Abstract
Cancer is a highly heterogeneous disease in term of molecular signature even though it is originated from the same tissue type. Cancer heterogeneity may occur during its development or treatment, which is the main cause resulting in drug resistance and recurrence. Precision medicine refers to matching the right medicine to the right patients based on their molecular signatures. Therefore, a thorough understanding of the mechanism of tumorigenesis and drug resistance is essential to precision medicine. CRISPR-cas9 system is a powerful tool for gene editing and CRISPR-based high-throughput screening has been widely applied especially in searching for tumor-driven or synergistic lethal genes aiming to overcome drug resistance. In this review, we describe the progress of CRISPR-cas9-based unbiased screening in precision medicine including identification of new drug targets, biomarkers and elucidation of mechanisms leading to drug resistance. The existing challenges as well as the future directions are also discussed.
Similar content being viewed by others
Introduction
The discovery of tumor-driven genes and the development of targeted drugs greatly improved the therapy of tumors. However, the existence of heterogeneity enables tumors to survive treatment with drugs, resulting in unfavorable response rates [1,2,3]. The aim of precision medicine is to treat the right patients with the right medicine. Thus, it is necessary to dissect the mechanism underlying tumorigenesis and search for the genes that cause drug resistance in tumors. Recently, the wide application of unbiased screening based on gene editing has made it possible to solve a variety of problems in cancer treatment, such as poor selectivity, drug resistance, recurrence, and limited targets.
High-throughput screening based on gene editing has developed rapidly. Small interfering RNA interference technology has become a mature method because of its advantages in inducing the rapid downregulation of gene expression. Short hairpin RNA libraries have been used for loss-of-function (LOF) screening by stably downregulating the expression of genes across the whole genome, but the application of this method is limited by incomplete gene knockdown and off-target effects [4, 5]. Engineered nucleases, such as zinc finger nucleases (ZFNs), transcription activator-like effector nucleases (TALENs) and mega-ribozymes, have been successfully utilized in genetic engineering. However, the application of these genome editing techniques has been limited by their inevitable deficiencies. ZFNs and TALENs rely on the specific recognition of DNA sequences by proteins, and the complexity of their assembly makes them inconvenient to operate [6,7,8,9]. Recently, the clustered regularly interspaced short palindromic repeats (CRISPR)-associated protein 9 (Cas9) system has profoundly boosted the progress of genome engineering due to its capability to modify, replace or insert DNA sequences efficiently and accurately. CRISPR has been widely applied to genetically modified animal models, gene therapy and high-throughput screening in the scientific community [10,11,12,13]. The Zhang lab became the first to adopt large-scale CRISPR/Cas9-based screening to study drug resistance in tumors. The simplicity and reliability of the CRISPR-Cas9 system make it possible to discover new drug targets, biomarkers and genes that contribute to drug resistance which are essential factors in precision medicine. In this review, we summarize the common strategies used for CRISPR-cas9 screening and its application to precision medicine. We also discuss the existing problems and challenges of targeted screening technology.
Mechanism of CRISPR-cas9
The CRISPR-Cas9 system evolved as an immune defense against foreign bacteriophage or plasmid infection in bacteria or archaea [14,15,16]. Exogenous DNA or RNA fragments are identified and recorded by integrating a repeat-spacer array into CRISPR. Then, the CRISPR precursor is transcribed and processed into mature CRISPR-derived RNA (crRNA). After hybridization between the crRNA spacer and complementary invasive nucleotide sequences, double-strand breaks (DSBs) adjacent to the protospacer adjacent motif (PAM) are made by the cas endonuclease. The DSBs are then repaired via nonhomologous end joining (NHEJ) and homology directed repair (HDR), which directly result in end joining, base insertion and deletion or directional mutation by using the homologous repair template [17,18,19,20,21] (Fig. 1).
The synthesized 20 nt sgRNA is complementary to the target sequence near the protospacer adjacent motif (PAM). The cas9 protein introduces double-strand break (DSB) through the endonuclease domains HNH and Ruvc and then mediates gene repair by nonhomologous end joining (NHEJ) and homology directed repair (HDR), which may lead to an indel mutation or gene knockout. Moreover, cas9 can also be transformed into dcas9 by inactivating the endonuclease domain, which could cause gene transcriptional repression or activation via fusion to a transcriptional activator or repressor.
CRISPR is currently classified into six different types (I–VI) that include 33 subtypes according to the classification of the CRISPR-cas loci [22, 23]. Although it employs the same mechanism to acquire exogenous spacers, type II CRISPR requires only the cas9 protein for gene editing with the endonuclease domains HNH and RuvC. Therefore, the crRNA and trans-activating RNA (tracrRNA) used in type II A CRISPR were transformed into a small guide RNA (sgRNA) to make the CRISPR-cas9 system more convenient to operate [24,25,26]. In addition to functional loss, cas9 can also be transformed into dead Cas9 (dcas9) by inactivating the endonuclease domain and introducing a coactivator protein that can activate gene transcription to achieve gain-of-function (GOF) [27]. Thus, CRISPR-based gene editing technology has been adopted for large-scale genomic manipulation or for screening to find new drug targets, genes inducing drug resistance and pathological mechanisms (Fig. 1). A variety of pooled libraries used for CRISPR-cas9-based large-scale screening have been developed by different research groups and can be purchased from Addgene (http://www.addgene.org/pooled-library/#screening).
Strategies for CRISPR-based high-throughput screening
Tumor cells carry a wide range of genetic mutations that play important roles in oncogenesis and tumor evolution. CRISPR-cas9 has emerged as a promising method to identify tumor-driving genes as drug targets or biomarkers through large-scale screening [28]. Functional screening in cultured cells has been conducted in two modes, arrayed screens and pooled screens. Arrayed screens are performed in multiwell plates with a single genetic perturbation in each well. Cellular phenotypes (e.g., survival and migration) are then determined using fluorescence or luminescence assays or other techniques. Although arrayed screening is able to investigate a much wider range of phenotypic changes caused by a single genetic disturbance, its application is limited by its cost, complexity of operation and the need for specific facilities [29, 30]. In pooled screens, oligonucleotides are synthesized to construct a plasmid library. Pooled libraries are integrated into the genomes of target cells using viral transduction. The readout is based on the comparison of the abundance of the genomically integrated transgene between samples and is determined by PCR and next-generation sequencing. Pooled screening is easy to operate and cost-effective, and it is a strong technology capable of identifying genes with pro-tumor or antitumor potential in a high-throughput manner [31, 32].
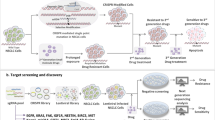
Pooled screening generally involves four steps. First, the plasmid libraries are created by synthesizing sgRNA oligonucleotides targeting genes of interest and cloning them into a lentiviral vector. Second, the plasmid pool is transduced into the target cells in the form of lentivirus or retrovirus, and tumor cells with stably edited gene(s) are identified by antibiotic selection. It is important that the multiplicity of infection (MOI) be maintained at a low level so that a single virus stably integrates a single cell in most cases [11, 27]. Then, the candidate genes potentially involved in drug resistance or tumorigenesis are identified by positive or negative screening for different research purposes. Finally, the genes are further verified in vitro and in vivo (Fig. 2).
A large number of sgRNAs were synthesized and cloned into a lentiviral vector to form pooled libraries. Meanwhile, cells stably expressing cas9/dcas9 were constructed. The viruses were then transduced at a low MOI, and the cells with stable perturbation remained after antibiotic selection. Positive or negative screening in vitro or in vivo was carried out according to different experimental purposes, and bioinformatics analysis was conducted according to the results of next-generation sequencing to identify the candidate genes.
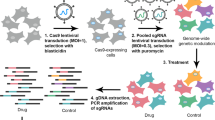
Positive or negative selection is usually utilized for unbiased screening (Fig. 3). In positive selection screening, cells with perturbations to induce pro-survival factors remain under long-term exposure to drugs. On the other hand, negative selection screening focuses on depleted cells with the selective pressure.
A CRISPR-cas9 screen can utilize either positive or negative screening according to the screening purpose. A positive screen focuses on surviving cells, whereas a negative screen focuses on dead cells. Screening can be further divided into loss-of-function (LOF knockout) or gain-of-function (GOF overexpression) screening according to the different functions of gene editing. Drug-resistance or suppressor genes are subsequently identified by the corresponding approach.
Progress in high-throughput screening with CRISPR-cas9 in precision medicine
Identification of new therapeutic targets
Oncogene addiction makes cancer cells vulnerable to therapies targeting specific driver genes. Due to the variety of driver genes, CRISPR-cas9 provides a powerful tool to identify these genes, which are potential drug targets. CRISPR-cas9 screening has been utilized to identify new targets in a variety of tumors in various cellular contexts [33,34,35]. Edwin et al. conducted a genome-wide LOF genetic screen in colorectal cancer HCT116 cells harboring mutant or wild-type KRAS [36]. They found that several genes involved in metabolism, such as NAD kinase or ketohexokinase, were potential therapeutic targets, and pharmacological inhibition of these enzymes attenuated the growth of colorectal cancer cells. Konstantinos el al. utilized the CRISPR-cas9 screening platform to identify genetic vulnerabilities in five types of acute myeloid leukemia (AML) cells (MOLM-13, MV4-11, HL-60, OCI-AML-2, and OCI-AML-3). As a result, they identified 492 AML-specific essential genes, including several established therapeutic targets, such as DOT1L, BCL2, and MEN1. KAT2A was proposed as a new therapeutic target for AML and validated in vitro and in vivo [37]. Recently, genome-scale CRISPR-CAS9 screening in 324 cancer cell lines from 30 cancer types was performed to develop a comprehensive database to prioritize candidate cancer therapeutic targets. For example, Werner syndrome ATP-dependent helicase (WRN) was identified as a promising new synthetic lethal target in microsatellite instability tumors [38].
Because CRISPR libraries are delivered by lentiviral vectors, it is difficult to operate in vivo. However, large-scale screening based on CRISPR-cas9 is not limited to the cellular system, and researchers managed to conduct screening in vivo [39]. Xu et al. first introduced piggyBac (PB) transposons for sgRNA delivery for in vivo screening [40]. PHD finger protein 5A was identified as a critical apoptotic suppressor involved in cancer progression and as a promising epigenetic target in breast cancer by using an in vivo CRISPR screen targeting RNA-binding proteins [41]. Although CRISPR screening has contributed significantly to the discovery of new targets, further improvements to reduce the false positive rate caused by the off-target effects of cas9 is warranted.
Identification of biomarkers in cancer therapy
Biomarkers are widely used for disease diagnosis, staging, and prediction or for monitoring the efficacy of therapeutics. Thus, the discovery of biomarkers is an important factor in precision medicine. Screening based on CRISPR-cas9 is very helpful in searching for biomarkers to identify populations sensitive to a given treatment [42, 43]. McCleland et al. found that BRD4 promotes the proliferation of colon cancer RKO cells through LOF screening [44]. Combined transcriptomic and genomic analysis further revealed that the long noncoding RNA (LncRNA) colon cancer-associated transcript 1 (CCAT1) was a biomarker that could indicate the sensitivity of colon cancer patients to BET inhibitors. Small cell lung cancer (SCLC) is generally considered to be associated with RB1 inactivation and TP53 mutations. CRISPR-cas9 screening revealed that RB1-negative SCLC NCI-H82 cells are hyperdependent on chromatin separation-related proteins, such as Aurora B kinase. Therefore, the loss of RB1 may be a potential biomarker for sensitivity to Aurora B kinase inhibitors [45]. Responses to the first-line chemotherapeutic drug cisplatin have not been improved in the treatment of bladder cancer. Researchers performed whole-genome CRISPR screening to confirm that MSH2, as the most significantly enriched gene, promoted resistance to cisplatin. Therefore, MSH2 is a potential biomarker that may indicate the response of bladder cancer patients to cisplatin [46]. A number of genes have been identified as potential biomarkers based on CRISPR-cas9 screening, which deserves further validation in clinical studies.
Identification of genes involved in drug resistance
Drug resistance is prone to occur during drug treatment and may ultimately lead to tumor recurrence. Therefore, elucidation of the resistance mechanism and discovery of combinatorial regimens are important strategies to overcome drug resistance. With the emergence of CRISPR-cas9 technology, researchers have been devoted to searching for essential genes causing drug resistance or synergistic lethality through large-scale screening [47, 48]. The Zhang lab constructed a Genome-Scale CRISPR Knock-Out library targeting 18 080 genes with 64,751 unique guide sequences to identify vemurafenib resistant genes in A375 melanoma cells [11]. Since then, unbiased LOF screening based on CRISPR-cas9 has been widely used in the search for synergistic lethal or resistant genes [49,50,51,52,53,54]. Gallipoli et al. utilized a genome-wide CRISPR-cas9 screen to determine that the knockout of glutaminase, which acts in the first step in glutamine metabolism, has a synergistic lethal effect when combined with FLT3-TKI inhibitors in FLT3ITD AML patients [55]. Thus, by inhibiting the feedback-activated survival signaling pathway, the recurrence of tumors caused by single drug therapy can be effectively prevented.
Genome-wide phenotypic changes can be achieved by transcriptional activation, and high-throughput screening based on GOF has also been introduced to identify genes inducing drug resistance [27, 56]. Konermann et al. synthesized a transcriptionally activated CRISPR library consisting of 70290 sgRNAs. They used a synergistic activation mediator system to screen genes potentially mediating resistance to BRAF inhibitors in melanoma A375 cells and patient-derived melanoma samples. Unlike pooled libraries used for knockout screening, sgRNAs in CRISPR activation (CRISPRa) libraries are designed to target promoter sites in genes of interest. Moreover, investigators have developed comprehensive genome-wide CRISPRa screens targeting 14 701 lncRNA genes. By screening these libraries, transcriptional activation of the GAS6-AS2 lncRNA was found to lead to Ara-C resistance in multiple cancers, including AML [57]. Although transcriptional activation CRISPR libraries avoid the deficiencies of cDNA libraries by capturing the complexity of transcript isoform variance and by utilizing cloning into size-limited viral expression vectors, function-acquired screening is still less widely used than knockout-based screening.
In recent years, cancer immunotherapy has achieved great success. However, the response rate is relatively low. In vivo CRISPR-cas9-based screening provides a powerful tool to identify genes mediating resistance to immunotherapy. Such an approach has been conducted in mice transplanted with melanoma, and defects in interferon-γ signaling have been identified to cause resistance to checkpoint blockade [58]. Recently, a genome-scale CRISPR-Cas9 screen was used to identify the mechanisms inducing resistance to the killing of tumor cells by cytotoxic T cells. The loss of PBAF function caused by inactivating genes, including Pbrm1, Arid2, and Brd7, sensitized mouse melanoma B16F10 cells to killing by T cells, and the inactivation of Pbrm1 rendered resistant tumors sensitive to immunotherapy [59]. In another study, the authors invented the CHIME (CHimeric IMmune Editing) method, which uses a CRISPR-CAS9 bone marrow delivery system that allows the rapid assessment of the genetic functions of innate and adaptive immune cells in vivo. This approach was employed to perform an in vivo pooled genetic screen, and Ptpn2 was identified as a negative regulator of CD8+ T cell-mediated responses to LCMV clone 13 viral infection [60]. Large-scale screening in vivo will be used primarily for the study of cancer immunotherapy, and the delivery system will be a critical factor for the efficient introduction of genetic alterations [61,62,63].
Challenges of CRISPR-cas9 screening in precision medicine
Large-scale screening based on CRISPR-cas9-mediated gene editing provides a powerful tool in the era of precision medicine. Although this approach has shown tremendous advantages, there are a few issues that need to be improved urgently. Minimization of the off-target effects of gene editing has been a significant challenge for the development of CRISPR technology. Several websites are available to help researchers design sgRNAs with improved specificity [64]. Scientists have also been working to optimize the design principles of sgRNAs to improve their specificity [65,66,67,68]. Strategies for designing sgRNAs targeting noncoding RNA are also being improved. A lentiviral paired-guide RNA library for the large-scale knockout of noncoding RNA has been developed [69]. Moreover, programmable single-base RNA editing has been realized by a RNA editing for specific C-to-U exchange system [70].
Genetic perturbations can be introduced by either LOF or GOF. Therefore, both strategies are utilized in large-scale screening. However, the GOF method requires the participation of coactivator proteins together with sgRNAs, which makes the method less accessible. To allow GOF screening to be more widely used, the multiple-plasmid system should be streamlined. Another concern is that the knockout or overexpression effects caused by CRISPR-cas9 may be compensated for by paracrine pathways [71]. Neighboring cells can compensate for functional knockout or gain by secreting growth-promoting cytokines, which would interfere with the functional readout. Furthermore, the phenotype and microenvironment may fail to adhere to the desired pathological conditions when cells are cultured in 2D [72]. Large-scale screening based on CRISPR experienced a process from in vitro to in vivo. The improvement of the delivery system and CRISPR-cas9 efficiency will be critical for improved screening in vivo. Alternatively, screening in organoids could better simulate the tumor microenvironment.
Finally, the analytical methods need to be optimized to reduce false positive or negative candidates during large-scale screening [48, 73,74,75]. A model-based analysis of genome-wide CRISPR/Cas9 knockout (MAGeCK) method (http://bitbucket.org/liulab/mageck-vispr) was created to control the quality of original sequencing results and to establish model analysis so that ‘seemingly relevant’ genes can be removed [76]. Researchers recently performed computational analysis of CRISPR screens using the MAGeCKFlute pipeline, which combines the MAGeCK and MAGeCK-VISPR algorithms and incorporates additional downstream analysis functionalities [77].
Conclusion
The discovery of CRISPR-cas9 not only represents a new method for gene editing but also provides a powerful tool in precision medicine for cancer treatment. The use of CRISPR-cas9 to construct cell or animal models has greatly facilitated cancer research. CRISPR-cas9-based screening accelerates the identification of new drug targets and biomarkers, which will promote the discovery and development of precise cancer treatments. By validating the mechanisms of drug resistance, new combinatorial strategies have been proposed to control recurrence and improve the survival rate of patients. Large-scale screening methods based on CRISPR-cas9 are highly variable, and researchers should adopt appropriate strategies according to their research purposes. Nevertheless, this editing method still needs to be improved in terms of the reduction of off-target effects and the improvement of the analytical methods. From the initial in vitro screening to ex vivo and in vivo screening, the technology has gradually been optimized to more closely reflect pathological conditions [78,79,80,81]. Due to the combined efforts of various research communities, CRISPR-cas9 will continue to play an increasingly important role in precision medicine for cancer treatment.
References
Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–74.
Greaves M, Maley CC. Clonal evolution in cancer. Nature. 2012;481:306–13.
McGranahan N, Swanton C. Clonal heterogeneity and tumor evolution: past, present, and the future. Cell. 2017;168:613–28.
Kampmann M, Horlbeck MA, Chen YW, Tsai JC, Bassik MC, Gilbert LA, et al. Next-generation libraries for robust RNA interference-based genome-wide screens. Proc Natl Acad Sci U S A. 2015;112:E3384–E91.
Fennell M, Xiang Q, Hwang A, Chen C, Huang CH, Chen CC, et al. Impact of RNA-guided technologies for target identification and deconvolution. J Biomol Screen. 2014;19:1327–37.
Urnov FD, Rebar EJ, Holmes MC, Zhang HS, Gregory PD. Genome editing with engineered zinc finger nucleases. Nat Rev Genet. 2010;11:636–46.
Joung JK, Sander JD. INNOVATION TALENs: a widely applicable technology for targeted genome editing. Nat Rev Mol Cell Biol. 2013;14:49–55.
Gaj T, Gersbach CA, Barbas CF. ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 2013;31:397–405.
Koo T, Lee J, Kim JS. Measuring and reducing off-target activities of programmable nucleases including CRISPR-Cas9. Mol Cells. 2015;38:475–81.
Heck D, Kowalczyk MS, Yudovich D, Belizaire R, Puram RV, McConkey ME, et al. Generation of mouse models of myeloid malignancy with combinatorial genetic lesions using CRISPR-Cas9 genome editing. Nat Biotechnol. 2014;32:941–6.
Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelsen TS, et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014;343:84–7.
Chen SD, Sanjana NE, Zheng KJ, Shalem O, Lee K, Shi X, et al. Genome-wide CRISPR screen in a mouse model of tumor growth and metastasis. Cell. 2015;160:1246–60.
Lin KH, Xie A, Rutter JC, Ahn YR, Lloyd-Cowden JM, Nichols AG, et al. Systematic dissection of the metabolic-apoptotic interface in AML reveals heme biosynthesis to be a regulator of drug sensitivity. Cell Metab. 2019;29:1217–31.
Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–21.
Heler R, Marraffini LA, Bikard D. Adapting to new threats: the generation of memory by CRISPR-Cas immune systems. Mol Microbiol. 2014;93:1–9.
Mojica FJM, Rodriguez-Valera F. The discovery of CRISPR in archaea and bacteria. FEBS J. 2016;283:3162–9.
Garneau JE, Dupuis ME, Villion M, Romero DA, Barrangou R, Boyaval P, et al. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature. 2010;468:67–71.
Gasiunas G, Barrangou R, Horvath P, Siksnys V. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc Natl Acad Sci U S A. 2012;109:E2579–E86.
Hale CR, Zhao P, Olson S, Duff MO, Graveley BR, Wells L, et al. RNA-guided RNA cleavage by a CRISPR RNA-Cas protein complex. Cell. 2009;139:945–56.
Jiang FG, Doudna JA. The structural biology of CRISPR-Cas systems. Curr Opin Struct Biol. 2015;30:100–11.
Lieber MR. The mechanism of double-strand dna break repair by the nonhomologous DNA end-joining pathway. Annu Rev Biochem. 2010;79:181–211.
Koonin EV, Makarova KS, Zhang F. Diversity, classification and evolution of CRISPR-Cas systems. Curr Opin Microbiol. 2017;37:67–78.
Jiang FG, Doudna JA. CRISPR-Cas9 structures and mechanisms. Annu Rev Biophys. 2017;46:505–29.
van der Oost J, Westra ER, Jackson RN, Wiedenheft B. Unravelling the structural and mechanistic basis of CRISPR-Cas systems. Nat Rev Microbiol. 2014;12:479–92.
Wiedenheft B, Sternberg SH, Doudna JA. RNA-guided genetic silencing systems in bacteria and archaea. Nature. 2012;482:331–8.
Zhan TZ, Rindtorff N, Betge J, Ebert MP, Boutros M. CRISPR/Cas9 for cancer research and therapy. Semin Cancer Biol. 2019;55:106–19.
Konermann S, Brigham MD, Trevino AE, Joung J, Abudayyeh OO, Barcena C, et al. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature. 2015;517:583–U332.
Ahmad G, Amiji M. Use of CRISPR/Cas9 gene-editing tools for developing models in drug discovery. Drug Discov Today. 2018;23:519–33.
Hasson SA, Kane LA, Yamano K, Huang CH, Sliter DA, Buehler E, et al. High-content genome-wide RNAi screens identify regulators of parkin upstream of mitophagy. Nature. 2013;504:291–5.
Shalem O, Sanjana NE, Zhang F. High-throughput functional genomics using CRISPR-Cas9. Nat Rev Genet. 2015;16:299–311.
Malina A, Mills JR, Cencic R, Yan YF, Fraser J, Schippers LM, et al. Repurposing CRISPR/Cas9 for in situ functional assays. Gene Dev. 2013;27:2602–14.
Zhou PH, Shaffer DR, Arias DAA, Nakazaki Y, Pos W, Torres AJ, et al. In vivo discovery of immunotherapy targets in the tumour microenvironment. Nature. 2014;506:52-7.
Hart T, Chandrashekhar M, Aregger M, Steinhart Z, Brown KR, MacLeod G, et al. High-resolution CRISPR screens reveal fitness genes and genotype-specific cancer liabilities. Cell. 2015;163:1515–26.
Munoz DM, Cassiani PJ, Li L, Billy E, Korn JM, Jones MD, et al. CRISPR screens provide a comprehensive assessment of cancer vulnerabilities but generate false-positive hits for highly amplified genomic regions. Cancer Discov. 2016;6:900–13.
Shi JW, Wang E, Milazzo JP, Wang ZH, Kinney JB, Vakoc CR. Discovery of cancer drug targets by CRISPR-Cas9 screening of protein domains. Nat Biotechnol. 2015;33:661–7.
Yau EH, Kummetha IR, Lichinchi G, Tang R, Zhang YL, Rana TM. Genome-wide CRISPR screen for essential cell growth mediators in mutant KRAS colorectal cancers. Cancer Res. 2017;77:6330–9.
Tzelepis K, Koike-Yusa H, De Braekeleer E, Li Y, Metzakopian E, Dovey OM, et al. A CRISPR dropout screen identifies genetic vulnerabilities and therapeutic targets in acute myeloid leukemia. Cell Rep. 2016;17:1193–205.
Behan FM, Iorio F, Picco G, Goncalves E, Beaver CM, Migliardi G, et al. Prioritization of cancer therapeutic targets using CRISPR-Cas9 screens. Nature. 2019;568:511.
Winters IP, Murray CW, Winslow MM. Towards quantitative and multiplexed in vivo functional cancer genomics. Nat Rev Genet. 2018;19:741–55.
Xu CL, Qi XL, Du XG, Zou HY, Gao F, Feng T, et al. piggyBac mediates efficient in vivo CRISPR library screening for tumorigenesis in mice. Proc Natl Acad Sci U S A. 2017;114:722–7.
Zheng YZ, Xue MZ, Shen HJ, Li XG, Ma D, Gong Y, et al. PHF5A epigenetically inhibits apoptosis to promote breast cancer progression. Cancer Res. 2018;78:3190–206.
Brunen D, de Vries RC, Lieftink C, Beijersbergen RL, Bernards R. PIM kinases are a potential prognostic biomarker and therapeutic target in neuroblastoma. Mol Cancer Ther. 2018;17:849–57.
Wang C, Jin HJ, Gao DM, Lieftink C, Evers B, Jin GZ, et al. Phospho-ERK is a biomarker of response to a synthetic lethal drug combination of sorafenib and MEK inhibition in liver cancer. J Hepatol. 2018;69:1057–65.
McCleland ML, Mesh K, Lorenzana E, Chopra VS, Segal E, Watanabe C, et al. CCAT1 is an enhancer-templated RNA that predicts BET sensitivity in colorectal cancer. J Clin Investig. 2016;126:639–52.
Oser MG, Fonseca R, Chakraborty AA, Brough R, Spektor A, Jennings RB, et al. Cells lacking the RB1 tumor suppressor gene are hyperdependent on Aurora B kinase for survival. Cancer Discov. 2019;9:230–47.
Goodspeed A, Jean A, Costello JC. A whole-genome CRISPR screen identifies a role of MSH2 in cisplatin-mediated cell death in muscle-invasive bladder cancer. Eur Urol. 2019;75:242–50.
Goncalves E, Behan FM, Louzada S, Arnol D, Stronach EA, Yang FT, et al. Structural rearrangements generate cell-specific, gene-independent CRISPR-Cas9 loss of fitness effects. Genome Biol. 2019;20:27. https://doi.org/10.1186/s13059-019-1637-z.
Sanson KR, Hanna RE, Hegde M, Donovan KF, Strand C, Sullender ME, et al. Optimized libraries for CRISPR-Cas9 genetic screens with multiple modalities. Nat Commun. 2018;9:5416. https://doi.org/10.1038/s41467-018-07901-8.
Zhang XY, Choi PS, Francis JM, Gao GF, Campbell JD, Ramachandran A, et al. Somatic superenhancer duplications and hotspot mutations lead to oncogenic activation of the KLF5 transcription factor. Cancer Discov. 2018;8:108–25.
Barazas M, Annunziato S, Pettitt SJ, de Krijger I, Ghezraoui H, Roobol SJ, et al. The CST complex mediates end protection at double-strand breaks and promotes PARP inhibitor sensitivity in BRCA1-deficient cells. Cell Rep. 2018;23:2107–18.
Zimmermann M, Murina O, Reijns MAM, Agathanggelou A, Challis R, Tarnauskaite Z, et al. CRISPR screens identify genomic ribonucleotides as a source of PARP-trapping lesions. Nature. 2018;559:285–9.
Kanarek N, Keys HR, Cantor JR, Lewis CA, Chan SH, Kunchok T, et al. Histidine catabolism is a major determinant of methotrexate sensitivity. Nature. 2018;559:632–6.
Shang WJ, Jiang Y, Boettcher M, Ding K, Mollenauer M, Liu ZY, et al. Genome-wide CRISPR screen identifies FAM49B as a key regulator of actin dynamics and T cell activation. Proc Natl Acad Sci U S A. 2018;115:E4051–E60.
Veo B, Fosmire S, Wang D, Madhavan K, Pierce A, Venkataraman S, et al. Functional CRISPR-Cas9 screen identifies druggable dependencies in Myc-driven medulloblastoma. Neuro Oncol. 2019;21(Suppl 2):105–6.
Gallipoli P, Giotopoulos G, Tzelepis K, Costa ASH, Vohra S, Medina-Perez P, et al. Glutaminolysis is a metabolic dependency in FLT3(ITD) acute myeloid leukemia unmasked by FLT3 tyrosine kinase inhibition. Blood. 2018;131:1639–53.
Wangensteen KJ, Wang YJ, Dou ZX, Wang AW, Mosleh-Shirazi E, Horlbeck MA, et al. Combinatorial genetics in liver repopulation and carcinogenesis with a in vivo CRISPR activation platform. Hepatology. 2018;68:663–76.
Bester AC, Lee JD, Chavez A, Lee YR, Nachmani D, Vora S, et al. An integrated genome-wide CRISPRa approach to functionalize lncRNAs in drug resistance. Cell. 2018;173:649–64.
Manguso RT, Pope HW, Zimmer MD, Brown FD, Yates KB, Miller BC, et al. In vivo CRISPR screening identifies Ptpn2 as a cancer immunotherapy target. Nature. 2017;547:413–8.
Pan D, Kobayashi A, Jiang P, de Andrade LF, Tay RE, Luoma AM, et al. A major chromatin regulator determines resistance of tumor cells to T cell-mediated killing. Science. 2018;359:770–5.
LaFleur MW, Nguyen TH, Coxe MA, Yates KB, Trombley JD, Weiss SA, et al. A CRISPR-Cas9 delivery system for in vivo screening of genes in the immune system. Nat Commun. 2019;10:1668. https://doi.org/10.1038/s41467-019-09656-2.
Yin H, Xue W, Anderson DG. CRISPR-Cas: a tool for cancer research and therapeutics. Nat Rev Clin Oncol. 2019;16:281–95.
Rogers ZN, McFarland CD, Winters IP, Naranjo S, Chuang CH, Petrov D, et al. A quantitative and multiplexed approach to uncover the fitness Landscape of tumor suppression in vivo. Nat Methods. 2017;14:737–42.
Chiou SH, Winters IP, Wang J, Naranjo S, Dudgeon C, Tamburini FB, et al. Pancreatic cancer modeling using retrograde viral vector delivery and in vivo CRISPR/Cas9-mediated somatic genome editing. Gene Dev. 2015;29:1576–85.
Ma J, Koster J, Qin Q, Hu SG, Li W, Chen CH, et al. CRISPR-DO for genome-wide CRISPR design and optimization. Bioinformatics. 2016;32:3336–8.
Ma XL, Zhang QY, Zhu QL, Liu W, Chen Y, Qiu R, et al. A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol Plant. 2015;8:1274–84.
Richardson CD, Ray GJ, DeWitt MA, Curie GL, Corn JE. Enhancing homology-directed genome editing by catalytically active and inactive CRISPR-Cas9 using asymmetric donor DNA. Nat Biotechnol. 2016;34:339–44.
Haeussler M, Schonig K, Eckert H, Eschstruth A, Mianne J, Renaud JB, et al. Evaluation of off-target and on-target scoring algorithms and integration into the guide RNA selection tool CRISPOR. Genome Biol. 2016;17:148. https://doi.org/10.1186/s13059-016-1012-2.
Doench JG, Fusi N, Sullender M, Hegde M, Vaimberg EW, Donovan KF, et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat Biotechnol. 2016;34:184–91.
Zhu SY, Li W, Liu JZ, Chen CH, Liao Q, Xu P, et al. Genome-scale deletion screening of human long non-coding RNAs using a paired-guide RNA CRISPR-Cas9 library. Nat Biotechnol. 2016;34:1279–86.
Abudayyeh OO, Gootenberg JS, Franklin B, Koob J, Kellner MJ, Ladha A, et al. A cytosine deaminase for programmable single-base RNA editing. Science. 2019;365:382–6.
Ford K, McDonald D, Mali P. Functional genomics via CRISPR-Cas. J Mol Biol. 2019;431:48–65.
Jacobson EF, Tzanakakis ES. Human pluripotent stem cell differentiation to functional pancreatic cells for diabetes therapies: innovations, challenges and future directions. J Biol Eng. 2017;11:21. https://doi.org/10.1186/s13036-017-0066-3.
Mohr SE, Hu YH, Ewen-Campen B, Housden BE, Viswanatha R, Perrimon N. CRISPR guide RNA design for research applications. FEBS J. 2016;283:3232–8.
Jagadevan S, Banerjee A, Banerjee C, Guria C, Tiwari R, Baweja M, et al. Recent developments in synthetic biology and metabolic engineering in microalgae towards biofuel production. Biotechnol Biofuels. 2018;11:185. https://doi.org/10.1186/s13068-018-1181-1.
Doench JG. Am I ready for CRISPR? A user’s guide to genetic screens. Nat Rev Genet. 2018;19:67–80.
Li W, Xu H, Xiao TF, Cong L, Love MI, Zhang F, et al. MAGeCK enables robust identification of essential genes from genome-scale CRISPR/Cas9 knockout screens. Genome Biol. 2014;15:554.
Wang BB, Wang M, Zhang WB, Xiao TF, Chen CH, Wu A, et al. Integrative analysis of pooled CRISPR genetic screens using MAGeCKFlute. Nat Protoc. 2019;14:756–80.
Fellmann C, Cowen BC, Lin PC, Doudna JA, Corn JE. Cornerstones of CRISPR-Cas in drug discovery and therapy. Nat Rev Drug Discov. 2017;16:89–100.
O’Neil NJ, Bailey ML, Hieter P. Synthetic lethality and cancer. Nat Rev Genet. 2017;18:613–23.
Kampmann M. CRISPRi and CRISPRa screens in mammalian cells for precision biology and medicine. ACS Chem Biol. 2018;13:406–16.
Chow RD, Chen SD. Cancer CRISPR screens in vivo. Trends Cancer. 2018;4:349–58.
Acknowledgements
This work was supported by the “Personalized Medicine Molecular Signature-based Drug Discovery and Development” Strategic Priority Research Program of the Chinese Academy of Sciences [XDA12020111], the National Science and Technology Major Project “Key New Drug Creation and Manufacturing Program” [2018ZX09711002-011-014 & 2018ZX09711002-004-011] and the National Natural Science Foundation of China [81773760]. It was also partially supported by the Fudan-SIMM Joint Research Fund [FU-SIMM20172005].
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Xing, H., Meng, Lh. CRISPR-cas9: a powerful tool towards precision medicine in cancer treatment. Acta Pharmacol Sin 41, 583–587 (2020). https://doi.org/10.1038/s41401-019-0322-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41401-019-0322-9
Keywords
This article is cited by
-
Biological roles of SLC16A1-AS1 lncRNA and its clinical impacts in tumors
Cancer Cell International (2024)
-
Multi-faceted CRISPR/Cas technological innovation aspects in the framework of 3P medicine
EPMA Journal (2023)
-
CRISPR based therapeutics: a new paradigm in cancer precision medicine
Molecular Cancer (2022)