Abstract
Long non-coding RNAs (lncRNAs) have been identified as essential mediators in neurological dysfunction. Our previous study shows that berberine (BBR) hampers the nuclear-to-cytosolic translocation of high-mobility group box 1 (HMGB1) in the process of poststroke inflammation. In this study, we explored the role of lncRNA metastasis‐associated lung adenocarcinoma transcript 1 (Malat1) in the process of BBR-induced inhibition of HMGB1 in ischemic brain. Before the 60-min MCAO surgery, the mice were pretreated with BBR (50 mg· kg−1 per day, ig) for 14 days or ICV injected with specific lentiviral vector or shRNA. We showed that MCAO caused marked increase in the expression Malat1 and HMGB1 in the ipsilateral cortex, which was significantly attenuated by pretreatment with BBR. Knockdown of Malat1 attenuated the inflammatory injury after brain ischemia, whereas overexpression of Malat1 exacerbated ischemic brain inflammation. Overexpression of Malat1 also reversed BBR-induced reduction of HMGB1 and proinflammatory cytokines. The above results suggested a potential correlation between Malat1 and stroke inflammation. Based on informatics analysis we predicted that HMGB1 was a direct downstream target of miR-181c-5p, whereas Malat1 acted as a competitive endogenous RNA (ceRNA) for miR-181c-5p targeted the 3′-UTR of HMGB1 to promote inflammation after ischemic stroke. Knockdown of Malat1 significantly decreased HMGB1 level, which could be abrogated by transfection with miR-181c-5p inhibitors. Taken together, our results demonstrate for the first time that Malat1/miR-181c-5p/HMGB1 axis may be a key pathway of BBR-induced antiinflammation effects in stroke, and they may provide a novel avenue for targeted therapy.
Similar content being viewed by others
Introduction
Ischemic stroke, which is caused by a permanent or transient local reduction in blood supply to the brain, represents one of the most widespread and principal causes of public health problems in modern society [1, 2]. Currently, the only clinically effective therapeutic drug approved by the FDA is tissue plasminogen activator, which is largely limited in use because of its narrow therapeutic window [3, 4]. Various neurological events have been implicated in ischemic injury, in which inflammation is a complex process that contributes to stroke onset and progression [5,6,7]. Consequently, there is a pressing need to explore relevant genes involved in the pathogenesis of the complex inflammatory microenvironment and develop effective therapeutic interventions for ischemic stroke.
Approximately 90% of the genome is transcribed into noncoding RNAs, which have no protein-encoding ability, including small noncoding RNAs (miRNAs, <200 nt) and long noncoding RNA (lncRNAs, >200 nt) [8, 9], which can play critical roles in gene expression regulation, chromatin architecture modulation, cell growth and development, and various disease pathologies [10]. Moreover, noncoding RNAs are potentially novel biomarkers that could be applied in the diagnosis, treatment, and prognosis of diseases [11, 12]. Within the past few decades, RNA-seq analyses have demonstrated that the abundant expression and mutations of lncRNAs are involved in pathological aspects of the central nervous system [13]. However, little is known about the roles and mechanisms of lncRNAs in stroke. Malat1 was initially found to be upregulated in various tumor tissues and was thought to participate in the development and metastasis of cancer [14]. Subsequently, a clinical study demonstrated that Malat1 was highly expressed in ischemic stroke patients among the 20 studied lncRNAs [15]. In addition, increasing evidence has also indicated that Malat1 is significantly upregulated in microglia and neural cells to accelerate the inflammatory response in stroke [16, 17]. Given that upregulated Malat1 expression is associated with poor overall clinical survival, elucidating the functions and mechanisms of Malat1 and its related miRNAs during ischemic insult may help to establish a better targeted treatment strategy.
Under ischemic and hypoxic physiological conditions, high-mobility group box 1 (HMGB1), acting as an endogenous danger-associated molecular pattern, can be released by necrotic cells or actively secreted by macrophages/monocytes into the extracellular environment, thereby triggering and amplifying inflammatory processes [18,19,20]. Our research group has demonstrated that berberine (BBR) pretreatment could significantly hamper the nuclear-to-cytosolic translocation of HMGB1 to alleviate cerebral ischemic–reperfusion inflammation through the TLR4/NF-κB pathway [21]. However, the underlying mechanisms by which BBR regulates HMGB1 remain unclear.
The miR-181 family is highly expressed in the brain and has been reported to associate with apoptosis and inflammation [22]. Among miRNAs in that family, miR-181c levels were markedly low in the plasma of stroke patients, which could inhibit proinflammatory mediators by suppressing NF-κB activation [23, 24]. Based on the existing findings, we hypothesized that Malat1 may act as a ceRNA for miR-181c-5p to downregulate the expression of HMGB1 in ischemic stroke progression after BBR treatment.
Materials and methods
Animals and groups
All experiments were approved by the Ethics Committee of Nanjing First Hospital, Nanjing Medical University. Animal experimental procedures were conducted in accordance with the ARRIVE (Animal Research: Reporting In Vivo Experiments) guidelines. Adult male C57BL/6 mice weighing 20–25 g were purchased from Vital River Laboratory Animal Company (Beijing, China) and were housed at a strictly controlled environmental temperature and relative humidity, all animals were housed under a 12/12-h dark/light cycle and were allowed free access to food pellets and water.
Mice were randomly divided and intragastric administered with 0.5% carboxymethylcellulose sodium or BBR (50 mg/kg) once a day for 14 consecutive days before MCAO or Intracerebroventricular (ICV)-injected with Lenti-Malat1, Lenti-miR-181c-5p, sh-Malat1, miR-181c-5p inhibitor, Lenti-NC, sh-NC, or NC inhibitor 48 h before MCAO. Mice underwent MCAO surgery as described previously [21] and then were reperfused after 60 min.
Intracerebroventricular injection
Lentiviral vectors that overexpress lncRNA Malat1 (Lenti-Malat1), miR-181c-5p (Lenti-miR-181c-5p), an shRNA that targets lncRNA Malat1 (sh-Malat1), or a miR-181c-5p inhibitor, and vectors expressing nontargeting controls (Lenti-NC, sh-NC, NC inhibitor) were prepared by GenePharma (Shanghai, China). Mice were anesthetized with 4% chloral hydrate and fixed by a stereotaxic instrument (David Kopf Instrument, Tujunga, CA, USA). Each mouse was given injection in the right lateral ventricle at 2 mm from the anterior iliac crest and 1.5 mm from the sagittal suture. A small hole was placed at this position with a microsyringe at a depth of 2.5 mm below the surface of the skull, and 2 μL of the virus was injected.
Immunohistochemistry (IHC)
Briefly, IHC procedures were performed as previously described to detect HMGB1 protein expression in brain tissues. Rabbit monoclonal antibody anti-HMGB1 (1:400, Abcam, Cambridge, MA, USA) was used. Three high-power fields (×400) of the ischemic region of the infarct were randomly selected, and slides were viewed and photographed with an Olympus CX23 microscope (Olympus, Tokyo, Japan).
Real-time PCR for mRNA expression
Total RNA was isolated from ipsilateral cortex tissues after 24 h of MCAO by TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. RNA samples were reverse transcribed into cDNA using a commercially available kit (ABM, Richmond, BC, Canada). Target genes were examined by a standard SYBR-Green method in the ABI 7500 sequence detection system (Applied Biosystems, Foster City, CA, USA). Thirty-five cycles were set as follows: 95 °C for 15 s and 60 °C for 60 s. All results were normalized to the expression levels of GAPDH or snRNA U6 and quantified by the comparative (2−△△Ct) method. The primer sequences used were as follows: Malat1 forward 5′-GGACTTGAGCTGAGGTGCTT-3′, reverse 5′-GCTTCACCACCACATCCGTA-3′; HMGB1 forward 5′-GGATGCTTCTGTCAACTTCTC-3′, reverse 5′-TTCATAACGAGCCTTGTCAG-3′; miR-181c-5p forward 5′-AACATTCAACCTGTCGGTGAGT-3′, reverse 5′-CATGTCAGCCAGTGTTGAATGTC-3′.
Western blot analysis
Cytoplasmic and nuclear protein extracted from the ipsilateral cortex tissues after 24 h of MCAO were lysed with a protein extraction kit (KeyGEN BioTECH, Nanjing, China) on ice and centrifuged at 16 000 × g at 4 °C for 15 min, and the quantity of protein was assessed by the BCA protein assay kit (KeyGEN BioTECH, Nanjing, China). Protein samples (30 μg each lane) were separated by 12% SDS-PAGE and then transferred onto PVDF membranes (Millipore, Shenzhen, China). The membranes were blocked with 5% fat-free milk powder in TBST buffer for 1 h and then incubated with anti-HMGB1 antibody (1:1000, Abcam, Cambridge, MA, USA), anti-β-actin antibody (1:1000, Abcam, Cambridge, MA, USA) and antihistone H3 antibody (1:1000, Abcam, Cambridge, MA, USA) at 4 °C overnight. Next, the membranes were incubated with the HRP-linked secondary antibodies for 1 h at room temperature. Protein signals were detected with Image Quant LAS 4000 mini system using a New Super ECL Assay (KeyGEN BioTECH, Nanjing, China).
Enzyme-linked immunosorbent assay (ELISA)
After 24 h of reperfusion, ipsilateral brain homogenates were prepared for ELISA. TNF-α, IL-1β, IL-6, and IL-10 levels were measured by a corresponding ELISA kit (Maijian Biotechnology Center, Beijing, China) according to the manufacturer’s instructions.
Fluorescence in situ hybridization (FISH) assay
LncRNA FISH was performed using Servicebio lncRNA FISH Probe Mix (Red) (G3016-3, Servicebio, Wuhan, China). Briefly, after preheating 4-μm paraffin-embedded tissue sections, 20 μg/mL proteinase K (G1205, Servicebio, Wuhan, China) was added to digest for 25 min. Next, the hybridization solution containing the Malat1 lncRNA probe was added at a concentration of 8 ng/μL, and the mixture was incubated at 37 °C overnight. The sections were observed under a Nikon upright fluorescence microscope (Nikon, Tokyo, Japan), and images were taken.
Cell culture methods
BV-2 microglial cells and HEK-293T cells were cultured at 37 °C in a 5% CO2 incubator in RPMI-1640 medium (Gibco, Thermo Fisher Scientific, Waltham, MA, USA) containing 10% fetal bovine serum (Everygreen, Tianhang Biotechnology, Hangzhou, China), 100 U/mL penicillin and 100 μg/mL streptomycin (Gibco, Thermo Fisher Scientific, Waltham, MA, USA). The medium was changed every 2 days. The cells were subcultured with 0.25% trypsin and 0.02% EDTA (Gibco, Thermo Fisher Scientific, Waltham, MA, USA) when they reached 80% confluence.
Luciferase reporter assays
The sequences of the luciferase reporter plasmids Malat1-WT and Malat1-Mut were verified, and HEK-293T cells were cotransfected with the luciferase reporter plasmids and Lenti-NC or Lenti-miR-181c-5p using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA). Then, the cells were collected and lysed 24 h after transfection, and the luciferase activity was measured with a Glomax 20/20 luminometer fluorescence detector (Promega, Beijing, China) by a luciferase assay kit (RG005, Beyotime, Shanghai, China). The same procedure was used to determine the combined effect of HMGB1 and miR-181c-5p.
mRNA stability assay
BV-2 cells were seeded in six-well plates and grown to ∼80% confluence. Then, cells were transfected with Lenti-NC or Lenti-Malat1 using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) in Opti-MEM. After 6 h of transfection, the cells were incubated with 0.1% FBS medium containing actinomycin D (Act D) (10 mM) for 0, 6, 12, and 24 h. Total RNA was isolated from cells at the indicated time points (0, 6, 12, and 24 h) and reverse transcribed into cDNA to perform semiquantitative PCR analysis to determine the HMGB1 degradation kinetics of each mRNA.
Statistical analysis
All data measurements were expressed as the means ± SD and differences in means across multiple groups were compared using one-way analysis of variance, followed by the LSD test. All tests were analyzed by SPSS 24.0 software (IBM, Armonk, New York, USA), and a P-value of less than 0.05 was considered statistically significant. All statistical analyses were performed with GraphPad Prism 7 (GraphPad Software, San Diego, CA, USA).
Results
Malat1 is remarkably decreased in cerebral ischemia/reperfusion after BBR treatment
In total, 220 mice were used for the experiments. The mortality rate was ∼30%. First, RT-PCR was conducted to detect Malat1 expression changes in I/R injury. As displayed in Fig. 1a, the mRNA expression of Malat1 was greatly increased at 24 h after MCAO, whereas the expression of Malat1 in the BBR group was significantly lower than that in the Model group (P < 0.05), indicating that Malat1 is a potential target of BBR in ischemic injury.
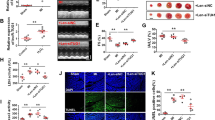
Malat1 knockdown ameliorates ischemic brain inflammation in a mouse MCAO model (n = 3). a Malat1 and HMGB1 were detected by the real-time polymerase chain reaction after 24 h of MCAO. b Enlarged images from representative immunohistochemistry photographs of HMGB1 in different treatment groups and the number of cells with positive staining for HMGB1. Scale bar, 20 μm. Magnification of the microphotograph, ×400. c Representative photographs of Western blots for nHMGB1, cyHMGB1 and HMGB1, and the ratio of cyHMGB1 to nHMGB1. β-Actin and histone H3 were used as loading controls. d Concentrations of inflammatory cytokines (TNF-α, IL-1β, IL-6, and IL-10) were measured by ELISA. Data are presented as the means ± SD. *P < 0.05, ***P < 0.001 versus the Sham group, #P < 0.05, ##P < 0.01, ###P < 0.001 versus the Model group or sh-NC group. nHMGB1: nuclear HMGB1; cyHMGB1: cytosolic HMGB1; nβ-actin: nuclear β-actin; cyβ-actin: cytosolic β-actin
Malat1 knockdown ameliorates ischemic brain inflammation in a mouse MCAO model
Given that Malat1 is highly downregulated by BBR in mouse cortex tissues, we further investigated the molecular mechanism of Malat1 upon ischemic stroke. With the knockdown of Malat1 by ICV injection in a mouse MCAO model, we found that the mRNA levels of Malat1 and HMGB1 were significantly lower in the Malat1 shRNA group than in the control shRNA group (P < 0.01, P < 0.001, respectively; Fig. 1a). Furthermore, there was no statistically significant difference in expression between the control shRNA group and the Model group. Therefore, we replaced the Model group with the sh-NC group in the subsequent experiments. As shown in Fig. 1b, the IHC results showed that the knockdown of Malat1 remarkably decreased the number of cells positive for HMGB1 in ischemic brain tissue at 24 h after MCAO (P < 0.05). Western blot analysis also revealed a low expression of cytoplasmic HMGB1 in the Malat1 knockdown group (P < 0.01, Fig. 1c). To further verify the effects of Malat1 on inflammation, ELISA was used to detect cytokines and the results implied that knockdown of Malat1 reduced the secretion of the proinflammatory cytokines TNF-α, IL-1β, and IL-6 (P < 0.01, P < 0.01, P < 0.01, respectively; Fig. 1d), while increasing the secretion of the antiinflammatory cytokine IL-10 (P < 0.01, Fig. 1d). Consequently, the knockdown of Malat1 could attenuate inflammatory injury after brain ischemia.
Malat1 overexpression exacerbates ischemic brain inflammation in a mouse MCAO model
We further sought to identify the role of Malat1 in inflammatory regulation, and Lenti-Malat1 was designed to enhance the expression of Malat1 in mice. As expected, the mRNA levels of Malat1 and HMGB1 were significantly upregulated after Lenti-Malat1 treatment (P < 0.01, P < 0.01, respectively; Fig. 2a). As shown in Fig. 2b, the IHC results showed that the overexpression of Malat1 remarkably increased the number of cells positive for HMGB1 in ischemic brain tissue at 24 h after MCAO (P < 0.05). Western blot analysis also revealed a greater activation of cytoplasmic HMGB1 after Lenti-Malat1 treatment (P < 0.05, Fig. 2c). Furthermore, ELISA results showed that Lenti-Malat1 treatment increased the secretion of the proinflammatory cytokines TNF-α, IL-1β, and IL-6 (P < 0.05, P < 0.05, P < 0.05, respectively; Fig. 2d) and decreased the secretion of the anti-inflammatory cytokine IL-10 (P < 0.05, Fig. 2d). As shown in Fig. 2e, the rate of HMGB1 degradation in response to Act D (10 mM) treatment for 24 h was lower in the Lenti-Malat1 BV-2 cells than in the Lenti-NC BV-2 cells, revealing that HMGB1 mRNA stability increases in a time-dependent manner in the Lenti-Malat1 BV-2 cells and that Malat1 plays a crucial role in HMGB1 mRNA degradation in these cells. Taken together, these results revealed that the overexpression of Malat1 could exacerbate inflammatory injury in the ischemic brain.
Malat1 overexpression exacerbates ischemic brain inflammation in mouse MCAO model (n = 3). a Malat1 and HMGB1 were detected by the real-time polymerase chain reaction after 24 h of MCAO. b Enlarged images from representative immunohistochemistry photographs of HMGB1 in different treatment groups and the number of cells with positive staining for HMGB1. Scale bar, 20 μm. Magnification of the microphotograph, ×400. c. Representative photographs of Western blots for nHMGB1, cyHMGB1 and HMGB1, and the ratio of cyHMGB1 to nHMGB1. β-actin and histone H3 were used as loading controls. d Concentrations of inflammatory cytokines (TNF-α, IL-1β, IL-6, and IL-10) were measured by ELISA. e The effect of Malat1 overexpression on HMGB1 mRNA stability was detected by the real-time polymerase chain reaction to determine the HMGB1 mRNA levels. Data are presented as the means ± SD. *P < 0.05, **P < 0.01, ***P < 0.001 versus the Sham group, #P < 0.05, ##P < 0.01 versus the Lenti-NC group. nHMGB1: nuclear HMGB1; cyHMGB1: cytosolic HMGB1; nβ-actin: nuclear β-actin; cyβ-actin: cytosolic β-actin
BBR exerts antiinflammatory effects through Malat1 after MCAO
To further explore whether BBR exerts antiinflammatory effects through Malat1, we used rescue experiments by enhancing Malat1 expression in the BBR administration group. As shown in Fig. 3a, Lenti-Malat1 reversed the reduction in Malat1 and HMGB1 mRNA levels by BBR. The results shown in Fig. 3b demonstrated that the inhibition of HMGB1 nuclear-to-cytosolic translocation by BBR could also be reversed by Lenti-Malat1. In addition, ELISA results implied that Lenti-Malat1 also reversed the cytokine changes induced by BBR (Fig. 3c). Therefore, these findings supported our hypothesis and suggested that the antiinflammatory effect of BBR was positively associated with Malat1 expression.
BBR exerts anti-inflammatory effects through Malat1 after MCAO (n = 3). a Malat1 and HMGB1 were detected by the real-time polymerase chain reaction after 24 h of MCAO. b Representative photographs of Western blots for nHMGB1, cyHMGB1 and HMGB1 and the ratio of cyHMGB1 to nHMGB1. β-actin and histone H3 were used as loading controls. c Concentrations of inflammatory cytokines (TNF-α, IL-1β, IL-6, and IL-10) were measured by ELISA. Data are presented as the means ± SD. **P < 0.01, ***P < 0.001 versus the Sham group, #P < 0.05, ##P < 0.01, ###P < 0.001 versus the Model group, NS versus the Model group. nHMGB1: nuclear HMGB1; cyHMGB1: cytosolic HMGB1; nβ-actin: nuclear β-actin; cyβ-actin: cytosolic β-actin
Malat1 binds to miR-181c-5p and functions as a ceRNA
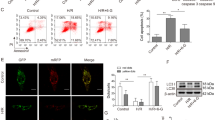
In order to facilitate subsequent functional studies of the mechanisms of Malat1, we analyzed the subcellular localization of Malat1 in the brain. RNA-FISH assay results revealed that Malat1 was located in the cytoplasm (Fig. 4a). To identify the target of HMGB1, Targetscan (http://www.targetscan.org/vert_72/) was used to perform a target prediction search, and the results implied that miR-181c-5p was the best candidate (Fig. 4b). Then, RNAhybrid (https://bibiserv.cebitec.uni-bielefeld.de/rnahybrid/) was used to evaluate whether miR-181c-5p has a strong ability to bind to Malat1 (Fig. 4c). Based on these results, we concluded that Malat1 could function as a ceRNA for miR-181c-5p. Next, RT-PCR was used to investigate whether miR-181c-5p could be negatively regulated by Malat1. As shown in Fig. 4d, e, miR-181c-5p was significantly upregulated by Malat1 knockdown but was greatly inhibited by Malat1 overexpression (P < 0.01, P < 0.05, respectively). To further investigate the binding ability between the two, we conducted a luciferase reporter assay. The results showed that miR-181c-5p mimic remarkably downregulated the luciferase activity of the Malat1-WT vector but not the Malat1-Mut vector (P < 0.001, Fig. 4f). Considering these results, we propose that miR-181c-5p can function as a molecular target of Malat1 in ischemic stroke.
Malat1 binds to miR-181c-5p and functions as a ceRNA (n = 3). a Malat1 subcellular localization results indicated that it was expressed in the cytoplasm. b Predicted binding sites for miR-181c-5p in HMGB1 sequences. c Binding sites of Malat1 on miR-181c-5p predicted by bioinformatics software. d-e RT-PCR was performed on RNA collected from the ipsilateral brain after 24 h of MCAO and showed the expression of miR-181c-5p in the sh-Malat1 and Lenti-Malat1 groups, respectively. f Luciferase reporter assay revealed that Lenti-miR-181c-5p decreased the luciferase activity of Malat1-WT, but not of Malat1-Mut. Data are presented as the means ± SD. **P < 0.01, ***P < 0.001 versus the Sham group, #P < 0.05, ##P < 0.01 versus the sh-NC or Lenti-NC group
HMGB1 is a direct downstream target of miR-181c-5p
To determine whether HMGB1 was involved in the activation of miR-181c-5p, RT-PCR was performed to examine the expression of miR-181c-5p, Malat1 and HMGB1. As shown in Fig. 5a, miR-181c-5p expression was increased and Malat1 and HMGB1 expression was decreased in the Lenti-miR-181c-5p group compared with the control group (P < 0.001, P < 0.05, P < 0.001, respectively), whereas, as seen in Fig. 6a, the above expression changes were reversed in the miR-181c-5p inhibitor group (P < 0.05, P < 0.05, P < 0.01, respectively). Western blot assays also showed the same trend in which cytoplasmic HMGB1 activation was decreased in the Lenti-miR-181c-5p group and increased in the miR-181c-5p inhibitor group (P < 0.01, P < 0.05, respectively; Figs. 5b and 6b). Furthermore, ELISA results showed that Lenti-miR-181c-5p treatment decreased the secretion of the proinflammatory cytokines TNF-α, IL-1β, and IL-6 (P < 0.01, P < 0.01, P < 0.01, respectively; Fig. 5c) and increased the secretion of the anti-inflammatory cytokine IL-10 (P < 0.01, Fig. 5c), whereas the above expression changes were reversed in the miR-181c-5p inhibitor group (Fig. 6c). To further investigate the interaction between HMGB1 and miR-181c-5p, we conducted a luciferase reporter assay. Our data showed that the miR-181c-5p mimic distinctly inhibited the luciferase activity of the HMGB1-WT vector but not the HMGB1-Mut vector (P < 0.001, Fig. 6d). Altogether, these results indicated that HMGB1 positively binds to miR-181c-5p.
miR-181c-5p overexpression ameliorates ischemic brain inflammation in a mouse MCAO model (n = 3). a RT-PCR was performed on mRNA collected from the ipsilateral brain after 24 h of MCAO and showed the expression of miR-181c-5p, Malat1 and HMGB1 in the Lenti-miR-181c-5p group. b Representative photographs of Western blot for nHMGB1, cyHMGB1 and HMGB1 and the ratio of cyHMGB1 to nHMGB1. β-actin and histone H3 were used as loading controls. c Concentrations of inflammatory cytokines (TNF-α, IL-1β, IL-6, and IL-10) were measured by ELISA. Data are presented as the means ± SD. *P < 0.05, **P < 0.01, ***P < 0.001 versus the Sham group, #P < 0.05, ##P < 0.01, ###P < 0.001 versus the Lenti-NC group. nHMGB1: nuclear HMGB1; cyHMGB1: cytosolic HMGB1; nβ-actin: nuclear β-actin; cyβ-actin: cytosolic β-actin
miR-181c-5p knockdown exacerbates ischemic brain inflammation in a mouse MCAO model (n = 3). a RT-PCR was performed on RNA collected from the ipsilateral brain after 24 h of MCAO and showed the expression of miR-181c-5p, Malat1 and HMGB1 in the miR-181c-5p inhibitor group. b Representative photographs of Western blots for nHMGB1, cyHMGB1 and HMGB1 and the ratio of cyHMGB1 to nHMGB1. β-actin and histone H3 were used as loading controls. c Concentrations of inflammatory cytokines (TNF-α, IL-1β, IL-6, and IL-10) were measured by ELISA. d Luciferase reporter assay revealed that Lenti-miR-181c-5p decreased the luciferase activity of HMGB1-WT but not of HMGB1-Mut. Data are presented as the means ± SD. *P < 0.05, **P < 0.01, ***P < 0.001 versus the Sham group, #P < 0.05, ##P < 0.01 versus the NC inhibitor group. nHMGB1: nuclear HMGB1; cyHMGB1: cytosolic HMGB1; nβ-actin: nuclear β-actin; cyβ-actin: cytosolic β-actin
miR-181c-5p inhibitor rescues HMGB1 inhibition induced by Malat1 downregulation
Finally, to further evaluate the specific crosstalk between Malat1 and HMGB1 through competitive binding of miR-181c-5p, we carried out rescue experiments. As shown in Fig. 7a, the expression of HMGB1 decreased (P < 0.01) when Malat1 was knocked down and increased (P < 0.01) when miR-181c-5p was knocked down, which was in line with our previous results. However, when Malat1 and miR-181c-5p were knocked down at the same time, there was no significant difference in the expression of HMGB1 compared with the Model group. In addition, Western blot assay showed coincident results (Fig. 7b). Overall, Malat1 upregulates the expression of HMGB1 by competitively binding to miR-181c-5p, thereby exacerbating inflammation after stroke.
miR-181c-5p inhibitor rescues HMGB1 inhibition induced by Malat1 downregulation (n = 3). a RT-PCR was performed on mRNA collected from the ipsilateral brain after 24 h of MCAO and showed the expression of miR-181c-5p and HMGB1 in the Lenti-miR-181c-5p or miR-181c-5p inhibitor groups. b Representative photographs of Western blots for nHMGB1, cyHMGB1 and HMGB1 and the ratio of cyHMGB1 to nHMGB1. β-actin and histone H3 were used as loading controls. Data are presented as the means ± SD. *P < 0.05, **P < 0.01, ***P < 0.001 versus the Sh-NC + NC inhibitor group. nHMGB1: nuclear HMGB1; cyHMGB1: cytosolic HMGB1; nβ-actin: nuclear β-actin; cyβ-actin: cytosolic β-actin
Discussion
In this study, the level of Malat1 was augmented in mouse brain after stroke, consistent with previous studies [15, 25]. More importantly, our results are the first to illustrate that BBR could exert anti-inflammatory effects via the Malat1/miR-181c-5p/HMGB1 signaling pathway after ischemic stroke (Fig. 8).
Ischemic stroke is a devastating disease with high mortality and long-term disability worldwide. The inflammatory response after ischemic stroke is characterized by increased permeability of the blood–brain barrier, cerebral edema, infiltration of peripheral leukocytes, and activation of microglia and astrocytes, releasing a large amount of tumor necrosis factors, interleukins, inflammatory chemokines, and potential cytotoxic factors [26, 27]. After stroke, a sterile inflammatory cascade is initiated, aggravating brain damage. Several studies have shown that HMGB1, a proinflammatory factor, is involved in aseptic inflammation in cerebral ischemia–reperfusion injury [28]. In addition, patients with acute cerebral ischemia have a significantly high level of serum inflammatory markers TNF-α, IL-1β, IL-6, and a high concentration of HMGB1 in the serum is closely related to poor prognosis in patients [29].
Recently, lncRNAs have been recognized as playing versatile roles in biological process and clinical outcomes. LncRNA SNHG1 can alleviate OGD-induced injury in BMECs via the miR-338/HIF-1α axis [30]. LncRNA SETD5-AS1 can stimulate neuron death in stroke by promoting PTRN expression [31]. LncRNA HOTTIP can alleviate OGD-induced neuronal injury by modulating the miR-143/hexokinase 2 pathways [32]. In light of the results from multiple investigations, Malat1, first recognized to be correlated with metastasis of lung tumors, is shown to be aberrantly expressed in most human tissues and is associated with human cardiovascular diseases, cancers, and CNS dysfunctions [33]. Accumulating evidence has shown that Malat1 is sensitive to inflammation induced by pathological stimuli. For example, Wang et al. reported that Malat1 promoted the HG-H/R-induced inflammatory response in the progression of DM-associated cerebral ischemic stroke [34]. Dong et al. found that Malat1 knockdown ameliorated diabetic retinopathy and inhibited MCP-1 release in the retinas of STZ-induced diabetic rats [17]. Zhang et al. showed that downregulation of Malat1 enhanced cell proliferation ability and reduced apoptosis in MCAO/R brains [15]. However, the molecular mechanisms of Malat1 and HMGB1 in ischemic stroke remain unknown. In our study, we discovered that Malat1 and cytoplasmic HMGB1 expression were increased in stroke brain tissue, while BBR pretreatment could downregulate Malat1 and cytoplasmic HMGB1 and then attenuate the inflammatory response. In addition, we systematically identified the link between Malat1 and HMGB1 by Malat1 knockdown or overexpression techniques and indicated that Malat1 presented a positive correlation with HMGB1 and the inflammatory response, in agreement with previous studies. The results of the rescue experiment also confirmed that Malat1 is a key target for BBR-mediated inhibition of HMGB1 nucleus release in stroke inflammation.
Although the role of Malat1 in ischemic stroke inflammation has been previously researched, the specific regulatory mechanism remains unknown. Salmena et al. first proposed a ceRNA hypothesis, describing that lncRNA acts as a natural miRNA sponge and could inhibit the function of the target mRNA [35]. For instance, lncRNA GAS5 promotes the progression of ischemic stroke by acting as a ceRNA for miR-137 [36]. LncRNA MEG3 functions as a ceRNA and competes with programmed cell death 4 (PDCD4) mRNA for directly binding to miR-21 to mediate ischemic neuronal death [37]. LncRNA SNHG1 promotes BMEC survival by acting as a ceRNA for miR-18a via HIF-1α/VEGF signaling [38]. In our work, we first confirmed that Malat1 was expressed in the cytoplasm by FISH assay. HMGB1 was predicted a miR-181c-5p target by Targetscan software. Additionally, RNAhybrid modeling showed that Malat1 could bind strongly to miR-181c-5p. A study has also proven that miR-181c is a potential candidate target in neuroinflammation [39]. Zhang et al. reported that miR-181c inhibited the inflammatory response in OGD microglia via the TLR4/NF-κB signaling pathway [24]. Li et al. found that miR-181c from hUCMSC-derived exosomes could suppress proinflammatory factors by downregulating TLR4 signaling [40]. Thus, to identify whether the levels of Malat1 and miR-181c-5p inversely correlated in ischemic stroke, knockdown and overexpression techniques were conducted in animal MCAO models. The results demonstrated that miR-181c-5p expression was negatively linked to Malat1. Then, we further investigated whether the levels of miR-181c-5p and HMGB1 were inversely correlated by transfection with Lenti-miR-181c-5p and miR-181c-5p inhibitor, and the results were completely as expected. Furthermore, the exact binding of Malat1 to miR-181c-5p and HMGB1 to miR-181c-5p were confirmed by a luciferase reporter assay. Taken together, the data validated that Malat1 regulated HMGB1 by competing for miR‐181c-5p binding in BBR-mediated ischemic stroke inflammation.
Lastly, how BBR downgrades Malat1 remains to be further explored. The mechanism could be possibly related to BBR inhibition of methyltransferase or Malat1 promoter methylation and promotion of acetylation. In addition, Malat1 plays a role in the regulation of HMGB1 in the cytoplasm. The expression of miR-181c-5p is also mainly located in the cytoplasm. However, the reason that a miR-181c-5p inhibitor can increase nuclear HMGB1 expression may be associated with the degradation of an unknown nuclear protein and needs to be explored further.
Conclusion
In summary, we illustrated that BBR could alleviate inflammation in ischemic stroke through the Malat1/miR-181c-5p/HMGB1 axis, which had a pivotal role in regulating inflammation. To understand this mechanism more comprehensively, future endeavors on whether pharmacological manipulation of the Malat1 gene affects stroke outcomes and neurological function will be carried out.
References
Pandian JD, Gall SL, Kate MP, Silva GS, Akinyemi RO, Ovbiagele BI, et al. Prevention of stroke: a global perspective. Lancet. 2018;392:1269–78.
Della-Morte D, Guadagni F, Palmirotta R, Testa G, Caso V, Paciaroni M, et al. Genetics of ischemic stroke, stroke-related risk factors, stroke precursors and treatments. Pharmacogenomics. 2012;13:595–613.
Zhang ZG, Chopp M. Neurorestorative therapies for stroke: underlying mechanisms and translation to the clinic. Lancet Neurol. 2009;8:491–500.
Li D, Wang C, Yao Y, Chen L, Liu G, Zhang R, et al. mTORC1 pathway disruption ameliorates brain inflammation following stroke via a shift in microglia phenotype from M1 type to M2 type. FASEB J. 2016;30:3388–99.
Brennan-Minnella AM, Won SJ, Swanson RA. NADPH oxidase-2: linking glucose, acidosis, and excitotoxicity in stroke. Antioxid Redox Signal. 2015;22:161–74.
Anrather J, Iadecola C. Inflammation and stroke: an overview. Neurotherapeutics. 2016;13:661–70.
Guo JM, Liu AJ, Su DF. Genetics of stroke. Acta Pharmacol Sin. 2010;31:1055–64.
Zampa F, Hartzell AL, Zolboot N, Lippi G. Non-coding RNAs: the gatekeepers of neural network activity. Curr Opin Neurobiol. 2019;57:54–61.
Chandran R, Mehta SL, Vemuganti R. Non-coding RNAs and neuroprotection after acute CNS injuries. Neurochem Int. 2017;111:12–22.
Ng SY, Lin L, Soh BS, Stanton LW. Long noncoding RNAs in development and disease of the central nervous system. Trends Genet. 2013;29:461–8.
Aprea J, Calegari F. Long non-coding RNAs in corticogenesis: deciphering the non-coding code of the brain. EMBO J. 2015;34:2865–84.
Lander ES, Linton LM, Birren B, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921.
Indolfi C, Iaconetti C, Gareri C, Polimeni A, Rosa SD. Non-coding RNAs in vascular remodeling and restenosis. Vasc Pharmacol. 2019;114:49–63.
Sun Y, Ma L. New insights into long non-coding RNA MALAT1 in cancer and metastasis. Cancers. 2019;11:E216.
Zhang T, Wang H, Li Q, Fu J, Huang J, Zhao Y. MALAT1 activates the p53 signaling pathway by regulating MDM2 to promote ischemic stroke. Cell Physiol Biochem. 2018;50:2216–28.
Zhou HJ, Wang LQ, Wang DB, Yu JB, Zhu Y, Xu QS, et al. Long noncoding RNA MALAT1 contributes to inflammatory response of microglia following spinal cord injury via the modulation of a miR-199b/IKKbeta/NF-kappaB signaling pathway. Am J Physiol Cell Physiol. 2018;315:C52–C61.
Dong N, Xu B, Shi H. Long noncoding RNA MALAT1 acts as a competing endogenous RNA to regulate Amadori-glycated albumin-induced MCP-1 expression in retinal microglia by a microRNA-124-dependent mechanism. Inflamm Res. 2018;67:913–25.
Chaudhry SR, Hafez A, Rezai Jahromi B, Kinfe TM, Lamprecht A, Niemela M, et al. Role of damage associated molecular pattern molecules (DAMPs) in aneurysmal subarachnoid hemorrhage (aSAH). Int J Mol Sci. 2018;19:E2035.
Gulke E, Gelderblom M, Magnus T. Danger signals in stroke and their role on microglia activation after ischemia. Ther Adv Neurol Disord. 2018;11:1–14.
Richard SA, Sackey M, Su Z, Xu H. Pivotal neuroinflammatory and therapeutic role of high mobility group box 1 in ischemic stroke. Biosci Rep. 2017;37:1–28.
Zhu JR, Lu HD, Guo C, Fang WR, Zhao HD, Zhou JS, et al. Berberine attenuates ischemia–reperfusion injury through inhibiting HMGB1 release and NF-κB nuclear translocation. Acta Pharmacol Sin. 2018;39:1706–15.
Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303:83–86.
Ma Q, Zhao H, Tao Z, Wang R, Liu P, Han Z, et al. MicroRNA-181c exacerbates brain injury in acute ischemic stroke. Aging Dis. 2016;7:705–14.
Zhang L, Li YJ, Wu XY, Hong Z, Wei WS. MicroRNA‐181c negatively regulates the inflammatory response in oxygen‐glucose‐deprived microglia by targeting Toll‐like receptor 4. J Neurochem. 2015;132:713–23.
Guo D, Ma J, Yan L, Li T, Li Z, Han X, et al. Down-regulation of Lncrna MALAT1 attenuates neuronal cell death through suppressing beclin1-dependent autophagy by regulating Mir-30a in cerebral ischemic stroke. Cell Physiol Biochem. 2017;43:182–94.
Yong HYF, Rawji KS, Ghorbani S, Xue M, Yong VW. The benefits of neuroinflammation for the repair of the injured central nervous system. Cell Mol Immunol. 2019;16:540–6.
Lambertsen KL, Finsen B, Clausen BH. Post-stroke inflammation-target or tool for therapy? Acta Neuropathol. 2019;137:693–714.
Chamorro Á, Dirnagl U, Urra X, Planas AM. Neuroprotection in acute stroke: targeting excitotoxicity, oxidative and nitrosative stress, and inflammation. Lancet Neurol. 2016;15:869–81.
Tsukagawa T, Katsumata R, Fujita M, Yasui K, Akhoon C, Ono K, et al. Elevated serum high-mobility group box-1 protein level is associated with poor functional outcome in ischemic stroke. J Stroke Cereb Dis. 2017;26:2404–11.
Yang X, Zi XH. LncRNA SNHG1 alleviates OGD induced injury in BMEC via miR-338/HIF-1alpha axis. Brain Res. 2019;1714:174–81.
Miao SY, Miao SM, Cui RT, Yu AL, Miao ZJ. SETD5-AS1 stimulates neuron death in stroke via promoting PTEN expression. Eur Rev Med Pharm Sci. 2018;22:6035–41.
Wang Y, Li G, Zhao L, Lv J. Long noncoding RNA HOTTIP alleviates oxygen-glucose deprivation-induced neuronal injury via modulating miR-143/hexokinase 2 pathway. J Cell Biochem. 2018;119:10107–17.
Zhang X, Hamblin MH, Yin KJ. The long noncoding RNA Malat1: its physiological and pathophysiological functions. RNA Biol. 2017;14:1705–14.
Wang LQ, Zhou HJ. LncRNA MALAT1 promotes high glucose-induced inflammatory response of microglial cells via provoking MyD88/IRAK1/TRAF6 signaling. Sci Rep. 2018;8:8346.
Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the rosetta stone of a hidden RNA language? Cell. 2011;146:353–8.
Chen F, Zhang L, Wang E, Zhang C, Li X. LncRNA GAS5 regulates ischemic stroke as a competing endogenous RNA for miR-137 to regulate the Notch1 signaling pathway. Biochem Biophys Res Commun. 2018;496:184–90.
Yan H, Rao J, Yuan J, Gao L, Huang W, Zhao L, et al. Long non-coding RNA MEG3 functions as a competing endogenous RNA to regulate ischemic neuronal death by targeting miR-21/PDCD4 signaling pathway. Cell Death Dis. 2017;8:3211.
Zhang L, Luo X, Chen F, Yuan W, Xiao X, Zhang X, et al. LncRNA SNHG1 regulates cerebrovascular pathologies as a competing endogenous RNA through HIF-1alpha/VEGF signaling in ischemic stroke. J Cell Biochem. 2018;119:5460–72.
Hutchison ER, Kawamoto EM, Taub DD, Lal A, Abdelmohsen K, Zhang Y, et al. Evidence for miR-181 involvement in neuroinflammatory responses of astrocytes. Glia. 2013;61:1018–28.
Li X, Liu L, Yang J, Yu Y, Chai J, Wang L, et al. Exosome derived from human umbilical cord mesenchymal stem cell mediates MiR-181c attenuating burn-induced excessive inflammation. EBioMedicine. 2016;8:72–82.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (grant No. 81773987), the Program of Nanjing Health and Family Planning Commission (YKK188107) and the Hospital Pharmaceutical Research Program of Nanjing Pharmaceutical Commission (2017YX001).
Author information
Authors and Affiliations
Contributions
JRZ and JGS conceived and designed the study. DWC, MML and RD performed the experiments. MML, RD and LN analyzed the data. DWC and WRF wrote the paper. JRZ, LN and JGS reviewed and edited the manuscript. All authors read and approved the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
These authors are co-first authors: Ding-wen Cao, Man-man Liu, Rui Duan
Rights and permissions
About this article
Cite this article
Cao, Dw., Liu, Mm., Duan, R. et al. The lncRNA Malat1 functions as a ceRNA to contribute to berberine-mediated inhibition of HMGB1 by sponging miR-181c-5p in poststroke inflammation. Acta Pharmacol Sin 41, 22–33 (2020). https://doi.org/10.1038/s41401-019-0284-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41401-019-0284-y
Keywords
This article is cited by
-
Brain alarm by self-extracellular nucleic acids: from neuroinflammation to neurodegeneration
Journal of Biomedical Science (2023)
-
Functions and mechanisms of lncRNA MALAT1 in cancer chemotherapy resistance
Biomarker Research (2023)
-
The role of long noncoding ribonucleic acids in the central nervous system injury
Molecular and Cellular Biochemistry (2023)
-
LncRNA MALAT1 and Ischemic Stroke: Pathogenesis and Opportunities
Molecular Neurobiology (2023)
-
LncRNA MALAT1 Promoted Neuronal Necroptosis in Cerebral Ischemia-reperfusion Mice by Stabilizing HSP90
Neurochemical Research (2023)