Macdonald D, et al. Multicenter phase II trial of temozolomide in patients with glioblastoma multiforme at first relapse. Ann Oncol. 2001;12:259–66.
Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJ, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N. Engl J Med. 2005;352:987–96.
Delsing DJ, Offerman EH, van Duyvenvoorde W, van Der Boom H, de Wit EC, Gijbels MJ, et al. Acyl-CoA:cholesterol acyltransferase inhibitor avasimibe reduces atherosclerosis in addition to its cholesterol-lowering effect in ApoE*3-Leiden mice. Circulation. 2001;103:1778–86.
Tardif JC, Gregoire J, L’Allier PL, Anderson TJ, Bertrand O, Reeves F, et al. Effects of the acyl coenzyme A:cholesterol acyltransferase inhibitor avasimibe on human atherosclerotic lesions. Circulation. 2004;110:3372–7.
Robertson DG, Breider MA, Milad MA. Preclinical safety evaluation of avasimibe in beagle dogs: an ACAT inhibitor with minimal adrenal effects. Toxicol Sci. 2001;59:324–34.
Insull W Jr., Koren M, Davignon J, Sprecher D, Schrott H, Keilson LM, et al. Efficacy and short-term safety of a new ACAT inhibitor, avasimibe, on lipids, lipoproteins, and apolipoproteins, in patients with combined hyperlipidemia. Atherosclerosis. 2001;157:137–44.
Yang W, Bai Y, Xiong Y, Zhang J, Chen S, Zheng X, et al. Potentiating the antitumour response of CD8+ T cells by modulating cholesterol metabolism. Nature. 2016;531:651–5.
Li W, Liu J, Fu W, Zheng X, Ren L, Liu S, et al. 3-O-acetyl-11-keto-beta-boswellic acid exerts anti-tumor effects in glioblastoma by arresting cell cycle at G2/M phase. J Exp Clin Cancer Res. 2018;37:132.
Borowicz S, Van Scoyk M, Avasarala S, Karuppusamy Rathinam MK, Tauler J, Bikkavilli RK, et al. The soft agar colony formation assay. J Vis Exp. 2014;92:51998.
Huang TC, Lee JF, Chen JY. Pardaxin, an antimicrobial peptide, triggers caspase-dependent and ROS-mediated apoptosis in HT-1080 cells. Mar Drugs. 2011;9:1995–2009.
Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–9.
Trapnell C, Hendrickson DG, Sauvageau M, Goff L, Rinn JL, Pachter L. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat Biotechnol. 2013;31:46–53.
Yang Y, Guan D, Lei L, Lu J, Liu JQ, Yang G, et al. H6, a novel hederagenin derivative, reverses multidrug resistance in vitro and in vivo. Toxicol Appl Pharmacol. 2018;341:98–105.
Liu H, Song Y, Guan J, Luo L, Zhuang Z. Inferring new indications for approved drugs via random walk on drug-disease heterogenous networks. BMC Bioinforma. 2016;17:539.
Maurice J. Teaching old drugs new tricks raises hopes of new treatment for lung cancer. Lancet Respir Med. 2013;1:677.
Viagra and Cialis for heart failure? Erectile dysfunction drugs may do what no others have done. Harv Heart Lett. 2012;23:5.
Okafor MC. Thalidomide for erythema nodosum leprosum and other applications. Pharmacotherapy 2003;23:481–93.
Wang J, Byers LA. Teaching an old dog new tricks: drug repositioning in small cell lung cancer. Cancer Discov. 2013;3:1333–5.
Lopez J, Tait SW. Mitochondrial apoptosis: killing cancer using the enemy within. Br J Cancer. 2015;112:957–62.
Um HD. Bcl-2 family proteins as regulators of cancer cell invasion and metastasis: a review focusing on mitochondrial respiration and reactive oxygen species. Oncotarget. 2016;7:5193–203.
Green DR, Llambi F. Cell death signaling. Cold Spring Harb Perspect Biol. 2015;7:a006080.
Galluzzi L, Vitale I, Abrams JM, Alnemri ES, Baehrecke EH, Blagosklonny MV, et al. Molecular definitions of cell death subroutines: recommendations of the Nomenclature Committee on Cell Death 2012. Cell Death Differ. 2012;19:107–20.
Taylor RC, Cullen SP, Martin SJ. Apoptosis: controlled demolition at the cellular level. Nat Rev Mol Cell Biol. 2008;9:231–41.
Bemlih S, Poirier M-D, Andaloussi AE, Acyl-coenzyme A. Cholesterol acyltransferase inhibitor Avasimibe affect survival and proliferation of glioma tumor cell lines. Cancer Biol Ther. 2014;9:1025–32.
Diaz-Moralli S, Tarrado-Castellarnau M, Miranda A, Cascante M. Targeting cell cycle regulation in cancer therapy. Pharmacol Ther. 2013;138:255–71.
Norbury C, Nurse P. Animal cell cycles and their control. Annu Rev Biochem. 1992;61:441–70.
Morgan DO. Principles of CDK regulation. Nature 1995;374:131–4.
Goel S, DeCristo MJ, McAllister SS, Zhao JJ. CDK4/6 inhibition in cancer: beyond cell cycle arrest. Trends Cell Biol. 2018;28:911–25.
Coqueret O. New roles for p21 and p27 cell-cycle inhibitors: a function for each cell compartment? Trends Cell Biol. 2003;13:65–70.
Waga S, Hannon GJ, Beach D, Stillman B. The p21 inhibitor of cyclin-dependent kinases controls DNA replication by interaction with PCNA. Nature. 1994;369:574–8.
Chen J. The cell-cycle arrest and apoptotic functions of p53 in tumor initiation and progression. Cold Spring Harb Perspect Med. 2016;6:a026104.
Harper JW, Adami GR, Wei N, Keyomarsi K, Elledge SJ. The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell. 1993;75:805–16.
Li Y, Jenkins CW, Nichols MA, Xiong Y. Cell cycle expression and p53 regulation of the cyclin-dependent kinase inhibitor p21. Oncogene. 1994;9:2261–8.
Roy A, Banerjee S. p27 and leukemia: cell cycle and beyond. J Cell Physiol. 2015;230:504–9.
Jin S, Antinore MJ, Lung FD, Dong X, Zhao H, Fan F, et al. The GADD45 inhibition of Cdc2 kinase correlates with GADD45-mediated growth suppression. J Biol Chem. 2000;275:16602–8.
Zhan Q, Antinore MJ, Wang XW, Carrier F, Smith ML, Harris CC, et al. Association with Cdc2 and inhibition of Cdc2/Cyclin B1 kinase activity by the p53-regulated protein Gadd45. Oncogene. 1999;18:2892.
Wang XW, Zhan Q, Coursen JD, Khan MA, Kontny HU, Yu L, et al. GADD45 induction of a G2/M cell cycle checkpoint. Proc Natl Acad Sci U S A. 1999;30:3706–11.
Zhu N, Shao Y, Xu L, Yu L, Sun L. Gadd45-alpha and Gadd45-gamma utilize p38 and JNK signaling pathways to induce cell cycle G2/M arrest in Hep-G2 hepatoma cells. Mol Biol Rep. 2009;36:2075–85.
Zou J, Luo S-D, Wei Y-Q, Yang SY. Integrated computational model of cell cycle and checkpoint reveals different essential roles of Aurora-A and Plk1 in mitotic entry. Mol Biosyst. 2011;7:169–79.
Macurek L, Lindqvist A, Lim D, Lampson MA, Klompmaker R, Freire R, et al. Polo-like kinase-1 is activated by aurora A to promote checkpoint recovery. Nature. 2008;455:119–23.
Goldenson B, Crispino JD. The aurora kinases in cell cycle and leukemia. Oncogene. 2015;34:537–45.
Marumoto T, Honda S, Hara T, Nitta M, Hirota T, Kohmura E, et al. Aurora-A kinase maintains the fidelity of early and late mitotic events in HeLa cells. J Biol Chem. 2003;278:51786–95.
Satinover DL, Brautigan DL, Stukenberg PT. Aurora-A kinase and inhibitor-2 regulate the cyclin threshold for mitotic entry in Xenopus early embryonic cell cycles. Cell Cycle. 2006;5:2268–74.
Seki A, Coppinger JA, Jang CY, Yates GR, Fang G. Bora and the kinase Aurora a cooperatively activate the kinase Plk1 and control mitotic entry. Science. 2008;320:1655–8.
Bruinsma W, Macůrek L, Freire R, Lindqvist A, Medema RH. Bora and Aurora-A continue to activate Plk1 in mitosis. J Cell Sci. 2014;127:801.
Uchiumi T, Longo DL, Ferris DK. Cell cycle regulation of the human polo-like kinase (PLK) promoter. J Biol Chem. 1997;272:9166–74.
Pezuk JA, Brassesco MS, Morales AG, de Oliveira JC, de Oliveira HF, Scrideli CA, et al. Inhibition of polo-like kinase 1 induces cell cycle arrest and sensitizes glioblastoma cells to ionizing radiation. Cancer Biother Radiopharm. 2013;28:516–22.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (81573454 for Jinhua Wang, 81703565 for Weiqi Fu and No. 81703536 for Wan Li) and supported by the Beijing Natural Science Foundation (7172142). This work was supported by Technology Major Projects for “Major New Drugs Innovation and Development” (2018ZX09711001-005-025). This work was also supported by the CAMS Innovation Fund for Medical Sciences (2016-I2M-3-007).
Author information
Authors and Affiliations
Contributions
JHW, CY, and GHD designed research; JYL and WQF performed most functional experiments; XJZ and WL analyzed data; JYL and LWR wrote the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Supplementary information
Rights and permissions
About this article
Cite this article
Liu, Jy., Fu, Wq., Zheng, Xj. et al. Avasimibe exerts anticancer effects on human glioblastoma cells via inducing cell apoptosis and cell cycle arrest. Acta Pharmacol Sin 42, 97–107 (2021). https://doi.org/10.1038/s41401-020-0404-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41401-020-0404-8
Keywords
This article is cited by
-
RAS signaling and immune cells: a sinister crosstalk in the tumor microenvironment
Journal of Translational Medicine (2023)
-
Exploring the inverse association of glioblastoma multiforme and Alzheimer’s disease via bioinformatics analysis
Medical Oncology (2022)
-
The cholesterol esterification inhibitor avasimibe suppresses tumour proliferation and metastasis via the E2F-1 signalling pathway in prostate cancer
Cancer Cell International (2021)
-
Simultaneously targeting SOAT1 and CPT1A ameliorates hepatocellular carcinoma by disrupting lipid homeostasis
Cell Death Discovery (2021)