Abstract
Ischemic heart diseases (IHDs) cause great morbidity and mortality worldwide, necessitating effective treatment. Salvianic acid A sodium (SAAS) is an active compound derived from the well-known herbal medicine Danshen, which has been widely used for clinical treatment of cardiovascular diseases in China. This study aimed to confirm the cardioprotective effects of SAAS in rats with myocardial infarction and to investigate the underlying molecular mechanisms based on proteome and transcriptome profiling of myocardial tissue. The results showed that SAAS effectively protected against myocardial injury and improved cardiac function. The differentially expressed proteins and genes included important structural molecules, receptors, transcription factors, and cofactors. Functional enrichment analysis indicated that SAAS participated in the regulation of actin cytoskeleton, phagosome, focal adhesion, tight junction, apoptosis, MAPK signaling, and Wnt signaling pathways, which are closely related to cardiovascular diseases. SAAS may exert its cardioprotective effect by targeting multiple pathways at both the proteome and transcriptome levels. This study has provided not only new insights into the pathogenesis of myocardial infarction but also a road map of the cardioprotective molecular mechanisms of SAAS, which may provide pharmacological evidence to aid in its clinical application.
Similar content being viewed by others
Introduction
Ischemic heart diseases (IHDs) cause great morbidity and mortality worldwide, calling for urgent solutions [1, 2]. With a long history of clinical use, herbal extracts and their derivatives have made great contributions to the treatment of IHDs [3]. Danshen, the dried root of Salvia miltiorrhiza, has been widely used for clinical treatment of cardiovascular diseases such as coronary heart disease, angina pectoris, hyperlipidemia, acute ischemic stroke, and cerebrovascular disease in China for thousands of years [4, 5]. Salvianic acid A sodium (SAAS, Fig. 1a) is an active compound derived from Danshen. Injectable SAAS is a new drug developed by our group that has been entered into phase I clinical trials for the treatment of coronary heart disease and stable angina pectoris conducted by the China Food and Drug Administration (CFDA). However, the underlying cardioprotective mechanisms of SAAS remain to be further elucidated to support its clinical use.
The development of high-throughput omics technologies, including genomics, transcriptomics, proteomics, and metabolomics, has provided a powerful approach to investigate the underlying pathogenesis of diseases and modes of drug action [6,7,8,9]. Although the genetic code remains constant over the lifetime of an animal, its expression from gene to mRNA to protein is highly dynamic and influenced by various factors, such as environmental, biological, and processing factors [10]. Proteomic and transcriptomic techniques have provided promising and powerful tools for mechanistic investigation, which has been reflected by the increasing number of studies emerging in the literature [11,12,13]. As a comprehensive and quantitative record of protein expression influenced by a series of biological perturbations, proteome, particularly, shows the link between the genomic and functional effects of a drug. Integration of data from transcriptome and proteome analysis enables a holistic view of drug modes of action at the center of biological systems.
In this study, Sprague-Dawley rats that underwent left anterior descending artery ligation (a model of acute myocardial infarction) or sham surgery were treated with or without SAAS (Fig. 1b). Proteome and transcriptome analysis of myocardial tissue was designed to gain insights into the underlying molecular networks of the cardioprotective mechanisms of SAAS. This study has provided not only new insights into the pathogenesis of myocardial infarction but also a road map of the cardioprotective molecular mechanisms of SAAS to aid in its clinical use. To the best of our knowledge, this is the first study in which the potential underlying mechanisms of myocardial infarction have been comprehensively investigated at both the transcriptome and proteome levels.
Materials and methods
Materials
SAAS was supplied by our group (purity ≥99%). Injectable SAAS (20 mg/mL) was produced by Changchun Puhua Pharmaceutical Co., Ltd. (Changchun, China), and dissolved in saline to various concentrations. Kits for determination of lactate dehydrogenase (LDH), creatine kinase (CK), creatine kinase isoenzyme (CK-MB), and cardiac troponin I (cTnI) were obtained from Jiancheng Bioengineering Institute (Nanjing, China). Antibodies against KCNN2, p63, HTSF1, and GAPDH were obtained from Abcam (Cambridge, MA, USA). Antibody against KC1A was acquired from Cell Signaling Technologies (Danvers, MA, USA). IRDye 800-conjugated anti-mouse and anti-rabbit secondary antibodies were obtained from Rockland Immunochemicals, Inc. (Philadelphia, PA, USA). All chemicals were purchased from Sigma-Aldrich (St. Louis, MO, USA) unless specifically noted.
Animals and establishment of the myocardial infarction model
All animal experiments were performed at the Centre of Laboratory Animals of the Second Military Medical University (Shanghai, China) in accordance with the relevant national legislation and local guidelines. Male Sprague-Dawley rats (140–160 g) were supplied by Shanghai Laboratory Animal Co. and given adaptive feeding for 24 h before the experiments. The animals were randomly divided into four groups: (1) a sham group, in which the rats underwent the same surgical procedures as the rats in the other groups without ligation of the left anterior descending coronary artery (n = 20); (2) a myocardial infarction (MI) model group, in which MI rats received saline at the same volume as that used for injection of SAAS (n = 30); (3) a 24 mg/kg SAAS group, in which MI rats received SAAS at 24 mg/kg every day (n = 30); and (4) a 96 mg/kg SAAS group, in which MI rats received SAAS at 96 mg/kg every day (n = 30). Before surgery, animals in the SAAS treatment groups were administered SAAS intraperitoneally at doses of 24 and 96 mg/kg for ten days, while animals in sham and model groups received the same volume of saline. The myocardial infarction model was induced by ligation of left anterior descending coronary artery as described previously [14]. Briefly, the rats were anesthetized with ether and placed on a warming pad for maintenance of body temperature. The thorax of each rat was shaved and disinfected with 75% alcohol. A left thoracotomy was performed in the intercostal space at the muscle layer between the third and fourth rib. The pectoralis muscle groups were separated transversely, exposing the rib cage. The pericardium was gently opened, and the left coronary artery was isolated. A 3/8 needle carrying a 5.0 suture was inserted approximately 2 mm below the root of the left atrial appendage, and the left anterior descending artery was rapidly ligated. The thorax was closed and the incisions were sutured layer by layer. Then the heart was gently massaged to recover the spontaneous breathing of the rat. For sham operation, animals underwent the same surgical procedure, and ligatures were placed in identical locations but the ligatures were not tied. All animals were administered 100 000 U of penicillin intraperitoneally for 3 days to prevent infection. There were no mortalities in the sham group, and the surgical mortality in MI groups was 27.8% (25/90) during 72 h observation period. After surgery, animals in each group received the same dose of SAAS or saline as before for 3 weeks.
Echocardiography
Echocardiographic studies were performed on the 21st day after the surgery. Eight rats from each group were lightly anesthetized with controlled inhalation of a mixture of 1.5% isoflurane and 0.5 L/min oxygen and placed in dorsal recumbency on a heated 37 °C platform to maintain body temperature throughout the recordings. The anterior chest was shaven, and a layer of acoustic coupling gel was applied to the thorax. M-mode echocardiography was measured using an Esaote MyLab™ One/Touch equipped with an SL3116 linear transducer (Esaote Biomedica, Genova, Italy).
Sample collection
Blood samples (1 mL) were taken from orbital venous plexus 24 h after the surgery. Serum was obtained from the whole blood after centrifugation at 3000 × g for 10 min at 4 °C and was stored at −80 °C for future use. After echocardiographic measurement, the rats were intraperitoneally anesthetized with 10% chloral hydrate (0.3 mL/100 g). The hearts were harvested, rinsed with cold PBS, and gently squeezed with forceps to minimize residual blood contamination. Fat, vessels, and other noncardiac tissue were trimmed. The atrium was resected along the coronary sulcus, and the left ventricle was harvested. The moisture was absorbed, and then the left ventricle was fixed in 10% neutral buffered formalin or snap-frozen in liquid nitrogen and stored at −80 °C for further analysis.
Determination of serum enzymes
The levels of LDH, CK, CK-MB, and cTnI were measured using commercialized enzyme-linked immunosorbent assay kits following the manufacturer’s instructions (Nanjing Jiancheng Bioengineering Institute, China), and the results were read on a microplate absorbance reader (Bio-Rad Instruments, USA).
Histological analysis
Heart tissues were fixed with 4% paraformaldehyde (PFA), embedded in paraffin, cut into 3–5 μm thick sections and stained with hemat oxylin and eosin (H&E) and Masson’s trichrome stain. All images were obtained at ×200 magnification under an inverted optical microscope (DP73; Olympus, Tokyo, Japan).
Protein extraction and proteome analysis
Protein extraction
Four myocardial tissue samples (50 mg of each from the same group) were mixed, ground in liquid nitrogen, sonicated in lysis buffer (8 M urea and 40 mM Tris-HCl with 1 mM PMSF, 2 mM EDTA and 10 mM DTT, pH 8.5), placed into a TissueLyser for 2 min at 50 Hz and centrifuged at 12 000 × g for 20 min at 4 °C. The supernatant was incubated with 10 mM dithiothreitol (DTT) at 56 °C for 1 h and alkylated with 55 mM iodoacetamide (IAM) in the dark at room temperature for 45 min. After centrifugation at 12 000 × g for 20 min at 4 °C, protein in the supernatant was quantified by the Bradford method.
Protein digestion
Protein samples (100 μg) with 8 M urea were diluted fourfold with 100 mM TEAB. Trypsin Gold (Promega, Madison, WI, USA) was used to digest the proteins with a protein:trypsin ratio of 40:1 at 37 °C overnight. After trypsin digestion, the peptides were desalted with a Strata X C18 column (Phenomenex) and vacuum-dried according to the manufacturer’s protocol.
Peptide labeling
The peptides were dissolved in 30 μL of 0.5 M TEAB with vortexing. After the iTRAQ labeling reagents had returned to ambient temperature, they were added to appropriate samples and mixed. Peptide labeling was performed with an iTRAQ Reagent 8-plex Kit according to the manufacturer’s protocol (AB Sciex, Foster City, CA, USA). The peptides labeled with different reagents were combined, desalted with a Strata X C18 column (Phenomenex), and vacuum-dried.
Fractionation
The peptides were separated on a Shimadzu LC-20AB HPLC pump system coupled with a high pH RP column. The peptides were reconstituted with buffer A (5% ACN, 95% H2O, pH adjusted to 9.8 with ammonia) to 2 mL and loaded onto a column containing 5-μm particles (Phenomenex). The peptides were separated at a flow rate of 1 mL/min with a gradient of 5% buffer B (5% H2O, 95% ACN, pH adjusted to 9.8 with ammonia) for 10 min, 5%–35% buffer B for 40 min, and 35%–95% buffer B for 1 min. The system was then maintained at 95% buffer B for 3 min and decreased to 5% within 1 min before equilibration for 10 min. Elution was monitored by measuring the absorbance at 214 nm, and fractions were collected every 1 min. The eluted peptides were pooled as 20 fractions and vacuum-dried. More details about the HPLC analysis, mass spectrometry detection, and bioinformatics analysis are given in the Supplementary Materials.
Transcriptome sample preparation and sequencing
Total RNA from 12 myocardium samples was extracted using TRIzol reagent (Invitrogen) and then quantified and qualified with an Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA, USA). Total RNA (1 μg) with an RIN value above 7 was used for library preparation according to the manufacturer’s protocol (NEBNext® Ultra™ RNA Library Prep Kit for Illumina®). Libraries with different indices were multiplexed and loaded on an Illumina HiSeq instrument according to the manufacturer’s instructions (Illumina, San Diego, CA, USA). Sequencing was carried out using a 2 × 150 bp paired-end (PE) configuration. Image analysis and base calling were conducted with HiSeq Control Software (HCS) + OLB + GAPipeline-1.6 (Illumina) on the HiSeq instrument. Raw sequencing reads were obtained by removing adapters and low-quality reads. The clean reads were obtained through strict quality control steps and subjected to further analysis according to the standard Ion Proton RNA-seq protocol, as described in the Supplementary Materials. A count table for differential expression analysis was prepared with HTSeq v0.6.1 software. The standalone script htseq-count counts how many aligned reads overlap the exons for each gene, and its input file contains a SAM/BAM file and a GTF or GFF file with gene models. The details of the transcriptomic and bioinformatics analysis are given in the Supplementary Materials.
Differential expression analysis
Differential expression analysis was performed with the DESeq Bioconductor package, a model based on negative binomial distribution. After the results were adjusted with Benjamini and Hochberg’s approach for controlling the false discovery rate, proteins/genes with P-values < 0.05 were regarded as differentially expressed and used for further bioinformatics analysis.
Bioinformatics analysis
GO-TermFinder was used to identify Gene Ontology (GO) terms that annotated a list of enriched proteins/genes with significant P-values less than 0.05. The Kyoto Encyclopedia of Genes and Genomes (KEGG) is a collection of databases associated with genomes, biological pathways, diseases, drugs, and chemical substances (http://en.wikipedia.org/wiki/KEGG). In-house scripts were used to analyze the enrichment of significant differentially expressed proteins (DEPs)/genes (DEPs/DEGs) in KEGG pathways. The Search Tool for the Retrieval of Interacting Genes (STRING) database was used to analyze the protein interactions and construct interaction networks of the DEPs/DEGs (http://string-db.org/). The input files were imported into Cytoscape v3.5.1 software to visualize complex networks and integrate the networks with any available types of attribute data (http://www.cytoscape.org/download.php).
Statistical analysis
Statistical analysis was performed with GraphPad Prism 5.0 software (GraphPad Software Inc., La Jolla, CA, USA). All experiments were performed at least three times in a parallel manner. The data are expressed as the mean ± standard deviation, and significant differences were estimated with Student’s t-test. Significance is indicated as follows: #P < 0.05 and ##P < 0.01 compared with the sham group; *P < 0.05 and **P < 0.01 compared with the myocardial infarction model group.
Results
SAAS improved cardiac function and protected against myocardial injury
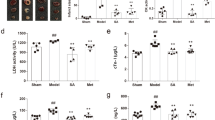
Since myocardial injury disrupts myocardial cell membrane integrity, LDH, CK, CK-MB, and cTnI in serum are often regarded as myocardial markers. Twenty-four hours after surgery, the serum contents of LDH, CK, CK-MB, and cTnI in the model group were markedly increased compared with those in the sham group. In the groups subjected to MI and treated with SAAS (24 or 96 mg/kg), the levels of these myocardial markers were significantly decreased compared to those in the MI model group (P < 0.01; Fig. 2a). The LDH levels were decreased by 9.2% and 28.9% (P < 0.01), CK release was decreased by 25.0% and 36.9% (P < 0.01), the CK-MB levels were decreased by 6.7% and 45.9% (P < 0.01), and cTnI was decreased by 37.3% and 59.2% (P < 0.05) in the 24 and 96 mg/kg SAAS groups, respectively, compared to the MI model group. During 72 h observation period after the surgery, there were no mortalities in the sham group, while the mortality percentages in the model, 24 and 96 mg/kg SAAS groups were 40% (12/30), 26.7% (8/30), and 16.7% (5/30), respectively.
Cardioprotective effects of SAAS in rats with myocardial infarction. a The contents of lactic dehydrogenase (LDH), creatine kinase (CK), creatine kinase isoenzyme (CK-MB), and cardiac troponin I (cTnI) in serum of rats. Data were expressed as the mean ± SD, with ##P < 0.01 compared with the sham group, and **P < 0.01 compared with myocardial infarction model group (n = 8). b The representative pictures of echocardiographic images (upper panel, n = 8) and histological analysis stained with hematoxylin & eosin (middle panel, n = 4) and Masson’s trichrome staining (lower panel, n = 4, ×200 magnification) ##P<0.01 compared with the sham group
Cardiac function was examined by echocardiography on the 21st day after coronary artery ligation. As shown in Table 1 and Fig. 2b (upper panel), the left ventricular internal diameter in diastole (LVIDd) was increased by 26.9% and the internal diameter in systole (LVIDs) was increased by 93.5% (P < 0.01) in the MI model group compared to the sham group. Concomitantly, in the MI model group, the left ventricular ejection fraction (LVEF) and fractional shortening (LVFS) were decreased by 47.7% and 57.0%, respectively, compared with those in the sham group (P < 0.01). SAAS treatment at doses of 24 and 96 mg/kg effectively improved left ventricular function compared with the model group. LVIDd was decreased by 12.8% and 17.1% (P < 0.01), LVIDs was decreased by 27.2% and 33.7% (P < 0.01), LVEF was increased by 47.8% and 67.6% (P < 0.01), and LVFS was increased by 59.5% and 98.6% (P < 0.01) in 24 and 96 mg/kg SAAS groups, respectively, compared to the model group.
Nitrotetrazolium blue chloride (NBT) staining showed obvious myocardial injury due to MI, while SAAS reduced the infarct size (Supplementary Fig. S1). H&E and Masson’s trichrome staining illustrated that the epicardium and endocardium of left ventricle in the sham group were intact, presenting neat myocardial cell arrangement with adjacent myofibrils. Conversely, in animals subjected to ligation surgery, myocardial cells were infarcted, swollen, and degenerated. These changes were accompanied by cardiac necrosis, loss of transverse striations, and thinning of the ventricular wall. SAAS therapy significantly decreased cardiac apoptosis and attenuated cardiac fibrosis in MI rats, another feature of ventricular remodeling (Fig. 2b, middle and lower panels).
Cardioprotective effects of SAAS against myocardial infarction at the proteome level
Proteomics analysis of eight myocardial tissue samples showed that a total of 5876 peptides and 1290 proteins were identified with a 1% false discovery rate (FDR). The proteins were quantified with IQuant software (BGI, Shenzhen, China), and the DEPs were defined as those with fold change >1.2 and P < 0.05. In total, 233 proteins were differentially expressed in the model-vs-sham comparison, 155 of which were upregulated and 78 of which were downregulated. SAAS therapy led to significant changes in 73 and 78 proteins in the 24 and 96 mg/kg SAAS groups, respectively, compared with the sham group (Fig. 3a and Supplementary Table S1). Cluster analysis using Euclidean distance and a hierarchical algorithm was used to show the protein expression patterns in each group (Fig. 3b).
Cardioprotective effects of SAAS on myocardial infarction at the proteome level. a Summary of differentially expressed proteins (DEPs). Red represents upregulation and blue represents downregulation. b Heat map of cluster analysis using Euclidean distance and hierarchical algorithm in each group. c KEGG pathway functional enrichment of DEPs. X-axis represents enrichment factor. Y-axis represents pathway name. The color indicates the P-value (high: blue; low: red), the lower P-value indicates the more significant enrichment. Point size indicates DEPs number. The bigger dots refer to larger amount. Rich factor refers to the value of enrichment factor, which is the quotient of foreground value (the number of DEPs) and background value (total protein amount). The larger the value, the more significant enrichment
The DEPs were assigned to GO categories to determine their functional classifications. The DEPs in the MI model group compared with the sham group were mainly associated with the single-organism process, cytoskeleton and calcium ion binding terms in the biological process, cellular component and molecular function categories, respectively. In the 96 mg/kg SAAS group compared to the model group, the most enriched GO terms were the regulation of nitrogen compound metabolic process, cilium and binding terms in the biological process, cellular component and molecular function categories, respectively (Supplementary Fig. S2 and Table S2).
KEGG enrichment analysis showed that the top five most enriched pathways in the model-vs-sham comparison were the regulation of actin cytoskeleton, phagosome, focal adhesion, tight junction, and apoptosis pathways. These results were highly consistent with those of the 96 mg/kg SAAS-vs-model comparison (Fig. 3c and Supplementary Table S3).
The DEPs were exported to the STRING database and Cytoscape v3.5.1 software to create a landscape for protein–protein interactions. STRING analysis showed that 36 DEPs in the model-vs-sham comparison were functionally linked (Supplementary Fig. S3) with Actb (actin), Rac1 (Ras-related C3 botulinum toxin substrate 1), Myh10 (myosin-10), Myh9 (myosin-9), Actn4 (alpha-actinin-4), Acta1 (actin, alpha skeletal muscle), and Ptk2 (focal adhesion kinase 1) among the most linked. Cytoscape v3.5.1 was used to visualize the protein interactions with topological parameters including the node degree, betweenness centrality (BC) index, network density, and average number of neighbors. The principal hub interactions considering the BC parameter were the Actb-Rac1, Actb-Actn4, and Actb-Myl9 interactions (Fig. 4a).
Network of protein–protein interactions and effects of SAAS on the expression of several proteins. a Visualized protein–protein interactions network of DEPs in model-vs-sham comparison groups using Cytoscape v3.5.1 software. The red dots refer to upregulated proteins and the blue dots refer to downregulated proteins. Thickness of the line represents strength of the correlation between the two proteins. b Protein levels of KCNN2, p63, ARL11, HTSF1, ATMIN, and KC1A expression by proteome analysis and c identified by Western blotting analysis
Additionally, SAAS effectively reversed the expression changes induced in an array of proteins by MI injury. KCNN2 (small conductance calcium-activated potassium channel protein 2), p63 (tumor protein 63), ARL11 (ADP-ribosylation factor-like protein 11), ATMIN (ATM interactor), and KC1A (casein kinase I isoform alpha) were upregulated in MI rats compared with sham rats, but SAAS therapy significantly attenuated these changes. Moreover, SAAS treatment upregulated HTSF1 (HIV Tat-specific factor 1 homolog), which was decreased by MI injury (Fig. 4b). The results were validated by Western blot analysis (Fig. 4c).
Cardioprotective effects of SAAS against myocardial infarction at the transcriptome level
Transcriptome profiling of mRNA samples extracted from cardiac tissues was performed to investigate changes at the gene level. On average, 6.74 Gb of sequenced nucleotides were obtained from each sample, with a genome mapping rate of 84.93% (Supplementary Tables S4 and S5). In total, 17,580 genes were identified, and DEseq2 algorithms were used to identify the differentially expressed genes (DEGs) (fold change > 1.2 and P < 0.05). The results showed that 345 genes were differentially expressed in the model-vs-sham comparison, including 269 upregulated and 76 downregulated genes, and that SAAS therapy caused the expression of 27 genes to change significantly in the treated groups compared with the sham group (Fig. 5a and Supplementary Table S6). Cluster analysis was used to show the gene expression patterns in each group (Fig. 5b).
Cardioprotective effects of SAAS on myocardial infarction at transcriptome level. a Summary of differentially expressed genes (DEGs). Red represents upregulation and blue represents downregulation. b Heat map of cluster analysis using Euclidean distance and hierarchical algorithm in each group. c KEGG pathway functional enrichment of DEGs. X-axis represents enrichment factor. Y-axis represents pathway name. The color indicates the P-value (high: white, low: blue), the lower P-value indicates the more significant enrichment. Point size indicates DEGs number. The bigger dots refer to larger amount. Rich Factor refers to the value of enrichment factor, which is the quotient of foreground value (the number of DEGs) and background value (total gene amount). The larger the value, the more significant enrichment
Notably, the most enriched GO terms in the model-vs-sham comparison were the same as those in the 24 and 96 mg/kg SAAS-vs-model comparisons. The cellular process, cell, and binding terms were the most common terms in the biological process, cellular component, and molecular function categories, respectively (Supplementary Fig. S4).
KEGG pathway enrichment analysis showed that the top two most enriched pathways in the model-vs-sham, 24 and 96 mg/kg SAAS-vs-model comparisons were the protein digestion and absorption, ECM-receptor interaction, tryptophan metabolism and ovarian steroidogenesis, Wnt signaling and spliceosome pathways (Fig. 5c).
Analysis of protein interactions using the STRING database indicated that 100 DEGs in the model-vs-sham comparison were functionally linked (Supplementary Fig. S5), with Col1a1, Myo16, Col3a1, Col5a2, Col18a1, Col1a2, Itga11, and Col8a1 among the most linked. According to the BC parameter in Cytoscape v3.5.1 software, the strongest interactions were the Bgn-Myo16, Comp-Itga11, Comp-Hadh, Islr-Myo16, Bgn-Col1a1, and Itga11-Col1a1 interactions (Supplementary Fig. S6). However, SAAS therapy had such a slight influence on gene expression that there were too few DEGs to generate a landscape map.
Integrative pathway map based on the proteome and transcriptome analysis
Among the 317 DEGs and the 205 DEPs in the model-vs-sham comparison, 14 presented the same trends, and the expression patterns were visualized by cluster analysis (Fig. 6a, b and Supplementary Table S7). Maoa (amine oxidase [flavin-containing] A), Ppic (peptidyl-prolyl cis–trans isomerase C), Anxa2 (annexin A2), Postn (periostin), Uap1 (UDP-N-acetylhexosamine pyrophosphorylase), Fbn1 (fibrillin-1), Anxa1 (annexin A1), Ptgis (prostacyclin synthase), Col1a1 (collagen alpha-1(I) chain), Mfap5 (microfibrillar-associated protein 5), Prelp (prolargin), and Vcan (versican core protein) were upregulated, while Macrod1 (O-acetyl-ADP-ribose deacetylase MACROD1) and Tmod4 (tropomodulin-4) were downregulated in the MI model group compared to the sham group at both the proteome and transcriptome levels. However, there were no proteins or genes that were consistently present in the 24 or 96 mg/kg SAAS-vs-model comparisons.
Correlated analysis of proteome and transcriptome profiling. a Number of DEGs and DEPs in the sham-vs-model comparison groups. b Cluster analysis of DEGs and DEPs in the sham-vs-model comparison groups present at the same trend. c Integrative pathway maps of DEGs and DEPs in each comparison group. X-axis represents the number of DEGs/DEPs. Y-axis represents pathway name
Given the KEGG annotations simultaneously enriched in both the proteome and transcriptome analysis, the protein and gene expression data, and the correlation results, integrated pathway maps were constructed according to the enrichment of DEPs and DEGs for each annotation. The results showed that in the model-vs-sham comparison, the top five frequently targeted pathways were the metabolic, regulation of actin cytoskeleton, phagosome, focal adhesion, and tight junction pathways. In the 24 and 96 mg/kg SAAS-vs-model comparisons, metabolic pathways, MAPK signaling, and Wnt signaling were markedly affected (Fig. 6c).
Discussion
Since the molecular pathogenesis of IHDs is quite complicated, natural products may provide potent alternative therapeutics. A large amount of evidence has demonstrated that SAAS could improve microcirculation, alleviate free radical damage, inhibit hypertensive fibrosis, and protect against myocardial ischemia injury in vitro and in vivo [15,16,17,18,19]. This study aimed to gain detailed insight into the cardioprotective mechanisms of SAAS in rats with myocardial infarction by global analysis of myocardial tissue at the proteome and transcriptome levels.
Myocardial infarction injury induced the differential expression of various proteins and genes. STRING analysis showed that 36 DEPs and 100 DEGs in the model-vs-sham comparison were functionally linked. Among these molecules were diverse regulators of the cytoskeleton, including Actb, Myh10, Myh9, Actn4, Acta1, Myl9, Col1a1, Col3a1, Col5a2, Col18a1, and Col1a2. Cellular myosin plays an important role in cytoskeletal reorganization, and studies have found that the absence of Myh10 leads to a decrease in the number of embryonic cardiac myocytes [20]. Cardiac fibrillary collagens determine the physical properties of the left ventricular chamber and integrate adjacent cardiomyocytes into a coordinated biologic pump for blood circulation, which is of vital importance for the development of heart disease [21, 22]. Collagens are regarded as fibroblast marker genes, and our results indicated clear MI injury induced by the ligation [23].
Fourteen molecules in the model-vs-sham comparison exhibited the same trends at the proteome and transcriptome levels, including Maoa, Ppic, Anxa2, Postn, Uap1, Fbn1, Anxa1, Ptgis, Col1a1, Mfap5, Prelp, Vcan, Macrod1, and Tmod4. As sources of oxidants in the heart, monoamine oxidases might be beneficial for therapy of cardiovascular pathologies [24]. The annexins are a group of calcium-dependent phospholipid-binding proteins closely associated with the progression and metastasis of certain cancers, and accumulated studies have indicated that some annexins might play roles in cardiovascular diseases [25]. Anxa2 has been identified as an MI drug target by network analysis, and Anxa1 is an important effector of inflammation [26, 27]. Postn has been reported to induce proliferation of differentiated cardiomyocytes and recruit activated fibroblasts through FAK-integrin signaling to promote cardiac repair, which is essential for cardiac healing after MI [28, 29]. In contrast, there is also evidence that Postn inhibition might contribute to cardiac remodeling after MI [30]. Fbn1 is an important constituent of the myocardium, and its increased expression may affect myocardial stiffness, contributing to tissue fibrosis and repair [31]. Microfibrillar-associated proteins belong to the fibrinogen-related protein superfamily and are important for the development of the cardiovascular system [32]. Reduced activity of the Ptgis gene could be a genetic marker for MI and lead to its pathogenesis [33]. Macrod1 has been found to be a biomarker for the prediction and detection of MI [34]. Tmod4 was found to be a critical gene in postnatal mouse myocardial proliferation and development by a genomics approach [35]. These 14 potential molecules could be novel biomarkers for the prediction and detection of MI-induced injury.
SAAS therapy seems to have greater effects on the proteome than on the transcriptome, as the percentages of differentially expressed proteins and genes among all proteins and genes in the SAAS-vs-model/sham comparisons were 64.8% (151/233) and 7.8% (27/345), respectively. SAAS effectively reversed the changes in the expression of an array of proteins induced by MI injury, including KCNN2, p63, ARL11, ATMIN, KC1A, and HTSF1. The KCNN2 gene has been found to be increased in atrial myocytes in the context of several cardiovascular diseases, such as arrhythmia, which is consistent with our results that KCNN2 was upregulated in MI rats [36, 37]. p63 has been reported to be essential for cardiogenesis and control of cell migration and invasion [38, 39]. ATMIN, an ATM interactor protein, is crucial for cell survival in response to oxidative stress-induced DNA damage, which is implicated in cardiovascular diseases [40, 41]. SAAS may exert its cardioprotective effects on MI rats mainly by targeting multiple pathways at the proteome level.
However, there were no proteins or genes consistently present in the SAAS-vs-model comparisons. Studied have found that the correlation between mRNA and corresponding protein expression is approximately 27%–40% [10]. In this animal tissue-based analysis, the correlation was much lower, which may have been due to individual differences among the rats.
GO annotation of the transcriptome profiles showed that in the model-vs-sham and SAAS-vs-model comparisons, the most enriched terms were the cellular process, cell, and binding terms in the biological process, cellular component, and molecular function categories, respectively, consistent with the proteomic results. Coincidentally, the top enriched pathways in both the model-vs-sham and SAAS-vs-model comparisons included the regulation of actin cytoskeleton, phagosome, focal adhesion, tight junction, apoptosis, MAPK signaling, and Wnt signaling pathways.
Regulation of the actin cytoskeleton is vital for cell migration and invasion, which plays a key role in numerous pathological conditions, including wound healing, cardiac infarction, and atherosclerosis [42]. Phagosomes participate in tissue remodeling, clear apoptotic cells, and restrict the spread of intracellular pathogens [43]. Autophagosomes are present in surviving cardiomyocytes during chronic stages of myocardial infarction to inhibit apoptosis and mitigate deleterious effects, which may alleviate cardiac dysfunction and enhance remodeling [44]. A number of studies have clearly established roles for focal adhesion kinase (FAK) in angiogenesis and the development of the cardiovascular system and possible roles in heart diseases, including cardiac hypertrophy [45]. Tight junctions are essential for establishing a selectively permeable barrier to diffusion through the paracellular space between neighboring cells [46]. MAPK signaling, which regulates a variety of cellular activities including proliferation, survival, differentiation, and death, has been reported to be active in cardiovascular health and diseases [47]. Wnt signaling is an essential regulator of cardiovascular differentiation, morphogenesis, and progenitor cell self-renewal [48].
Conclusions
SAAS, which is derived from the well-known herbal medicine Danshen, was found to effectively improve cardiac function and protect against myocardial injury due to ligation of the left anterior descending coronary artery in rats. Numerous findings indicated the existence of coordinated regulation of important structural molecules, receptors, transcription factors, and cofactors and suggested that these molecules were mainly involved in regulation of actin cytoskeleton, phagosome, focal adhesion, tight junction, apoptosis, MAPK signaling, and Wnt signaling pathways at both the transcriptome and proteome levels. Taken together, the results indicate that SAAS may exert its cardioprotective effects by targeting multiple pathways at the proteome, transcriptome, and probably metabolome levels, which may provide pharmacological evidence to aid in its clinical application.
References
Akhtar S. Ischemic heart disease. Anesth Clin. 2006;24:461–85.
Piepoli MF, Hoes AW, Agewall S, Albus C, Brotons C, Catapano AL, et al. European Guidelines on cardiovascular disease prevention in clinical practice: The Sixth Joint Task Force of the European Society of Cardiology and Other Societies on Cardiovascular Disease Prevention in Clinical Practice (constituted by representatives of 10 societies and by invited experts) Developed with the special contribution of the European Association for Cardiovascular Prevention & Rehabilitation (EACPR). Eur Heart J. 2016;37:2315–81.
Xiong X, Borrelli F, de Sa Ferreira A, Ashfaq T, Feng B. Herbal medicines for cardiovascular diseases. Evid Based Complement Altern Med. 2014;2014:809741.
Cheng TO. Cardiovascular effects of Danshen. Int J Cardiol. 2007;121:9–22.
Zhou L, Zuo Z, Chow MS. Danshen: an overview of its chemistry, pharmacology, pharmacokinetics, and clinical use. J Clin Pharmacol. 2005;45:1345–59.
Chen R, Mias GI, Li-Pook-Than J, Jiang L, Lam HY, Chen R, et al. Personal omics profiling reveals dynamic molecular and medical phenotypes. Cell. 2012;148:1293–307.
Li M, Wang IX, Li Y, Bruzel A, Richards AL, Toung JM, et al. Widespread RNA and DNA sequence differences in the human transcriptome. Science. 2011;333:53–8.
Wu JQ, Habegger L, Noisa P, Szekely A, Qiu C, Hutchison S, et al. Dynamic transcriptomes during neural differentiation of human embryonic stem cells revealed by short, long, and paired-end sequencing. Proc Natl Acad Sci USA. 2010;107:5254–9.
Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, et al. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–6.
Muers M. Gene expression: transcriptome to proteome and back to genome. Nat Rev Genet. 2011;12:518.
Savitski MM, Wilhelm M, Hahne H, Kuster B, Bantscheff M. A scalable approach for protein false discovery rate estimation in large proteomic data sets. Mol Cell Proteomics. 2015;14:2394–404.
Tarun AS, Peng X, Dumpit RF, Ogata Y, Silva-Rivera H, Camargo N, et al. A combined transcriptome and proteome survey of malaria parasite liver stages. Proc Natl Acad Sci USA. 2008;105:305–10.
Zheng PZ, Wang KK, Zhang QY, Huang QH, Du YZ, Zhang QH, et al. Systems analysis of transcriptome and proteome in retinoic acid/arsenic trioxide-induced cell differentiation/apoptosis of promyelocytic leukemia. Proc Natl Acad Sci USA. 2005;102:7653–8.
Wang H, Cao C, Hui L, Liu T, Wang Y, Gao S, et al. A Study of myocardial ischemia model induced by left coronary artery ligation in rats. World J Cardiovasc Dis. 2016;6:133–42.
Song Q, Chu X, Zhang X, Bao Y, Zhang Y, Guo H, et al. Mechanisms underlying the cardioprotective effect of Salvianic acid A against isoproterenol-induced myocardial ischemia injury in rats: possible involvement of L-type calcium channels and myocardial contractility. J Ethnopharmacol. 2016;189:157–64.
Meng Y, Li WZ, Shi YW, Zhou BF, Ma R, Li WP. Danshensu protects against ischemia/reperfusion injury and inhibits the apoptosis of H9c2 cells by reducing the calcium overload through the p-JNK-NF-kappaB-TRPC6 pathway. Int J Mol Med. 2016;37:258–66.
Fan G, Yu J, Asare PF, Wang L, Zhang H, Zhang B, et al. Danshensu alleviates cardiac ischaemia/reperfusion injury by inhibiting autophagy and apoptosis via activation of mTOR signalling. J Cell Mol Med. 2016;20:1908–19.
Yu J, Wang L, Akinyi M, Li Y, Duan Z, Zhu Y, et al. Danshensu protects isolated heart against ischemia reperfusion injury through activation of Akt/ERK1/2/Nrf2 signaling. Int J Clin Exp Med. 2015;8:14793–804.
Yin Y, Guan Y, Duan J, Wei G, Zhu Y, Quan W, et al. Cardioprotective effect of Danshensu against myocardial ischemia/reperfusion injury and inhibits apoptosis of H9c2 cardiomyocytes via Akt and ERK1/2 phosphorylation. Eur J Pharmacol. 2013;699:219–26.
Dey D, Han L, Bauer M, Sanada F, Oikonomopoulos A, Hosoda T, et al. Dissecting the molecular relationship among various cardiogenic progenitor cells. Circ Res. 2013;112:1253–62.
Eyre DR, Wu J. Collagen cross-links. Top Curr Chem. 2005;247:207–29.
Díez J, López B, González A, Querejeta R. The Role of myocardial collagen network in hypertensive heart disease. Curr Hypertens Rev. 2007;3:1–7.
Wang J, Huang W, Xu R, Nie Y, Cao X, Meng J, et al. MicroRNA-24 regulates cardiac fibrosis after myocardial infarction. J Cell Mol Med. 2012;16:2150–60.
Kaludercic N, Mialet-Perez J, Paolocci N, Parini A, Di Lisa F. Monoamine oxidases as sources of oxidants in the heart. J Mol Cell Cardiol. 2014;73:34–42.
Cederholm A, Frostegard J. Annexin A5 in cardiovascular disease and systemic lupus erythematosus. Immunobiology. 2005;210:761–8.
Wang RS, Loscalzo J. Illuminating drug action by network integration of disease genes: a case study of myocardial infarction. Mol Biosyst. 2016;12:1653–66.
Perretti M, D’Acquisto F. Annexin A1 and glucocorticoids as effectors of the resolution of inflammation. Nat Rev Immunol. 2009;9:62–70.
Shimazaki M, Nakamura K, Kii I, Kashima T, Amizuka N, Li M, et al. Periostin is essential for cardiac healing after acute myocardial infarction. J Exp Med. 2008;205:295–303.
Kuhn B, del Monte F, Hajjar RJ, Chang YS, Lebeche D, Arab S, et al. Periostin induces proliferation of differentiated cardiomyocytes and promotes cardiac repair. Nat Med. 2007;13:962–9.
Iekushi K, Taniyama Y, Azuma J, Katsuragi N, Dosaka N, Sanada F, et al. Novel mechanisms of valsartan on the treatment of acute myocardial infarction through inhibition of the antiadhesion molecule periostin. Hypertension. 2007;49:1409–14.
Bouzeghrane F, Reinhardt DP, Reudelhuber TL, Thibault G. Enhanced expression of fibrillin-1, a constituent of the myocardial extracellular matrix in fibrosis. Am J Physiol Heart Circ Physiol. 2005;289:H982–91.
van Rooij E, Sutherland LB, Thatcher JE, DiMaio JM, Naseem RH, Marshall WS, et al. Dysregulation of microRNAs after myocardial infarction reveals a role of miR-29 in cardiac fibrosis. Proc Natl Acad Sci USA. 2008;105:13027–32.
Nakayama T, Soma M, Saito S, Honye J, Yajima J, Rahmutula D, et al. Association of a novel single nucleotide polymorphism of the prostacyclin synthase gene with myocardial infarction. Am Heart J. 2002;143:797–801.
Friede KA, Ginsburg GS, Voora D. Gene expression signatures and the spectrum of coronary artery disease. J Cardiovasc Transl Res. 2015;8:339–52.
Wang R, Su C, Wang X, Fu Q, Gao X, Zhang C, et al. Global gene expression analysis combined with a genomics approach for the identification of signal transduction networks involved in postnatal mouse myocardial proliferation and development. Int J Mol Med. 2018;41:311–21.
Kim JJ, Park YM, Yoon D, Lee KY, Seob Song M, Doo Lee H, et al. Identification of KCNN2 as a susceptibility locus for coronary artery aneurysms in Kawasaki disease using genome-wide association analysis. J Hum Genet. 2013;58:521–5.
Zhang Q, Timofeyev V, Lu L, Li N, Singapuri A, Long MK, et al. Functional roles of a Ca2+-activated K+ channel in atrioventricular nodes. Circ Res. 2008;102:465–71.
Rouleau M, Medawar A, Hamon L, Shivtiel S, Wolchinsky Z, Zhou H, et al. TAp63 is important for cardiac differentiation of embryonic stem cells and heart development. Stem Cells. 2011;29:1672–83.
Senoo M, Pinto F, Crum CP, McKeon F. p63 is essential for the proliferative potential of stem cells in stratified epithelia. Cell. 2007;129:523–36.
Kanu N, Zhang T, Burrell RA, Chakraborty A, Cronshaw J, DaCosta C, et al. RAD18, WRNIP1 and ATMIN promote ATM signalling in response to replication stress. Oncogene. 2016;35:4020.
Yan S, Sorrell M, Berman Z. Functional interplay between ATM/ATR-mediated DNA damage response and DNA repair pathways in oxidative stress. Cell Mol Life Sci. 2014;71:3951–67.
Yamaguchi H, Condeelis J. Regulation of the actin cytoskeleton in cancer cell migration and invasion. Biochim Biophys Acta. 2007;1773:642–52.
Garin J, Diez R, Kieffer S, Dermine JF, Duclos S, Gagnon E, et al. The phagosome proteome: insight into phagosome functions. J Cell Biol. 2001;152:165–80.
Lavandero S, Troncoso R, Rothermel BA, Martinet W, Sadoshima J, Hill JA. Cardiovascular autophagy: concepts, controversies, and perspectives. Autophagy. 2013;9:1455–66.
Vadali K, Cai X, Schaller MD. Focal adhesion kinase: an essential kinase in the regulation of cardiovascular functions. IUBMB Life. 2007;59:709–16.
Zihni C, Mills C, Matter K, Balda MS. Tight junctions: from simple barriers to multifunctional molecular gates. Nat Rev Mol Cell Biol. 2016;17:564–80.
Muslin AJ. MAPK signalling in cardiovascular health and disease: molecular mechanisms and therapeutic targets. Clin Sci (Lond). 2008;115:203–18.
Cohen ED, Tian Y, Morrisey EE. Wnt signaling: an essential regulator of cardiovascular differentiation, morphogenesis and progenitor self-renewal. Development. 2008;135:789–98.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (Grant Nos. 81503039, 81573396, and 81773683) and the Discipline Leader Training Program of Pudong New Area Health System of Shanghai (PWRd2014-11). D.J. would like to give special thanks to the BGI-Shenzhen Corporation for its help in the experiments.
Author information
Authors and Affiliations
Contributions
CZ, JH, ZYZ, CZZ, YQ, and DJ designed the experiments; DJ, CZZ, YQ, and XFC performed the experiments and analyzed the data; DJ, YQ, and LJ wrote the manuscript; and CZ, JH, ZYZ, YFC, and AFC reviewed the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Rights and permissions
About this article
Cite this article
Jia, D., Zhang, Cz., Qiu, Y. et al. Cardioprotective mechanisms of salvianic acid A sodium in rats with myocardial infarction based on proteome and transcriptome analysis. Acta Pharmacol Sin 40, 1513–1522 (2019). https://doi.org/10.1038/s41401-019-0265-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41401-019-0265-1
Keywords
This article is cited by
-
Danshensu alleviates pseudo-typed SARS-CoV-2 induced mouse acute lung inflammation
Acta Pharmacologica Sinica (2022)