Abstract
Urinary bladder cancer (UBC) is characterized by frequent recurrence and metastasis despite the standard chemotherapy with gemcitabine and cisplatin combination. Histone modifiers are often dysregulated in cancer development, thus they can serve as an excellent drug targets for cancer therapy. Here, we investigated whether G9a, one of the histone H3 methyltransferases, was associated with UBC development. We first analyzed clinical data from public databases and found that G9a was significantly overexpressed in UBC patients. The TCGA Provisional dataset showed that the average expression level of G9a in primary UBC samples (n = 408) was 1.6-fold as much as that in normal bladder samples (n = 19; P < 0.001). Then we used small interfering RNA to knockdown G9a in human UBC T24 and J82 cell lines in vitro, and observed that the cell viability was significantly decreased and cell apoptosis induced. Next, we choosed UNC0642, a small molecule inhibitor targeting G9a, with low cytotoxicity, and excellent in vivo pharmacokinetic properties, to test its anticancer effects against UBC cells in vitro and in vivo. Treatment with UNC0642 dose-dependently decreased the viability of T24, J82, and 5637 cells with the IC50 values of 9.85 ± 0.41, 13.15 ± 1.72, and 9.57 ± 0.37 μM, respectively. Furthermore, treatment with UNC0642 (1−20 μM) dose-dependently decreased the levels of histone H3K9me2, the downstream target of G9a, and increased apoptosis in T24 and J82 cells. In nude mice bearing J82 engrafts, administration of UNC0642 (5 mg/kg, every other day, i.p., for 6 times) exerted significant suppressive effect on tumor growth without loss of mouse body weight. Moreover, administration of UNC0642 significantly reduced Ki67 expression and increased the level of cleaved Caspase 3 and BIM protein in J82 xenografts evidenced by immunohistochemistry and western blot analysis, respectively. Taken together, our data demonstrated that G9a may be a promising therapeutic target for UBC, and an epigenetics-based therapy by UNC0642 is suggested.
Similar content being viewed by others
INTRODUCTION
Urinary bladder cancer (UBC) is one of the most common urologic neoplasms worldwide, with 430,000 new cases diagnosed and 165,000 deaths per year [1]. The majority of UBC cases are of the urothelial carcinoma histologic type, 75% of which are nonmuscle invasive bladder cancer (NMIBC), and the remaining 25% are muscle-invasive bladder cancer (MIBC) [2, 3]. More than half of all NMIBC patients experience tumor recurrence within 5 years, and 10–30% develop malignant progression to MIBC [4]. Hence, patients with NMIBC should be monitored by cystoscopy for many years, and multiple treatment rounds are always needed. Furthermore, MIBC patients have an unfavorable prognosis, with a 5-year survival rate of <50%. Unfortunately, currently, no high-grade MIBC therapies exist, and many patients choose radical cystectomy [5, 6]. Cisplatin-based chemotherapy administered before or after surgery in a neoadjuvant or adjuvant manner can partially reduce tumor recurrence and metastasis and improve survival time. In spite of taking these approaches, MIBC patients have an unfavorable prognosis, with a 5-year survival rate of <50%. Therefore, development of new therapeutic strategies and drugs for UBC is necessary and urgent.
G9a is a histone methyltransferase that specifically mediates mono- and di-methylation of histone H3 lysine 9 (H3K9) [7, 8]. Lysine methylation of histone H3 by G9a is associated with transcriptional repression. Accumulating studies demonstrate that G9a participates in many biological processes, including cell proliferation, survival, and invasion [9,10,11,12,13,14]. G9a is overexpressed in various cancer types, such as gastric, ovarian, and esophageal cancer [10, 15,16,17]. Knockdown of the G9a gene induces chromosome instability and suppresses cancer cell proliferation and invasion, supporting the notion that G9a is a promising drug target [18,19,20]. G9a inhibitors, such as UNC0638 and BIX-01294, can inhibit cell proliferation by inducing cell cycle arrest, triggering apoptosis, or inducing autophagic cell death [21,22,23]. However, UNC0638 is not suitable for animal studies due to its poor pharmacokinetic properties [24]. Recently, UNC0642, another selective G9a inhibitor, was demonstrated to have high in vitro cellular potency and excellent selectivity, along with improved in vivo pharmacokinetic properties [24, 25].
In this study, we found that G9a was overexpressed in UBC patients and regulated cell proliferation and apoptosis in UBC cells. Targeting of G9a activity by UNC0642 suppressed UBC cell growth and induced apoptosis in vitro and in vivo. These findings indicate that G9a may be a promising therapeutic target for UBC, and epigenetics-based therapy with UNC0642 is suggested.
MATERIALS AND METHODS
Cell lines and reagents
Human UBC T24, J82, and 5637 cell lines were purchased from Cell Bank of Type Culture Collection, Chinese Academy of Sciences (Shanghai, China). All cells were maintained in RPMI 1640 medium (Life Technologies, Carlsbad, CA, USA) containing 10% fetal bovine serum (HyClone, Logan, UT, USA) at 37 °C in a humidified incubator containing 5% CO2. The compound UNC0642 with 99.89% purity was purchased from MedChemExpress (HY-13980, Monmouth Junction, NJ, USA).
G9a expression in human UBC samples
TCGA portal was used to obtain normalized G9a mRNA expression data in Fragments Per Kilobase Million (FPKM) from UBC studies. In TCGA Provisional dataset, 19 normal bladder tissues and 408 primary UBC samples were included. GEO profiling was used to obtain G9a mRNA expression data (via count) for UBC studies. In the GSE3167 202326_at dataset, 14 normal bladder tissues and 46 UBC samples were included. The correlation between the G9a mRNA level and cumulative overall survival (OS) in human UBC samples was analyzed using the Kaplan–Meier method in the TCGA Provisional dataset (n = 405). Median, tertiles, quartiles, and quintiles were determined as the cutoff points for G9a expression, and differences in the survival rates between curves were assessed using a log-rank test. P <0.05 was considered statistically significant.
RNA interference
Cells were cultured in six-well plates at a density of 1 × 105 cells/well. After 24 h, the cells were transfected with G9a siRNA using Lipofectamine RNAiMAX (Life Technologies) according to the manufacturer’s instructions. Then, the cells were cultured for another 72 h and harvested for further analysis. The oligonucleotide sequences of the two siRNAs used to target G9a were siG9a-1: 5ʹ-CGCACAGAGAAGAUCAUCUTT-3ʹ and siG9a-2: 5ʹ-GGUGUCCAAUGACACAUCUTT-3ʹ, and that of the control siRNA (siNC) was 5ʹ-TTCTCCGAACGTGTCACGTTT-3ʹ.
Apoptosis analysis
Cells were cultured in six-well plates at a density of 1 × 105 cells/well. After 24 h, the cells were treated with DMSO (vehicle) or the indicated concentration of UNC0642 for 72 h. Then, both adherent and floating cells were harvested and washed with cold PBS. Prior to FACS analysis, cells were resuspended in 500 μL of binding buffer containing 5 μL of Annexin V and 5 μL of propidium iodide solution (PI; cat #: 556547, BD Biosciences, Piscataway, NJ, USA) and were stained for 15 min at room temperature in the dark. Then, apoptosis analysis was performed using a FACS Calibur flow cytometer (BD). The data were analyzed using CELLQuest software (BD).
Cytotoxicity assay
Cells were seeded in 96-well plates at 3000–5000 cells/well in triplicate. After 24 h, the cells were treated with different concentrations of UNC0642 and incubated at 37 °C for another 72 h. Then, the cells were fixed with 10% trichloroacetic acid overnight and stained with 4 mg/mL sulforhodamine B (SRB; Sigma, St Louis, MO, USA) in 1% acetic acid. The SRB in the cells was dissolved in 10 mM Tris-HCl and measured at 560 nm. IC50 values were analyzed using GraphPad Prism software (version 6.01, GraphPad Software Inc., La Jolla, CA, USA).
Western blotting
Protein extracts were prepared with SDS-lysis buffer (50 mM Tris-HCl, pH 7.4, 2% SDS). Cell lysates were boiled for 10 min at 100 °C and centrifuged at 14,000 r/min for 5 min at 4 °C. The supernatant was collected and subsequently resolved by SDS-PAGE, transferred to nitrocellulose membranes, probed with the appropriate primary antibodies and then incubated with horseradish peroxidase-conjugated secondary antibodies. The immunoreactive proteins were detected using an ECL detection reagent (Pierce, Rockford, IL, USA) and imaged with a chemiluminescence instrument (LAS-4000, GE Healthcare, Piscataway, NJ, USA). Primary antibodies targeting the following proteins were used: Histone H3 (#4499, Cell Signaling Technology (CST), Danvers, MA, USA, 1:5,000), di-methyl-Histone H3 (Lys9) (#4658, CST, 1:5,000), cleaved Caspase-3 (#9664, CST, 1:1,000), Caspase-3 (#9665, CST, 1:1,000), cleaved PARP (#5625, CST, 1:1,000), PARP (#9532, CST, 1:1000), BIM (#2933, CST, 1:1,000), and β-actin (60008-1-Ig, Proteintech, 1:10,000).
Gene expression microarray analysis
T24 cells were treated with siG9a-1 or siNC for 48 h, and total RNA was then extracted with TRIzol reagent (Life Technologies), purified, amplified, and labeled to obtain biotin-labeled cRNA. Affymetrix PrimeView GeneChip hybridization was performed according to the manufacturer’s protocol (Affymetrix, Carlsbad, CA, USA). Differentially expressed genes (exhibiting at least 1.5-fold change in expression) were screened via moderated-t test (P < 0.001). GSEA of gene sets was performed against MSigDB v5.1 with 105 permutations. The original and normalized microarray data from this study can be accessed at GSE115341 in NCBI GEO Datasets.
Quantitative real-time PCR
Quantitative real-time PCR was performed using a Bio-Rad Real-Time PCR detection system (Bio-Rad, Berkeley, CA, USA). Total RNA was extracted with TRIzol reagent and subjected to reverse transcription with HiScript II Q RT SuperMix (#R222-01, Vazyme, Nanjing, China). PCRs were performed with SYBR Green Master Mix (#Q121-02, Vazyme). The primers used were as follows: G9a: forward, 5ʹ-TCCAATGACACATCTTCGCTG-3ʹ and reverse, 5ʹ-CTGATGCGGTCAATCTTGGG-3ʹ Ampka2: forward, 5ʹ-GTGAAGATCGGACACTACGTG-3ʹ and reverse, 5ʹ-CTGCCACTTTATGGCCTGTTA-3ʹ ELL2: forward, 5ʹ-CATCACCGTACTGCATGTGAA-3ʹ and reverse, 5ʹ-ACTGGATTGAAGGTCGAAAAGG-3ʹ Bim: forward, 5ʹ-TAAGTTCTGAGTGTGACCGAGA-3ʹ and reverse, 5ʹ-GCTCTGTCTGTAGGGAGGTAGG-3ʹ.
In vivo xenograft study
The animal protocol was approved by the Association for Assessment & Accreditation of Laboratory Animal Care international of Model Animal Research Center of Nanjing University. J82 cells (2 × 106) were mixed with 100 μL of Matrigel (cat #: 356234, BD; 1:1 ratio) and then were injected subcutaneously into 8-week-old male nude mice, which were obtained from the Model Animal Research Center of Nanjing University. Seven days later, nude mice bearing palpable xenografts were randomly divided into two groups. One group was treated with vehicle (DMSO), and the other was treated with UNC0642 (5 mg/kg) via intraperitoneal (i.p.) injection every other day. Tumor volumes were measured using a caliper and calculated as length × width2/2. At the endpoint, mice were killed, and tumors were weighed and processed for further analysis.
Immunohistochemical (IHC) staining
Briefly, 5-μm-thick paraffin sections were stained using an UltraSensitive S-P IHC staining kit (cat #: 9707, MAXIN, Fujian, China). The primary antibodies were rabbit polyclonal antibodies for di-methyl-Histone H3 (Lys9) (#9753, CST; 1:400) and Ki67 (VP-RM04, Vector Laboratories, Burlingame, CA, USA; 1:10,000), which were incubated with sections overnight at 4 °C. The streptavidin–peroxidase method was used, and the signals were detected with an enhanced DAB substrate kit (cat #: DAB-2031, MAXIN), followed by counterstaining with hematoxylin.
Statistical analysis
Statistical analyses were performed using GraphPad Prism Software. Comparisons between groups were performed with two-tailed Student’s t test, and differences were considered statistically significant at P< 0.05.
RESULTS
G9a is overexpressed in UBC patients and is positively correlated with cancer progression
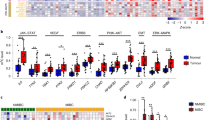
To test whether G9a is associated with UBC development, the expression level of G9a in UBC patients was analyzed using the public TCGA database and GEO Profiles database. In the dataset (TCGA Provisional) from TCGA database, the normalized mRNA expression level (FPKM) of G9a was analyzed. The average expression level of G9a in primary UBC samples (n = 408) was 1.6-fold higher than that in normal bladder samples (n = 19; P < 0.001; Fig. 1a). Next, correlations between the FPKM of G9a and tumor histology (Fig. 1b) or individual cancer stage (Fig. 1c) was investigated. Both papillary (P < 0.001; n = 271) and nonpapillary UBC samples (P < 0.001; n = 132) exhibited G9a overexpression compared with normal bladder samples (n = 19). However, there was no significant difference between papillary and nonpapillary UBC samples (Fig. 1b). In addition, UBC samples at different cancer stages, including T1 (P < 0.05; n = 3), T2 (P < 0.001; n = 119), T3 (P < 0.001; n = 194), and T4 (P < 0.01; n = 58), exhibited upregulation of G9a compared with normal bladder samples (n = 19). However, correlations between the FPKM of G9a and individual cancer stages were not observed (Fig. 1c). Next, the correlation between the FPKM of G9a and molecular subtype (Fig. 1d) was examined. All UBC samples, including neuronal (n = 20), basal squamous (n = 142), luminal (n = 26), luminal infiltrated (n = 78), and luminal papillary (n = 142) subtypes, expressed a higher level of G9a than normal bladder samples (n = 19; P < 0.05). Notably, G9a expression in the neuronal subtype, which behaves similarly to neuroendocrine neoplasms at other tissue sites and is associated with particularly poor survival [26, 27], was significantly higher than that in the other four UBC subtypes (P < 0.05). The relative expression level of G9a in another independent dataset (GSE3167, 202326_at) from GEO Profiles database was further analyzed. The average level of G9a in UBC samples (n = 46) was twofold higher than that in normal bladder samples (n = 14) (P < 0.001; Fig. 1e). However, a correlation between high G9a expression and poor overall survival could not be detected in UBC patients from the TCGA Provisional dataset (n = 405; Fig. S1). The above data demonstrate that G9a is overexpressed in human UBC patients and thus a potential therapeutic target in UBC patients.
G9a overexpression is associated with UBC progression. a Analysis of G9a mRNA expression level (FPKM) in normal bladder (n = 19) and primary UBC (n = 408) samples from TCGA provisional database. b Analysis of G9a mRNA expression level (FPKM) in normal bladder (n = 19), papillary UBC (n = 271) and nonpapillary (n = 132) samples in TCGA provisional database. c Analysis of G9a mRNA expression level (FPKM) in normal bladder (n = 19) samples and in samples of UBC growing into the layer of connective tissue under the lining of the bladder wall (T1; n = 3), UBC growing into the muscle layer of the bladder wall (T2; n = 119), UBC growing into the layer of fatty tissue that surrounds the bladder (T3; n = 194), and UBC spreading into the prostate, uterus, or vagina and growing into the pelvic or abdominal wall (T4; n = 58); all data were derived from TCGA provisional database. d Analysis of G9a mRNA expression level (FPKM) in normal bladder (n = 19), neuronal UBC (n = 20), basal squamous UBC (n = 142), luminal UBC (n = 26), luminal infiltrated UBC (n = 78) and luminal papillary UBC (n = 142) samples from TCGA provisional database. e Analysis of relative expression G9a level in normal bladder (n = 14) and primary UBC (n = 46) samples from GEO Profiles database. Bars represent the mean ± SD. *P < 0.05; **P < 0.01; ***P < 0.001; ###P < 0.001
G9a is required for UBC cell survival and regulates the expression of genes mediating cell viability and apoptosis
To test whether G9a can regulate UBC cell survival, we knocked down G9a expression with two different siRNAs in T24 and J82 cells. After 72 h, cell viability was assessed using an SRB assay. Downregulation of G9a decreased the methylation level of H3K9 according to a western blotting assay (Fig. 2a) and reduced cell viability (P < 0.01; Fig. 2b) in both cell lines. Annexin V-FITC/PI double staining showed that apoptosis occurred in >40% of T24 cells and >15% of J82 cells treated with siRNAs targeting the G9a gene, whereas the rate of apoptotic cells was 11% in T24 cells and 7% in J82 cells treated with negative control siRNA (siNC) (P < 0.05; Fig. 2c, d). Consistently, the expression of apoptotic markers, including cleaved Caspase-3 and its substrate cleaved PARP, was increased in G9a-depleted UBC cells (Fig. 2e).
Knockdown of G9a reduced cell viability and induced cell apoptosis of UBC cells. T24 and J82 cells were treated with G9a siRNAs for 72 h. a T24 and J82 cell lysates were analyzed by western blotting with the indicated antibodies. b Viability of T24 and J82 cells was measured using an SRB assay. The assays were performed in triplicate, and the results are presented as the mean ± SD. c, d Apoptosis of T24 and J82 cells was determined with Annexin V/PI assays (c). Cells in the first and forth quadrants were defined as apoptosis cells. The assays were performed in triplicate, and the results are presented as the mean ± SD (d). e T24 cell lysates were analyzed via western blotting with the indicated antibodies. *P < 0.05; **P < 0.01; ***P < 0.001
Because G9a is a histone methyltransferase that mediates gene transcription, a gene expression array was performed to determine the downstream effectors of the G9a gene, which may regulate UBC cell survival. PrimeView™ Human Gene Chips, comprising more than 530,000 probes covering more than 36,000 transcripts and variants, were chosen to analyze the mRNA expression profiles of T24 cells treated with G9a siRNA or siNC. As shown in Fig. 3a, 316 upregulated and 370 downregulated genes (exhibiting an at least 1.5-fold change in expression; P < 0.001) were identified. A Gene Set Enrichment Analysis (GSEA) also determined a set of genes related to apoptosis with statistically significant, concordant differences between siG9a and siNC-treated cells (Fig. 3b), such as downregulation of genes involved in cell proliferation (Cyclin D1 and ENO2) and upregulation of genes involved in apoptosis (Bim, Ampkα2, and ELL2). Quantitative real-time PCR was further applied, and the expression changes in the above genes were validated in two siG9a groups compared with the siNC group (P < 0.05; Fig. 3c). Our results show that G9a plays an important role in cell survival and that targeting of G9a with siRNA can induce apoptosis in UBC cells.
G9a regulated the expression of proliferation and apoptosis-related genes in UBC. a, b T24 cells were treated with G9a siRNA, and then, gene expression profiling was performed. Differentially expressed genes are shown in a heatmap (a). GSEA presented the hallmarks of apoptosis (b). c T24 cells were treated with G9a siRNAs. The expression levels of G9a, Cyclin D1, ENO2, Ampkα2, ELL2, and Bim were determined with qRT-PCR. *P < 0.05; **P < 0.01; ***P < 0.001
Targeting of G9a with UNC0642 reduced cell viability and induced apoptosis in UBC cells
UNC0642 is a novel inhibitor of G9a with high cellular potency and excellent selectivity in various cancer cell lines (Fig. 4a) [24], and thus, we chose it for further targeting of G9a of UBC cells both in vitro and in vivo. We applied UNC0642 at different concentrations (ranging from 0 to 20 µM) to three human UBC cell lines (T24, J82, and 5637) for 72 h and found that UNC0642 reduced cell viability of all three lines in a dose-dependent manner based on an SRB assay (Fig. 4b). The IC50 values of UNC0642 in T24, J82, and 5637 cells were 9.85 ± 0.41 μM, 13.15 ± 1.72 μM, and 9.57 ± 0.37 μM, respectively. Western blotting analysis validated the specific decrease in the global level of histone H3K9me2 with UNC0642 treatment (Fig. 4c). A stable G9a-knockdown T24 cell line (shG9a) was constructed as a control to test whether pharmacological inhibition of G9a by UNC0642 was specific (Fig. S2a). We found that T24 mock cells (shCTL) were more sensitive to UNC0642 than T24 shG9a cells (Fig. S2b), indicating that cell death induced by UNC0642 was partially dependent on G9a activity.
Cell viability of human UBC cells treated with UNC0642. a Chemical structure of UNC0642. b Cytotoxicity of UNC0642 in the human UBC cell lines T24, J82, and 5637 after treatment for 72 h. The assays were performed in triplicate, and the results are presented as the mean ± SD. c UNC0642 suppressed the activity of G9a protein according to detection of the level of H3K9me2. T24 and J82 cells were treated with UNC0642 for 24 h at the indicated concentration. β-Actin was used as the loading control
To examine whether UNC0642 could induce apoptosis, we carried out Annexin V-FITC/PI double staining of UBC cell lines treated with UNC0642 (Fig. 5a). Flow cytometry analysis showed that the ratio of apoptotic cells was increased in a dose-dependent manner in three UBC cell lines (Fig. 5b). The expression levels of apoptosis markers, such as cleaved Caspase-3 and cleaved PARP, were consistently raised in UBC cells upon UNC0642 treatment according to western blotting analysis (Fig. 5c). Meanwhile, we examined whether the expression of apoptosis-related genes, including Bim, Ampkα2, and ELL2, was also affected by UNC0642 treatment. Our results revealed that the mRNA levels of the Ampkα2, ELL2, and Bim genes were upregulated in T24 cells treated with UNC0642 at concentrations of 10 μM and 20 μM (Fig. 5d), consistent with the siG9a treatment results (Fig. 3c). These results indicate that targeting of G9a with UNC0642 reduces cell viability and induces apoptosis in UBC cells.
UNC0642 induces apoptosis in UBC cells. a–c T24, J82, and 5637 cells were treated with UNC0642 at the indicated concentrations for 72 h. Apoptosis was determined with an Annexin V/PI assay (a). Cells in the first and forth quadrants were defined as apoptotic cells. The assays were performed in triplicate, and the results are presented as the mean ± SD (b). Cell lysates were analyzed via western blotting with the indicated antibodies (c). d T24 cells were treated with UNC0642 at the indicated concentrations for 48 h. The expression levels of Ampkα2, ELL2, and Bim were determined with qRT-PCR. *P < 0.05; **P < 0.01; ***P < 0.001
Targeting of G9a with UNC0642 suppressed tumor growth in vivo
To test the in vivo effects of G9a inhibition, J82 cells were subcutaneously injected into nude mice. One week after xenografts became palpable, UNC0642 (5 mg/kg) was administered via i.p. injection every other day for 11 days. UNC0642 inhibited tumor growth during the treatment window (P < 0.05; Fig. 6a) without a significant effect on body weight compared with the vehicle-treated group (Fig. 6b). At the endpoint of the experiment, xenografts were harvested, weighed, and processed for further IHC study. The average tumor weight in the UNC0642 group (1.15 g) was approximately half of that in the vehicle treatment group (2.30 g, P < 0.05; Fig. 6c, d). Moreover, IHC staining demonstrated that UNC0642 treatment strikingly decreased the histone H3K9me2 level in the nuclei of cancer cells in J82 xenografts (Fig. 6e). Ki67 (cell proliferation marker)-positive staining and cleaved Caspase 3 (apoptosis marker)-positive staining in cells were quantified (Fig. 6f, g). Targeting of G9a in vivo reduced cell proliferation (from 61.73% in the vehicle group to 22.20% in the UNC0642 group; P < 0.01) and induced apoptosis (from 0.36% in the vehicle group to 4.89% in the UNC0642 group; P < 0.05). In addition, the level of the proapoptotic protein BIM was increased in the UNC0642 treatment group, consistent with the in vitro data (Fig. 6h). Thus, targeting of G9a with UNC0642 suppressed UBC tumor growth and induced apoptosis in vivo.
UNC0642 suppressed the tumorigenicity of J82 xenografts. The tumor volume (a) of the J82 xenografts and the body weights of nude mice bearing J82 xenografts (b) are presented. Arrows indicate the timepoints of administration of UNC0642 or vehicle. c The xenografts were harvested at the end of the experiment and photographed. d The tumor weights are plotted in the graph. e–g IHC staining to determine H3K9me2, cleaved Caspase 3 and Ki67 (proliferation index) levels in J82 xenografts treated with vehicle or UNC0642 (5 mg/kg). Scale bar, 20 μm (e). Quantification of the cells positive for Ki67 (f) and cleaved Caspase 3 (g) staining between the vehicle and UNC0642 treatment groups. h Western blotting and quantification of BIM protein levels in UNC0642 and vehicle treatment groups. *P < 0.05, **P < 0.01; ***P < 0.001
DISCUSSION
G9a has been shown to be overexpressed in multiple cancer types [10, 15,16,17]. Knockdown of G9a via RNA interference in breast or gastric cancer cells inhibited cell proliferation and invasion, indicating that G9a overexpression is required for cancer cell survival and aggressiveness [19, 20]. Although G9a overexpression was not correlated with poor prognosis in UBC patients from TCGA Provisional dataset (n = 405, P>0.05), we still found that G9a was highly expressed in human UBC patients (n = 454, P < 0.05), especially in patients with neuronal UBC, which is the most malignant type of UBC and is presently incurable [27]. Because G9a is a promising drug target, thus far, several small molecule inhibitors have been identified with great in vitro inhibitory effects against G9a activity. However, to date, few G9a pharmacological inhibitors have demonstrated excellent in vivo efficacy and pharmacological kinetics [24]. A recent study reported that one G9a-specific inhibitor, UNC0642, exhibited high in vitro cellular potency, excellent selectivity and greatly improved in vivo pharmacokinetic properties [24, 25]. In this study, we applied UNC0642 to target the G9a gene, which was overexpressed in human UBC patients. UNC0642 reduced cell viability and induced apoptosis in UBC cells in a dose-dependent manner. In vivo data also showed that UNC0642 suppressed the tumorigenicity of UBC xenografts.
BIX01294 is the first selective inhibitor of G9a/GLP discovered via high-throughput screening. Depletion of G9a or inhibition of its activity by BIX-01294 reduced the expression levels of anti-apoptotic proteins, such as Bcl-2 and Mcl1 [22, 28, 29], and induced the expression of proapoptotic proteins, such as BAX and BAD [28, 30]. In addition, BIX-01294 induces autophagy in breast cancer, head and neck squamous cell carcinoma, and neuroblastoma cells [31,32,33]. However, BIX01294 exhibits selectivity for GLP over G9a, and thus, more potent G9a inhibitors, including UNC0224, UNC0321, and E72, were developed based on the crystal structure of the protein complex [34]. Further, this series of compounds were optimized for discovery of more potent and selective G9a inhibitors, specifically, UNC0638 and UNC0642. UNC0642 has been demonstrated to possess excellent selectivity and improved in vivo PK properties compared with other G9a-specific inhibitors, such as UNC0638, which makes it suitable for animal studies [23, 24]. We found that UNC0642 inhibited G9a activity, decreased the level of histone H3K9me2 and induced the expression of the proapoptotic protein BIM, leading to tumorigenicity suppression in UBC xenografts. Notably, the body weights of nude mice did not show significant changes between the vehicle control and UNC0642 treatment groups, indicating that the dosage regimen of UNC0642 used in this study was relatively safe.
In conclusion, the present study is the first to report that G9a regulates UBC cell survival by mediating cell apoptosis and that targeting of G9a by UNC0642 significantly inhibits UBC cell proliferation and survival in vitro and in vivo. Thus, we have confirmed that UNC0642 has potential as a therapeutic option for UBC patients.
References
Antoni S, Ferlay J, Soerjomataram I, Znaor A, Jemal A, Bray F. Bladder cancer incidence and mortality: a global overview and recent trends. Eur Urol. 2017;71:96–108.
Cheung G, Sahai A, Billia M, Dasgupta P, Khan MS. Recent advances in the diagnosis and treatment of bladder cancer. BMC Med. 2013;11:13.
Knowles MA, Hurst CD. Molecular biology of bladder cancer: new insights into pathogenesis and clinical diversity. Nat Rev Cancer. 2015;15:25–41.
Kamat AM, Hahn NM, Efstathiou JA, Lerner SP, Malmstrom PU, Choi W, et al. Bladder cancer. Lancet. 2016;388:2796–810.
Giacalone NJ, Shipley WU, Clayman RH, Niemierko A, Drumm M, Heney NM, et al. Long-term outcomes after bladder-preserving tri-modality therapy for patients with muscle-invasive bladder cancer: an updated analysis of the massachusetts general hospital experience. Eur Urol. 2017;71:952–60.
Fang T, Liu DD, Ning HM, Liu D, Sun JY, Huang XJ. et al. Modified citrus pectin inhibited bladder tumor growth through downregulation of galectin-3. Acta Pharmacol Sin. 2018;39:1885–93.
Rada M, Vasileva E, Lezina L, Marouco D, Antonov AV, Macip S, et al. Human EHMT2/G9a activates p53 through methylation-independent mechanism. Oncogene. 2017;36:922–32.
Olcina MM, Leszczynska KB, Senra JM, Isa NF, Harada H, Hammond EM. H3K9me3 facilitates hypoxia-induced p53-dependent apoptosis through repression of APAK. Oncogene. 2016;35:793–9.
Li F, Zeng J, Gao Y, Guan Z, Ma Z, Shi Q, et al. G9a inhibition induces autophagic cell death via AMPK/mTOR pathway in bladder transitional cell carcinoma. PLoS ONE. 2015;10:e0138390.
Casciello F, Windloch K, Gannon F, Lee JS. Functional role of G9a histone methyltransferase in cancer. Front Immunol. 2015;6:487.
Yuan Y, Tang AJ, Castoreno AB, Kuo SY, Wang Q, Kuballa P, et al. Gossypol and an HMT G9a inhibitor act in synergy to induce cell death in pancreatic cancer cells. Cell Death Dis. 2013;4:e690.
Agarwal P, Jackson SP. G9a inhibition potentiates the anti-tumour activity of DNA double-strand break inducing agents by impairing DNA repair independent of p53 status. Cancer Lett. 2016;380:467–75.
Cho HS, Kelly JD, Hayami S, Toyokawa G, Takawa M, Yoshimatsu M, et al. Enhanced expression of EHMT2 is involved in the proliferation of cancer cells through negative regulation of SIAH1. Neoplasia. 2011;13:676–84.
Tachibana M, Sugimoto K, Nozaki M, Ueda J, Ohta T, Ohki M, et al. G9a histone methyltransferase plays a dominant role in euchromatic histone H3 lysine 9 methylation and is essential for early embryogenesis. Genes Dev. 2002;16:1779–91.
Hu L, Zang MD, Wang HX, Zhang BG, Wang ZQ, Fan ZY, et al. G9A promotes gastric cancer metastasis by upregulating ITGB3 in a SET domain-independent manner. Cell Death Dis. 2018;9:278.
Hua KT, Wang MY, Chen MW, Wei LH, Chen CK, Ko CH, et al. The H3K9 methyltransferase G9a is a marker of aggressive ovarian cancer that promotes peritoneal metastasis. Mol Cancer. 2014;13:189.
Zhong X, Chen X, Guan X, Zhang H, Ma Y, Zhang S, et al. Overexpression of G9a and MCM7 in oesophageal squamous cell carcinoma is associated with poor prognosis. Histopathology. 2015;66:192–200.
Kondo Y, Shen L, Ahmed S, Boumber Y, Sekido Y, Haddad BR, et al. Downregulation of histone H3 lysine 9 methyltransferase G9a induces centrosome disruption and chromosome instability in cancer cells. PLoS One. 2008;3:e2037.
Dong C, Wu Y, Yao J, Wang Y, Yu Y, Rychahou PG, et al. G9a interacts with Snail and is critical for Snail-mediated E-cadherin repression in human breast cancer. J Clin Invest. 2012;122:1469–86.
Lin X, Huang Y, Zou Y, Chen X, Ma X. Depletion of G9a gene induces cell apoptosis in human gastric carcinoma. Oncol Rep. 2016;35:3041–9.
Wang L, Dong X, Ren Y, Luo J, Liu P, Su D, et al. Targeting EHMT2 reverses EGFR-TKI resistance in NSCLC by epigenetically regulating the PTEN/AKT signaling pathway. Cell Death Dis. 2018;9:129.
Guo AS, Huang YQ, Ma XD, Lin RS. Mechanism of G9a inhibitor BIX01294 acting on U251 glioma cells. Mol Med Rep. 2016;14:4613–21.
Ding J, Li T, Wang X, Zhao E, Choi JH, Yang L, et al. The histone H3 methyltransferase G9A epigenetically activates the serine-glycine synthesis pathway to sustain cancer cell survival and proliferation. Cell Metab. 2013;18:896–907.
Liu F, Barsyte-Lovejoy D, Li F, Xiong Y, Korboukh V, Huang XP, et al. Discovery of an in vivo chemical probe of the lysine methyltransferases G9a and GLP. J Med Chem. 2013;56:8931–42.
Kim Y, Lee HM, Xiong Y, Sciaky N, Hulbert SW, Cao X, et al. Targeting the histone methyltransferase G9a activates imprinted genes and improves survival of a mouse model of Prader-Willi syndrome. Nat Med. 2017;23:213–22.
McConkey DJ, Choi W. Molecular Subtypes of Bladder Cancer. Curr Oncol Rep. 2018;20:77.
Robertson AG, Kim J, Al-Ahmadie H, Bellmunt J, Guo G, Cherniack AD, et al. Comprehensive Molecular Characterization of Muscle-Invasive Bladder. Cancer Cell. 2018;174:1033.
Huang Y, Zou Y, Lin L, Ma X, Huang X. Effect of BIX-01294 on proliferation, apoptosis and histone methylation of acute T lymphoblastic leukemia cells. Leuk Res. 2017;62:34–9.
Cui J, Sun W, Hao X, Wei M, Su X, Zhang Y, et al. EHMT2 inhibitor BIX-01294 induces apoptosis through PMAIP1-USP9X-MCL1 axis in human bladder cancer cells. Cancer Cell Int. 2015;15:4.
Choi JY, Lee S, Hwang S, Jo SA, Kim M, Kim YJ, et al. Histone H3 lysine 27 and 9 hypermethylation within the Bad promoter region mediates 5-Aza-2’-deoxycytidine-induced Leydig cell apoptosis: implications of 5-Aza-2’-deoxycytidine toxicity to male reproduction. Apoptosis. 2013;18:99–109.
Kim Y, Kim YS, Kim DE, Lee JS, Song JH, Kim HG, et al. BIX-01294 induces autophagy-associated cell death via EHMT2/G9a dysfunction and intracellular reactive oxygen species production. Autophagy. 2013;9:2126–39.
Li KC, Hua KT, Lin YS, Su CY, Ko JY, Hsiao M, et al. Inhibition of G9a induces DUSP4-dependent autophagic cell death in head and neck squamous cell carcinoma. Mol Cancer. 2014;13:172.
Ke XX, Zhang D, Zhu S, Xia Q, Xiang Z, Cui H. Inhibition of H3K9 methyltransferase G9a repressed cell proliferation and induced autophagy in neuroblastoma cells. PLoS ONE. 2014;9:e106962.
Wang DY, Kosowan J, Samsom J, Leung L, Zhang KL, Li YX, et al. Inhibition of the G9a/GLP histone methyltransferase complex modulates anxiety-related behavior in mice. Acta Pharmacol Sin. 2018;39:866–74.
Acknowledgements
We thank the Institutional Technology Service Center of Shanghai Institute of Materia Medica, Xiao-hong Yu, and Zheng-nan Huang for technical support. This work was supported by the National Natural Science Foundation of China (81771890 to RH and 81772712 to XY), One Hundred Talent Program of Chinese Academy of Sciences (to RH) and China Postdoctoral Science Foundation (no. 2018M630484 to JS).
Author Contributions
JY, RH, and XY conceived and designed the experiments; YC, JS, ML, YD, and YZ performed the experiments; YC, JS, ML, and JY analyzed the data; YC, JS, ML, JY, and RH wrote the paper.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Cao, Yp., Sun, Jy., Li, Mq. et al. Inhibition of G9a by a small molecule inhibitor, UNC0642, induces apoptosis of human bladder cancer cells. Acta Pharmacol Sin 40, 1076–1084 (2019). https://doi.org/10.1038/s41401-018-0205-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41401-018-0205-5
Keywords
This article is cited by
-
Targeting histone modifiers in bladder cancer therapy — preclinical and clinical evidence
Nature Reviews Urology (2024)
-
TRAIL-mediated signaling in bladder cancer: realization of clinical efficacy of TRAIL-based therapeutics in medical oncology
Medical Oncology (2023)
-
Dynamic de novo heterochromatin assembly and disassembly at replication forks ensures fork stability
Nature Cell Biology (2023)
-
Role of HOXA9 in solid tumors: mechanistic insights and therapeutic potential
Cancer Cell International (2022)
-
De novo methylation of histone H3K23 by the methyltransferases EHMT1/GLP and EHMT2/G9a
Epigenetics & Chromatin (2022)