Abstract
Triple-negative breast cancer (TNBC) is a heterogeneous disease with a poor prognosis due to the lack of an effective targeted therapy. Histone lysine methyltransferases (KMTs) have emerged as attractive drug targets for cancer therapy. However, the function of the majority of KMTs in TNBC has remained largely unknown. In the current study, we found that KMT nuclear receptor binding SET domain protein 2 (NSD2) is overexpressed in TNBC tumors and that its overexpression is associated with poor survival of TNBC patients. NSD2 regulates TNBC cell survival and invasion and is required for tumorigenesis and tumor growth. Mechanistically, NSD2 directly controls the expression of EGFR and ADAM9, a member of the ADAM (a disintegrin and metalloproteinase) family that mediates the release of growth factors, such as HB-EGF. Through its methylase activity, NSD2 overexpression stimulates EGFR-AKT signaling and promotes TNBC cell resistance to the EGFR inhibitor gefitinib. Together, our results identify NSD2 as a major epigenetic regulator in TNBC and provide a rationale for targeting NSD2 alone or in combination with EGFR inhibitors as a targeted therapy for TNBC.
Similar content being viewed by others
Introduction
Triple-negative breast cancer (TNBC) is a heterogeneous disease with tumors lacking the expression of estrogen receptor-alpha (ER), progesterone receptor, and HER-2. Although representing approximately 15% of all breast cancer cases, TNBC is considered one of the most aggressive subtypes of breast cancer. Most TNBC patients relapse from chemotherapy and suffer a poor outcome largely due to lack of effective targeted therapy [1]. Based largely on gene expression profiling, TNBC tumors can be classified into several subgroups. One major group is basal-like and defined by tumor expression of genes characteristic of normal basal/myoepithelial cells including specific cytokeratins and EGFR. EGFR, an actionable target, is frequently overexpressed (over 50% in some studies) in TNBC tumors. However, clinical trials with therapeutics targeting EGFR have yet to demonstrate significant benefits in patients, presumably because of multiple activation mechanisms of EGFR [2, 3].
Elevated EGFR expression and activation plays important roles in cancer cell survival, growth, invasion and tumor metastasis in TNBCs as well as other cancers [3, 4]. Transcriptional deregulation of EGFR has been well documented in different tumors [5, 6]. Early studies have shown that transcription factors such as Sp1, TGF-beta inducible early gene 1 and receptor interacting protein play important roles in the control of EGFR transcription [7,8,9]. A recent report demonstrated that EGFR is repressed by a Kruppel C2H2 zinc-finger protein ZNF516 through recruitment of the CtBP-LSD1-CoREST corepressor complex [10]. However, the mechanisms by which the EGFR gene is activated through epigenetic factors at the chromatin level are still poorly understood. Nuclear receptor-binding SET domain protein 1, 2, and 3 (NSD1, NSD2 and NSD3) are members of a SET histone methyltransferase family, which possesses a SET domain with lysine methyltransferase activity [11]. NSD2, also known as Wolf-Hirschhorn syndrome candidate 1 (WHSC1) or MMSET (multiple myeloma SET domain), catalyzes di- and trimethylation of lysine 36 on histone H3 (H3K36me2/3). Its overexpression increases global H3K36me2/3 in cells. H3K36me2 is an epigenetic mark that is commonly associated with transcriptionally active loci [12]. Therefore, NSD2 has been characterized as a transcriptional activator, although previous reports have shown that NSD2 can interact with histone deacetylase HDAC1 and histone demethylase LSD1 to suppress gene expression in specific contexts [12,13,14,15]. The NSD2 gene is fused to the IgH locus via t(4;14) translocation in 15–20% of multiple myeloma (MM) cases, and its overexpression is likely responsible for the tumorigenic expansion of MM cells [13, 16, 17]. Studies by our group and others have found that the NSD2 protein is highly overexpressed in several types of human tumors, including prostate cancer, neuroblastoma, carcinomas of the stomach and colon, small-cell lung cancers, and bladder cancers, and that its overexpression is associated with tumor aggressiveness [11, 18, 19]. However, whether NSD2 plays a role in TNBC remains unclear.
In the current study, we found that NSD2 protein is overexpressed in TNBC tumors and that its overexpression is associated with poor survival. We also demonstrated that NSD2 regulates TNBC cell survival and invasion and tumor growth by directly controlling the expression and signaling of ADAM9 and EGFR. Our results thus suggest NSD2 as new therapeutic target for TNBC.
Materials and methods
Cell culture and reagents
Two TNBC cell lines, MDA-MB-231(MB-231) and MDA-MB-436 (MB-436), were obtained from ATCC. MDA-MB-436 (MB-436) cells were cultured in RPMI medium (Gibco, Grand Island, NY, USA). MDA-MB-231(MB-231) cells were cultured in DMEM medium (Gibco, Grand Island, NY, USA). Cell culture medium was supplemented with 1% penicillin/streptomycin and 10% fetal bovine serum (Gemini Bio Products, West Sacramento, CA, USA) at 37 ℃ under 5% CO2 in a humidified incubator. Antibodies against the following proteins were used with the sources and dilution ratios indicated in parentheses: NSD2 (29D1, Abcam; ab75359; 1:2000); EGFR (Cell Signaling; #4267; 1:2000); β-actin (Santa Cruz; sc-47778; 1:2000); phosphor-EGFR (Tyr1068, Cell Signaling; #2236; 1:1,000); AKT (Cell Signaling; #9272; 1:1000); pAKT (Tyr473, Cell Signaling; #4051; 1:1000); ERK1/2 (Epitomics; #1171; 1:1000); pERK1/2 (Thr202/Tyr204, #4370; Cell Signaling; 1:500); STAT3 (Cell Signaling; #9132; 1:1,000); pSTAT3 (Tyr705, Cell Signaling; #9131; 1:1000); ADAM9 (Santa Cruz; sc-135822; 1:500); GAPDH (Cell Signaling; #2118; 1:500); H3K36me2 (Active motif; #39255; 1:2000); H3K36me3 (Active motif; #61021; 1:2000); H3 (Active motif; #39163; 1:2000). All chemicals were purchased from Sigma-Aldrich (St. Louis, MO) unless specified otherwise.
Apoptosis and cell growth assays
For apoptosis, terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) was performed using an in situ cell death detection kit (Roche) as previously described [20]. The results are expressed as a percentage of the apoptotic cell number/total cell number. For cell growth, cells were seeded in 6-well plates at 2 × 105 per well and treated as indicated. Total survival cell numbers were counted using a Coulter cell counter. The assays were performed in triplicate, and the experiments were repeated more than three times.
Immunohistochemistry (IHC) and statistical analysis
IHC was performed as previously described [11, 20] with the following modifications. The slides were incubated with anti-NSD2 monoclonal antibody (29D1; Abcam) at 1:50 dilutions overnight at 4 °C, followed by biotinylated secondary antibody and ABC reagents in the Vectastain Elite kit and counter-stained with hematoxylin. NSD2 IHC was performed on tissue microarrays containing specimens from 234 cases of informative breast cancer collected at UC Davis Cancer center. The percentage of positive nuclear staining was scored as follows: 0; <5%, score 0; 5; <10%, score 1; 10; 50%, score 2; >50%, score 3. Differences and correlations in immunostaining among groups were analyzed with the χ2 test.
Analysis of tumor NSD2 mRNA expression association with clinical outcome
Kaplan–Meier curves were obtained for relapse-free survival times of patients (n = 255) with their TNBC tumors stratified by NSD2 expression levels, using an online survival analysis tool (http://kmplot.com), as described [21]. Statistical significance was determined by the log-rank test.
Chromatin immunoprecipitation (ChIP) assay
ChIP was performed essentially as described previously [11] with the following modifications. Cells were lysed and sonicated using Sonic Dismembrator 550 (Fisher). The crude chromatin solutions were first cleared with protein A beads (Invitrogen) that had been pre-coated with normal IgG for 2 h at 4 °C. Then, the pre-cleared solutions were incubated at 4 °C overnight with antibodies against H3K36me2 (Abcam), H3K36me3 (Abcam) and NSD2 prior to precipitation with protein A beads that had been preblocked with BSA and sonicated salmon sperm DNA, and normal IgG was used as a negative control. The precipitated DNA was analyzed by real-time PCR with SYBR green on an iCycler instrument. Enrichment of genomic DNA was presented as the percent recovery relative to the input. The primers used were as follows: EGFR chip Forward 5′-GCTGGATGAGGAGGAAAGATGTAAGG-3′, EGFR chip Reverse 5′-GGGCTGCAGGAACTTGAGCTATATTC-3′; ADAM9 chip Forward 5′-TACTCTCCTTGGGCTAAAGGG-3′, ADAM9 chip Reverse 5′-GGTCGTTCCGGAGGCAAATAT-3′.
qRT-PCR and immunoblot analysis
Total RNA was isolated, and cDNA was prepared, amplified and detected in the presence of SYBR as previously described [11]. The fluorescent values were collected, and a melting curve analysis was performed. The fold difference was calculated as described previously [11]. Cell lysates were analyzed by immunoblotting with the indicated antibodies. The PCR primers used were as follows: EGFR Forward 5′-CAGCGCTACCTTGTCATTCA-3′, EGFR Reverse 5′-TGCACTCAGAGAGCTCAGGA-3′; ADAM9 Forward 5′-TGGGGGTGCTGGTGATGTGC-3′, ADAM9 Reverse 5′-CGCCTGCGTGGCTCCTTGAA-3′.
Colony formation
For the colony formation assay, 800 cells per well were seeded in a 6-well plate and cultured for 10–14 days in a 37 °C incubator changing the medium every 3 days. The cells were fixed with 10% formalin and stained with 0.2% crystal violet (in 10% formalin) for 15 min. The numbers of cell colonies were counted after washes. The assays were performed in triplicate, and the entire experiments were repeated more than three times.
Xenograft tumor models
For the breast cancer MDA-MB231 xenograft model, 5 to 6-wk-old BALB/c nu/nu athymic mice (Harlan, Madison, WI) were injected unilaterally with 2.5 × 106 cells (infected with lentivirus shControl or shNSD2) in 100 μL PBS/Matrigel (1:1) into the abdominal fat pad by subcutaneous injection at the base of the nipple. Tumor growth was monitored by calipers, and the volume was calculated using the following equation: (π/6 (length × width2)). At the end of the studies, the mice were sacrificed and tumors dissected. Tumors were harvested and immediately stored in liquid nitrogen for later tissue lysate preparation. The procedures were approved by the Institutional Animal Care and Use Committee of the University of California Davis.
Statistical analysis
The data are presented as the mean values ± SD from a minimum of three independent experiments. Data analysis was performed using two-tailed Student’s t tests, and the P values are shown. *P < 0.05; **P < 0.01; N.S. not significant.
Results
Histone methyltransferase NSD2 overexpression in TNBC tumors is significantly correlated with poor survival
Aberrant NSD2 expression is observed in solid tumors of several types of human cancer, including lung, gastric, bladder, colon and prostate cancer [11, 18, 19, 22]. Our recent IHC analysis of a cohort of more than 450 estrogen receptor (ER)-positive breast cancers demonstrated that NSD2 overexpression in ER-positive tumors is strongly associated with early relapse from tamoxifen treatment and poor survival [23]. However, its expression in TNBC tumors has not yet been investigated. We carried out an IHC analysis of NSD2 with breast cancer specimens from a total of 234 cases. Among them, 56 were TNBC tumors. Consistent with our recent study, we found that while normal or hyperplastic tissue displayed no or occasionally positive nuclear staining by anti-NSD2 antibody, more than 30% of the tumors examined displayed NSD2 protein overexpression with nuclear staining mostly in tumor epithelial cells (Fig. 1a). When tumors of low or high grade were compared, a significant correlation (P < 0.0001) between NSD2 protein overexpression and a high grade was observed (Fig. 1a). Interestingly, we found that 35 of 56 cases (62%) of TNBC tumors showed strong NSD2 expression, while 40 of 148 non-TNBC tumors (27%) displayed strong positive staining (P < 0.0001) (Fig. 1a). Kaplan Meier analysis showed that NSD2 protein overexpression in TNBC was significantly associated with earlier disease-related death (Fig. 1b). Furthermore, analysis of published tumor datasets also showed that NSD2 transcripts were overexpressed in TNBC and that high NSD2 mRNA levels were significantly correlated (P = 0.027) with poor survival of TNBC patients (Fig. 1c, d). Taken together, the above results suggest that overexpression of NSD2 mRNA and protein in TNBC tumors is significantly associated with poor survival of TNBC patients.
Histone methyltransferase NSD2 is overexpressed in TNBCs. a Representative images from NSD2 IHC of normal breast or a cohort of TNBC breast cancer tumor specimens (top) and statistical analysis of the association of NSD2 protein levels with pathological parameters. b Kaplan–Meier analysis of overall patient survival from death due to TNBC were stratified by tumor NSD2 protein levels determined by IHC. c Kaplan–Meier survival curve analysis of the probability of overall survival in TNBC patients stratified by NSD2 expression. d NSD2 transcript levels from four Gene Expression Omnibus (GEO) data sets were queried for association with disease status
NSD2 is required for the proliferation and survival of TNBC cells
Given the high levels of NSD2 expression in TNBC tumors, we first examined the expression and function of NSD2 in TNBC cells. In agreement with results from our tumor analysis, we found that NSD2 was overexpressed in breast cancer cell lines compared with normal human breast epithelial cells. Interestingly, TNBC cell lines appeared to express higher levels of NSD2 than the ER-positive cells (Fig. 2a). Knockdown of NSD2 with siRNAs showed that NSD2 silencing significantly inhibited the proliferation of both MB-231 and MB-436 TNBC cell lines (Fig. 2b). To determine whether the decrease in cell growth was partly due to apoptotic cell death, we performed a TUNEL assay and found that NSD2 depletion resulted in a marked increase in cell apoptosis (Fig. 2c). Taken together, these data indicate that high levels of NSD2 are required for the proliferation and survival of TNBC cells.
NSD2 is required for the proliferation and survival of TNBC cells. a Immunoblotting analysis of NSD2 protein in cell cultures of breast cancer cells and normal human breast epithelial cells (HMEC). b MB-231 and MB-436 cells were transfected with NSD2 or control siRNA. Three days later, the cells were harvested for immunoblotting with NSD2 and GAPDH antibodies. c MB-231 and MB-436 cells were transfected with NSD2 or control siRNA. At the indicated times, cells were harvested for determining cell growth by counting viable cells. Cells were harvested for immunoblotting with NSD2 and GAPDH antibodies. d MB-231 and MB-436 cells were transfected with NSD2 or control siRNA. After 4 days, TUNEL-positive apoptotic cells were counted and expressed as the percentage of total cells. Data are shown as the mean ± SD. Significance was calculated using Student’s t-test. **P < 0.01
NSD2 silencing suppresses TNBC cell invasion and migration
TNBC tumors are often highly invasive and display a rapid onset of distant metastasis. To examine the potential function of NSD2 in TNBC cell invasion and migration, we first performed Matrigel transwell assays to assess the effect of NSD2 silencing on the TNBC cell invasion capacity. Our results showed that NSD2 silencing significantly inhibited the invasion of both MB-231 and MB-436 cells (Fig. 3a). Similarly, NSD2 silencing also inhibited the migration of TNBC cells, as assessed by a wound-healing assay (Fig. 3b). These data suggest that NSD2 may play an important role in TNBC metastasis.
NSD2 silencing suppresses cell invasion and migration. a The invasive ability of MB-231 and MB-436 was assayed using a Matrigel-coated transwell. Cells were transfected with NSD2 or control siRNA, and after 48 h, the cells were plated in transwells and incubated for another 24 h. Cells that had invaded the Matrigel were quantified. b Cells were transfected with NSD2 or control siRNA, and after 48 h, the migration of MB-231 and MB-436 cells was evaluated using wound healing assays. Images from the same area were captured at time 0 and 72 h after wounding. Data are shown as the mean ± SD. Significance was calculated using Student’s t-test. **P < 0.01
NSD2 controls the expression and function of EGFR and ADAM9
To investigate the molecular pathways controlled by NSD2 in TNBC cells, we carried out a gene expression microarray analysis of MB-231 and MB-436 cells treated with control or NSD2 siRNA. A comparative analysis of genes with significantly altered expression in the two TNBC cell lines showed that receptor tyrosine kinase-mediated signaling represented one of the major pathways impacted by NSD2 silencing in both TNBC cells (Fig. 4a–c). Specifically, kinase signaling genes involved in cell survival and invasion, including ADAM9 and EGFR, were inhibited more than two-fold by NSD2 silencing (Fig. 4a). Our qRT-PCR analysis verified that the expression of ADAM9 and EGFR was indeed significantly inhibited by NSD2 knockdown in TNBC cells (Fig. 4b). The metalloproteases of a disintegrin and metalloprotease domain (ADAM) family were identified as the sheddases required for proteolytic processing and release of membrane proteins and growth factors, such as proheparin–binding epidermal growth factor-like growth factor (pro-HB-EGF) ligand precursors [24]. ADAM9 cleaves the transmembrane -proHB-EGF- precursor to yield HB-EGF, a soluble EGFR ligand, to activate EGFR and its intracellular signaling cascades, such as the AKT and STAT3 pathways, and thereby promote cell survival and invasion [24,25,26,27]. We thus focused our analysis on ADAM9 and EGFR. Indeed, Western blotting analysis showed that NSD2 silencing significantly inhibited EGFR and ADAM9 protein expression and reduced both the total and Tyr1068- phosphorylated/activated form of EGFR (Fig. 4c). As expected, NSD2 silencing resulted in a marked reduction of AKT activation, as demonstrated by deceased phosphorylation in both TNBC cells (Fig. 4c). Intriguingly, in TNBC cells, silencing also reduced the activation of STAT3 (Fig. 4c). These results suggest that NSD2 is required for the overexpression and/or hyperactivation of ADAM9 and the EGFR-AKT/STAT3 signaling pathway in TNBC cells. To assess the clinical significance of our findings from cancer cells, we interrogated published tumor gene expression datasets. As shown in Fig. 4d, the expression of NSD2 was significantly correlated with that of EGFR in two different cohorts of TNBC tumors. Likewise, NSD2 expression was also significantly correlated with that of ADAM9 in TNBC tumors (Fig. 4e).
NSD2 silencing inhibits ADAM9-EGFR-AKT/STAT3 signaling in TNBC cells. a The number of genes with significantly altered expression upon NSD2 knockdown in MB-231 and MB-436 cell lines is shown (left). The heatmap displays the fold decrease in gene expression detected by microarray in MB-231 and MB-436 cells treated with NSD2 siRNA for 48 h, compared to control siRNA (right). Genes involved in cell survival and invasion with a fold decrease > 2 in both cell lines are displayed. b qRT-PCR analysis of ADAM9 and EGFR expression in MB-231 and MB-436 cells transfected with NSD2 or control siRNA. Data are shown as the mean ± SD. Significance was calculated using Student’s t-test. **P < 0.01. c MB-231 and MB-436 cells were transfected with control or NSD2 siRNA. Three days later, cell lysates were collected for immunoblotting for the indicated proteins. d, e Scatter plots showing the correlation between NSD2 and EGFR (d) or ADAM9 (e) gene expression in TNBC patient tumors with expression data from four different clinical data sets
NSD2 promotes EGFR signaling through its histone methylase activity
Next, we examined whether EGFR signaling was one of the major pathways controlled by NSD2 in TNBC cells. First, we tested whether activation of EGFR signaling by exogenous EGF ligand could block the effects of NSD2 silencing on TNBC cell survival and invasion. The results shown in Fig. 5a showed that exogenous EGF was effective in the complete protection of TNBC cells against the growth and invasion-inhibitory effects of NSD2 silencing.
NSD2 promotes EGFR signaling through its histone methylase activity. a EGF treatment rescues NSD2 silencing-mediated growth inhibition (left) or invasion suppression (right) of MB-231 and MB-436 cells. b MB-231 cells were transfected as with control or NSD2 siRNA. Two days later, the cells were treated with vehicle or gefitinib (10 µM) for another 3 days before viable cells were counted. c MB-231 cells were infected with control lentiviruses or lentiviruses that mediate ectopic expression of NSD2 or NSD2 Y1118A. Two days later, the cells were treated with gefitinib for another 3 days and then harvested for cell proliferation measurement by counting viable cells. d MB-231 cells were infected with lentiviruses as in (C). Three days later, cell lysates were prepared for immunoblotting for the indicated proteins. e MB-231 cells were transfected with control or NSD2 siRNA. Three days later, cell lysates were prepared for immunoblotting with the indicated proteins. Data are shown as the mean ± SD. Significance was calculated using Student’s t-test. **P < 0.01
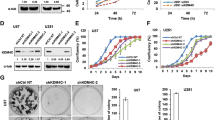
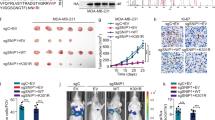
To further investigate the function of NSD2 in the control of EGFR signaling, we then examined whether alteration of NSD2 expression or function could affect the response of TNBC cells to the EGFR inhibitor gefitinib. As shown in Fig. 5b, NSD2 knockdown significantly enhanced the growth inhibitory effect of gefitinib in MB231 cells. Although it is well documented that the methylase activity of NSD2 is crucial for NSD2 function in cancer, major success has not yet been achieved in the development of NSD2 inhibitors that are highly potent and exhibit specific inhibition of NSD2 methylase activity. In a recent screening study, a few hit compounds were identified. However, most of them, excluding 3-deazaneplanocin A (DZNep), were compounds with previously described inhibitory activities against methylation-unrelated targets such as CDC25 phosphatase, a LPA and mineralocorticoid receptor [28]. Thus, we used DZNep, an inhibitor of SAM-dependent methylases, as a pharmacological tool to probe the importance of NSD2 methylase activity in EGFR signaling. Our results from the co-treatment of MB-231 cells with DZNep and gefitinib showed that DZNep significantly sensitized TNBC cells to gefitinib-induced growth inhibition (Supplementary Figure S1). In contrast, overexpression of wild-type NSD2, but not its methylase-defective mutant (NSD2-Y1118A), resulted in strong resistance of MB231 cells to high concentrations of gefitinib (Fig. 5c). Western blotting analysis showed that overexpressed wild-type NSD2, but not the mutant, increased the protein expression of EGFR and ADAM9, as well as the global H3K36me2/3 mark, and that NSD2 silencing decreased the global H3K36me2/3 mark (Fig. 5d and Fig. 5e). Taken together, the results suggest that NSD2 controls EGFR signaling in TNBC cells and that its histone methylase activity is critical for this function.
NSD2 directly regulates EGFR and ADAM9 gene expression by modulating histone methylation at target chromatin
NSD2 is a histone methyltransferase that is selective for di- and tri-methylation of residues lys36 of histone 3 tail (H3K36). We reasoned that NSD2 might be recruited to ADAM9 and EGFR gene promoters to mediate histone methylation to regulate their expression. Thus, we performed a chromatin immunoprecipitation (ChIP) assay with anti-NSD2 antibody. The immunoprecipitated ADAM9 and EGFR promoter sequences were quantified by qPCR to measure the binding of NSD2. As shown in Fig. 6a, b, anti-NSD2 ChIP indeed demonstrated that NSD2 was associated with the promoter region of EGFR and ADAM9 and that NSD2 knockdown could significantly decrease this binding. To further investigate the role of NSD2 in the regulation of ADAM9 and EGFR, we examined the H3K36me2/3 status on the ADAM9 and EGFR gene promoter. The results showed that NSD2 silencing effectively decreased the H3K36me2 mark and H3K36me3 mark at the gene promoter (Fig. 6a, b). These data suggest that NSD2 directly regulates ADAM9 and EGFR gene expression through binding to their promoters and increasing the local H3K36me2/3 marks.
NSD2 directly regulates ADAM9 and EGFR gene expression through histone methylation. a MB-231 cells were transfected with control or NSD2 siRNA. Three days later, ChIP was performed with siRNA-treated cells. NSD2 occupancy and H3K36me2/3 marks at the EGFR promoter were measured by qPCR analysis of ChIP DNA. b Cells were treated as above in a, and ChIP-qPCR analysis was performed for NSD2 occupancy and H3K36me2/3 marks at the ADAM9 promoter. Data are shown as the mean ± SD. Significance was calculated using Student’s t-test. *P < 0.05; **P < 0.01
Targeting NSD2 inhibits TNBC tumorigenesis and tumor growth
We next assessed the function of NSD2 in TNBC tumorigenesis. We first examined the effect of NSD2 silencing on colony formation of TNBC cells and found that NSD2 silencing significantly decreased the colony formation of both MB-231 and MB-436 cells (Fig. 7a). Consistent with the in vitro results, NSD2 knockdown strongly blocked tumorigenesis because at 9 weeks after the implantation of MB-231 cells, most of the mice implanted with shNSD2-treated cells did not develop measurable, tumors while mice with shControl-treated cells developed tumors with a size > 400 mm3 (Fig. 7b). Moreover, at 10 weeks, some of the mice with shNSD2-treated cells developed tumors, but their tumor sizes were barely measurable. Western blotting analysis of the tumors revealed that tumors with NSD2 silencing had decreased expression of ADAM9 and EGFR proteins. Therefore, the above data indicate that targeting NSD2 through silencing its expression can effectively block tumorigenesis and inhibit tumor growth and that one underlying mechanism is likely through the suppression of EGFR and ADAM9 expression.
Inhibition of NSD2 blocks TNBC tumorigenesis and tumor growth. a Colony formation of MB-231 and MB-436 cells infected with lentiviruses expression NSD2 or control shRNA. Data are shown as the mean ± SD. Significance was calculated using Student’s t-test. **P < 0.01. b MDA-MB231 cells were infected with control or NSD2 shRNA lentivirus. The next day, 2.5 × 106 cells from each treatment were injected into the mammary fat pad of each mouse. Tumor growth and tumor volume were measured every week. Each data point is the mean value ± s.e.m obtained from 5 to 6 mice. *P < 0.05; **P < 0.01. c Immunoblotting analysis of NSD2, ADAM9 and EGFR protein expression in tumors using lysates prepared from MB-231 xenograft tumor tissues
Discussion
Histone modification has emerged as a major determinant of chromatin function and gene activity. Its deregulation is linked to cancer development [29]. Among the enzymes that modify histones, histone methyltransferases are attractive cancer therapy targets [30, 31]. However, the role of histone methyltransferases in TNBC remains poorly understood. In this study, we found that the histone methyltransferase NSD2 is overexpressed in TNBC cell lines and patient tumors and that elevated NSD2 plays a key role in TNBC cell survival, invasion and tumorigenesis. Furthermore, our results demonstrated, in vitro and in vivo, that one major mechanism of NSD2 function in TNBC is its stimulation of the expression and activity of the ADAM9-EGFR-AKT signaling pathway.
EGFR is frequently activated by mutation and overexpression in a variety of cancers. EGFR plays a key role in cancer development and growth, making it an attractive target for pharmacological intervention [26]. Two classes of EGFR antagonists, anti-EGFR monoclonal antibodies and small-molecule EGFR tyrosine kinase inhibitors, are now in clinical use for the treatment of metastatic non–small cell lung cancer (NSCLC), colorectal cancer, squamous cell carcinoma of the head and neck, and pancreatic cancer [26, 32]. Studies have also shown that EGFR is overexpressed in a large percentage of TNBC tumors and plays a key role in TNBC cell survival and invasion [33, 34], suggesting that EGFR could be a biomarker and/or a target for TNBC. Although EGFR antagonists significantly improve the prognosis of some cancer patients compared to that of those treated with standard chemotherapy, de novo and acquired resistance appears inevitable [26]. The development of additional activating mutations in EGFR and elevated levels of HB-EGF in the tumors are two major causes of treatment resistance to EGFR inhibitors. In this study, we show that NSD2 can directly activate the expression of EGFR and ADAM9. ADAM9 is a metalloprotease and plays an important function in processing and releasing the active form of HB-EGF, a soluble ligand of EGFR, and other growth factor polypeptides. Therefore, stimulation of ADAM9 expression by NSD2 will likely further enhance the signaling by EGFR. Moreover, our finding that NSD2 is a positive regulator of both EGFR and its upstream factor ADAM9 underscores the crucial role played by NSD2 in the deregulation of EGFR signaling. In contrast, although this study focused on transcriptional regulation of EGFR expression, we cannot rule out the possibility that NSD2 may control EGFR expression at post-transcriptional levels. The stability of EGFR protein, particularly its activated form, is subject to intricate regulation [35]. Promoting EGFR degradation is regarded as an attractive strategy for EGFR-targeted cancer therapy. Therefore, it will be important to examine whether NSD2 is also involved in the control of EGFR stability or degradation. Nevertheless, given that NSD2 exhibits aberrant expression and function in many types of cancers, it is tempting to speculate that the NSD2-ADAM9-EGFR axis defined in this study may operate in other cancers, such as NSCLC, where EGFR signaling is a major oncogenic driver.
In this study, we also demonstrate that NSD2 overexpression confers TNBC cell resistance to EGFR inhibitors, while NSD2 silencing or inhibition of its methylase activity with a tool compound sensitizes TNBC cells to EGFR inhibitors. Therefore, these results provide proof-of-principle evidence for the development of NSD2-selective, potent inhibitors that can be used either as a stand-alone therapy or in combination with other drugs, such as those that target EGFR, for the effective treatment of TNBC and other types of malignancy.
References
Bianchini G, Balko JM, Mayer IA, Sanders ME, Gianni L. Triple-negative breast cancer: challenges and opportunities of a heterogeneous disease [Review Article]. Nat Rev Clin Oncol. 2016;13:674.
Carey LA, et al. TBCRC 001: randomized phase II study of cetuximab in combination with carboplatin in stage IV triple-negative breast cancer. J Clin Oncol. 2012;30:2615–23.
Nakai K, Hung M-C, Yamaguchi H. A perspective on anti-EGFR therapies targeting triple-negative breast cancer. Am J Cancer Res. 2016;6:1609–23.
Costa R, et al. Targeting epidermal growth factor receptor in triple negative breast cancer: new discoveries and practical insights for drug development. Cancer Treat Rev. 2017;53:111–19.
Blanchot-Jossic F, et al. Up-regulated expression of ADAM17 in human colon carcinoma: co-expression with EGFR in neoplastic and endothelial cells. J Pathol. 2005;207:156–63.
Al-Ejeh F, et al. Treatment of triple-negative breast cancer using anti-EGFR–directed radioimmunotherapy combined with radiosensitizing chemotherapy and PARP inhibitor. J Nucl Med. 2013;54:913–21.
Zhao Y., et al. TGF-β transactivates EGFR and facilitates breast cancer migration and invasion through canonical Smad3 and ERK/Sp1 signaling pathway. Molecular Oncology 2018;12:305–321.
Jin W, et al. TIEG1 inhibits breast cancer invasion and metastasis by inhibition of epidermal growth factor receptor (EGFR) transcription and the EGFR signaling pathway. Mol Cell Biol. 2012;32:50–63.
Ramnarain DB, et al. RIP1 links inflammatory and growth factor signaling pathways by regulating expression of the EGFR [Original Paper]. Cell Death Differ. 2007;15:344.
Li L, et al. ZNF516 suppresses EGFR by targeting the CtBP/LSD1/CoREST complex to chromatin. Nat Commun. 2017;8:691.
Yang P, et al. Histone methyltransferase NSD2/MMSET mediates constitutive NF-kappaB signaling for cancer cell proliferation, survival, and tumor growth via a feed-forward loop. Mol Cell Biol. 2012;32:3121–31.
Kuo AJ, et al. NSD2 links dimethylation of histone H3 at lysine 36 to oncogenic programming. Mol Cell. 2011;44:609–20.
Marango J, et al. The MMSET protein is a histone methyltransferase with characteristics of a transcriptional corepressor. Blood. 2008;111:3145–54.
Martinez-Garcia E, et al. The MMSET histone methyl transferase switches global histone methylation and alters gene expression in t(4;14) multiple myeloma cells. Blood. 2011;117:211–20.
Nimura K, et al. A histone H3 lysine 36 trimethyltransferase links Nkx2-5 to Wolf-Hirschhorn syndrome. Nature. 2009;460:287–91. eng.
Lauring J, et al. The multiple myeloma associated MMSET gene contributes to cellular adhesion, clonogenic growth, and tumorigenicity. Blood. 2008;111:856–64. eng.
Huang Z, et al. NSD2 is recruited through its PHD domain to oncogenic gene loci to drive multiple myeloma. Cancer Res. 2013;73:6277–88.
Hudlebusch HR, et al. MMSET is highly expressed and associated with aggressiveness in neuroblastoma. Cancer Res. 2011;71:4226–35. eng.
Hudlebusch HR, et al. The histone methyltransferase and putative oncoprotein MMSET is overexpressed in a large variety of human tumors. Clin Cancer Res. 2011;17:2919–33.
Kalashnikova EV, et al. ANCCA/ATAD2 overexpression identifies breast cancer patients with poor prognosis, acting to drive proliferation and survival of triple-negative cells through control of B-Myb and EZH2. Cancer Res. 2010;70:9402–12.
Zou JX, et al. Kinesin family deregulation coordinated by bromodomain protein ANCCA and histone methyltransferase MLL for breast cancer cell growth, survival, and tamoxifen resistance. Mol Cancer Res. 2014;12:539.
Toyokawa G, et al. Histone lysine methyltransferase Wolf-Hirschhorn syndrome candidate 1 is involved in human carcinogenesis through regulation of the Wnt pathway. Neoplasia. 2011;13:887–98.
Wang J, et al. Reprogramming metabolism by histone methyltransferase NSD2 drives endocrine resistance via coordinated activation of pentose phosphate pathway enzymes. Cancer Lett. 2016;378:69–79.
Fischer OM, Hart S, Gschwind A, Prenzel N, Ullrich A. Oxidative and osmotic stress signaling in tumor cells is mediated by ADAM proteases and heparin-binding epidermal growth factor. Mol Cell Biol. 2004;24:5172–83.
Liu R, et al. DNMT1–microRNA126 epigenetic circuit contributes to esophageal squamous cell carcinoma growth via ADAM9–EGFR–AKT signaling. Clin Cancer Res. 2015;21:854–63.
Ciardiello F, Tortora G. EGFR antagonists in cancer treatment. New Engl J Med. 2008;358:1160–74.
Itabashi H, et al. Angiotensin II and epidermal growth factor receptor cross-talk mediated by a disintegrin and metalloprotease accelerates tumor cell proliferation of hepatocellular carcinoma cell lines. Hepatol Res. 2008;38:601–13.
Coussens NP, et al. High-throughput screening with nucleosome substrate identifies small-molecule inhibitors of the human histone lysine methyltransferase NSD2. J Biol Chem. 2018;293:13750–65.
Copeland RA, Solomon ME, Richon VM. Protein methyltransferases as a target class for drug discovery. Nat Rev Drug Discov. 2009;8:724–32.
Tian X, et al. Histone lysine-specific methyltransferases and demethylases in carcinogenesis: new targets for cancer therapy and prevention. Curr Cancer Drug Targets. 2013;13:558–79.
Wagner T, Jung M. New lysine methyltransferase drug targets in cancer. Nat Biotech. 2012;30:622–23.
Harandi A, Zaidi AS, Stocker AM, Laber DA. Clinical efficacy and toxicity of anti-EGFR therapy in common cancers. J Oncol. 2009;2009:567486.
Williams CB, Soloff AC, Ethier SP, Yeh ES. Chapter seven—perspectives on epidermal growth factor receptor regulation in triple-negative breast cancer: ligand-mediated mechanisms of receptor regulation and potential for clinical targeting. Advances in cancer research. 2015;127:253–281
Nagaya T, et al. Near infrared photoimmunotherapy targeting EGFR positive triple negative breast cancer: optimizing the conjugate-light regimen. PLoS One. 2015;10:e0136829.
Sun S, Zhou X, Zhang W, Gallick Gary E, Kuang J. Unravelling the pivotal role of Alix in MVB sorting and silencing of the activated EGFR. Biochem J. 2015;466:475–87.
Acknowledgements
This work was supported in part by grants from the NIH R01DK060019 and R01CA206222 to Hong-Wu Chen, the National Natural Science Foundation of China (No. 81673037) and the Department of Education, Guangdong Government under the Top-tier University Development Scheme for Research and Control of Infectious Diseases (No. 2015017) to Jian-zhen Xu, the National Natural Science Foundation of China (No. 81373655) to Hai-bin Wang and National Natural Science Foundation of China (No. 81872891) to Jun-jian Wang.
Author contributions
H.-W.C. and J.W. conceived the research. H.-W.C. and J.W. designed the research. J.W., P.C., Z.D., H.W. and J.X.Z. performed research. H.-W.C., J.W., J.X.Z. and H.-B.W. analyzed the data. H.-W.C., J.W., P.L. and J.X.Z. wrote and/or edited the manuscript.
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Wang, JJ., Zou, J.X., Wang, H. et al. Histone methyltransferase NSD2 mediates the survival and invasion of triple-negative breast cancer cells via stimulating ADAM9-EGFR-AKT signaling. Acta Pharmacol Sin 40, 1067–1075 (2019). https://doi.org/10.1038/s41401-018-0199-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41401-018-0199-z
Keywords
This article is cited by
-
The role of NSD1, NSD2, and NSD3 histone methyltransferases in solid tumors
Cellular and Molecular Life Sciences (2022)
-
Circular RNA circ_001422 promotes the progression and metastasis of osteosarcoma via the miR-195-5p/FGF2/PI3K/Akt axis
Journal of Experimental & Clinical Cancer Research (2021)
-
NSD2 promotes tumor angiogenesis through methylating and activating STAT3 protein
Oncogene (2021)
-
Identification of histone methyltransferase NSD2 as an important oncogenic gene in colorectal cancer
Cell Death & Disease (2021)