Abstract
The fibroblast growth factor receptors (FGFRs) are increasingly considered attractive targets for therapeutic cancer intervention due to their roles in tumor metastasis and angiogenesis. Here, we identified a new selective FGFR inhibitor, C11, and assessed its antitumor activities. C11 was a selective FGFR1 inhibitor with an IC50 of 19 nM among a panel of 20 tyrosine kinases. C11 inhibited cell proliferation in various tumors, particularly bladder cancer and breast cancer. C11 also inhibited breast cancer MDA-MB-231 cell migration and invasion via suppression of FGFR1 phosphorylation and its downstream signaling pathway. Suppression of matrix metalloproteinases 2/9 (MMP2/9) was associated with the anti-motility activity of C11. Furthermore, the anti-angiogenesis activity of C11 was verified in endothelial cells and chicken chorioallantoic membranes (CAMs). C11 inhibited the migration and tube formation of HMEC-1 endothelial cells and inhibited angiogenesis in a CAM assay. In sum, C11 is a novel selective FGFR1 inhibitor that exhibits potent activity against breast cancer metastasis and angiogenesis.
Similar content being viewed by others
Introduction
The fibroblast growth factor receptor (FGFR) family comprises 4 members: FGFR1–4 [1]. Similar to other receptor tyrosine kinases (RTKs), FGFR1–4 proteins are single-pass transmembrane receptors with extracellular ligand-binding domains and an intracellular tyrosine kinase domain. On the cell surface, the interaction between FGF and FGFR is stabilized by heparin sulphate proteoglycans (HSPGs) via formation of a ternary complex. Dimerization of the ternary FGF:FGFR:HPSG complex leads to kinase activation and autophosphorylation of intracellular tyrosine residues. The subsequent phosphorylation of the intracellular receptor substrates FGFR substrate 2 (FRS2) and phospholipase Cγ activates two main downstream signaling pathways, the Ras-dependent mitogen-activated protein kinase (MAPK) and Ras-independent phosphoinositide 3-kinase (PI3K)-AKT signaling pathways [2,3,4]. In normal cells, the FGFR signaling pathway plays significant roles in key cell behaviors, including proliferation, differentiation, migration, and survival, and is a key regulator of angiogenesis and wound healing in adults [5, 6]. However, aberrant FGFR signaling is implicated in the pathogenesis of a broad range of solid tumors, including non-small cell lung cancer (NSCLC), breast cancer, and bladder cancer [7, 8]. Moreover, dysregulation of FGFRs is linked with poor prognosis, metastatic progression, and drug resistance due to cytotoxic and targeted cancer treatment [1].
Great efforts have been directed toward blockade of FGF/FGFR signaling as an antitumor therapeutic approach [9,10,11]. Present FGFR inhibitors can be divided into two groups: nonselective FGFR TKIs and selective FGFR TKIs. The first-generation FGFR TKIs typically inhibit a broad range of additional kinases (e.g., VEGFRs and PDGFRs), and approved multitarget FGFR TKIs examined in clinical trials include dovitinib (TKI258) [12], lucitanib (E-3810) [13], lenvatinib (E7080) [14], ponatinib (AP24534) [15], and nintedanib (BIBF1120) [16]. The FGFR inhibition activity of these compounds is often relatively weak, and they exert antitumor efficacy primarily by targeting RTKs rather than FGFRs. In addition, the inhibition of multiple tyrosine kinases results in increased side effects, which limits the ability to achieve doses required for effective FGFR inhibition. Therefore, there is a need to develop more selective inhibitors to suppress tumors through selective targeting of FGFRs. Recently, a second generation of FGFR inhibitors with greater FGFR selectivity has been reported. Representative compounds include AZD4547 (AstraZeneca, phase III) [17], BGJ398 (Novartis, phase II) [18], JNJ-42756493 (Janssen, phase II) [19], Debio-1347 (Debiopharm, phase I) [20] and LY2874455 (Eli Lilly, phase II) [21], These agents exhibited variable antitumor capacity in FGFR-dysfunctional xenograft models. In comparison with VEGFR inhibitors, blood pressure in tumor xenograft models was not elevated with efficacious doses of AZD4547 or LY2874455. However, there are still no FGFR targeting therapies approved for cancer treatment. Lack of selectivity for FGFR family members might be one cause for the slow progress of recent second-generation FGFR inhibitors. Therefore, it is of great importance to develop novel selective FGFR inhibitors with new scaffolds.
The 1-oxo-1H-phenalene derivatives (previously assigned as acenaphtho[1,2-b]pyrrole) have been reported by our group to be a series of potent FGFR1 inhibitors [22]. 9-Allylnaphtho[1,8-ef]isoindole-7,8,10(9 H)-trione (C11, Fig. 1a) is a representative compound in this series and was synthesized by introduction of an allyl group to naphtho[1,8-ef]isoindole-7,8,10(9H)-trione. C11 was noted for its facile synthesis, high yield and potent antitumor capacity. Herein, we report our research evaluating the inhibitory activity of the selective FGFR1 inhibitor C11 against breast cancer cell metastasis and angiogenesis.
Materials and methods
Compounds
Compounds were dissolved at 10 mM in DMSO as stock solutions for in vitro studies and stored at −20 °C. The compounds were diluted in normal saline before each experiment. The final DMSO concentration did not exceed 0.1% (v/v), and 0.1% DMSO in normal saline was used as a vehicle control for all the experiments.
Molecular simulation
The crystal structure of FGFR1 was retrieved from the Protein Data Bank (PDB entry: 2FGI, Chain B) [23]. Protein model preparation, after removal of all water molecules, the addition of hydrogen atoms and ionization consideration, was performed using the Glide 4.0 module in Maestro v7.5 (Schrödinger, Inc.) [24]. Considering the essential hydrogen bond interaction in the kinase hinge region, we set two hydrogen bond constraints of heteroatoms in residues Ala 564 and Glu 562 so that each compound was required to have at least one hydrogen bond interaction in the kinase hinge region. The grid-enclosing box was formed based on the ligand in the complex structure and constraints set in the last step. Three-dimensional (3D) structures of C11 were drawn using the Builder tool and generated with the Ligprep module. C11 was docked into the ATP-binding site of FGFR1 using Glide 4.0 with a standard precision (SP) approach, and a scaling factor of 1.0 was set for the VDW radii of the protein atoms with the partial atomic charges less than 0.25. The best configuration ranked by GlideScore among the top 10 binding configurations was chosen as the preliminary model for the next minimization, which was sequentially minimized via PCRG (Polak-Ribiere Conjugate Gradient) methods by the MacroModel module in Maestro v7.5. OPLS 2005 was selected as the force field, and the convergence criteria were set with max iterations of 500 and a gradient of 0.05 kcal/mol.
Cell lines and cell culture
HL60, K562, BEL-7402, T24, and 786-O cells were from Shanghai Institute of Biochemistry and Cell Biology (Shanghai, China). SMMC-7721 cells were obtained from Second Military Medical University (Shanghai, China). SGC-7901 cells were obtained from Renji Hospital (Shanghai, China). HepG-2, MDA-MB-468, MCF-7, A549, HeLa, BxPC-3, HCT-116, PC-3, KB, A431, and HMEC-1 cells were obtained from the American Type Culture Collection (Manassas, VA, USA). MDA-MB-231 and SKOV3 cells were obtained from Fudan University Shanghai Cancer Center (Shanghai, China). MKN-45 cells were obtained from the Institute of Basic Medical Sciences of the Chinese Academy of Medical Sciences (Beijing, China). HL60, K562, SMMC-7721, BEL-7402, SGC-7901, MKN45, BxPC-3 and 786-O cells were cultured in RPMI-1640 (GIBCO, Grand Island, NY, USA) supplemented with 10% fetal bovine serum (FBS) (GIBCO, Grand Island, NY, USA). HepG-2, MCF-7, HeLa, and KB cells were cultured in ATCC-formulated Eagle’s Minimum Essential Medium (MEM) supplemented with 10% FBS. MDA-MB-468 and MDA-MB-231 human breast cancer cells were cultured in Leiboviz’s L15 medium supplemented with 10% FBS. A549 and PC3 cells were cultured in Ham’s F12 supplemented with 10% FBS. HCT116 and T24 cells were cultured in McCoy’s 5 A supplemented with 10% FBS. SKOV-3 and A431 cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% FBS. Human microvascular endothelium HMEC-1 cells were propagated in MCDB131 medium (GIBCO, Grand Island, NY, USA) supplemented with 20% FBS.
Kinase ELISAs
The kinase domains of VEGFR2, c-Kit, RET, c-Src, EphA2, EphB2, c-Met, RON and IGF1R were expressed using a Bac-to-Bac baculovirus expression system (Invitrogen, Carlsbad, CA, USA) and purified on Ni-NTA columns (QIAGEN Inc., Valencia, CA, USA) as previously described [25, 26]. FGFR1, VEGFR1, PDGFRα, PDGFRβ, EGFR2 and Abl were purchased from Upstate Biotechnology Inc. (Charlottesville, VA, USA). Poly (Glu-Tyr)4:1, the substrate of tyrosine kinase, was obtained from Sigma-Aldrich Corp. (St. Louis, MO, USA). Monoclonal anti-phosphotyrosine (PY) antibody (PY99) was purchased from Santa Cruz Biotechnology, Inc. The tyrosine kinase activities were determined in 96-well ELISA plates (Corning, NY, USA) precoated with 20 μg/mL Poly (Glu, Tyr)4:1. First, 80 μL of 5 μM ATP solution diluted in kinase reaction buffer (50 mM HEPES pH 7.4, 20 mM MgCl2, 0.1 mM MnCl2, 0.2 mM Na3VO4, and 1 mM DTT) was added to each well. Various concentrations of compounds diluted in 10 μL of 1% DMSO (v/v) were then added to each reaction well, with 1% DMSO (v/v) being used as the negative control. Subsequently, the kinase reaction was initiated by the addition of tyrosine kinase protein diluted in 10 μL of kinase reaction buffer solution. Experiments at each concentration were performed in duplicate. After incubation for 60 min at 37 °C, the wells were washed three times with phosphate-buffered saline (PBS) containing 0.1% Tween 20 (T-PBS). Next, 100 μL anti-phosphotyrosine antibody (PY99, 1:1000 dilution) diluted in T-PBS containing 5 mg/mL BSA was added. After 30 min of incubation at 37 °C, the wells were washed three times. Horseradish peroxidase-conjugated goat anti-mouse IgG (100 μL) diluted 1:2000 in T-PBS containing 5 mg/mL BSA was added. The plate was reincubated at 37 °C for 30 min, and then, the wells were washed with PBS. Finally, 100 μL of a solution containing 0.03% H2O2 and 2 mg/mL o-phenylenediamine in 0.1 M citrate buffer, pH 5.5, was added, and the samples were incubated at room temperature until color emerged. The reaction was terminated by the addition of 50 μL of 2 M H2SO4, and the plate was read using a multiwell spectrophotometer (MAX190™, Molecular Devices, Sunnyvale, USA) at 490 nm. The inhibition rate (%) was calculated using the following equation: Inhibition rate = [1 − (A490 treated / A490 control)] × 100%. IC50 values were determined from the results of at least three independent tests and calculated from the inhibition curves. The accuracy of this in vitro screening assay was confirmed by the measured IC50 value of the positive control, which was similar to the reported value.
Cell proliferation assay
Cells were seeded in a 96-well plate, incubated overnight for attachment, and then treated in triplicate with gradient concentrations of C11 at 37 °C for 72 h. Cell proliferation was assessed using a sulforhodamine B (SRB; Sigma, St. Louis, MO, USA) assay. The IC50 values were determined by the four-parameter logit method. The data are expressed as the mean ± SD of three independent experiments.
Cell migration assay
Cell migration assays were performed in a transwell Boyden Chamber (6.5 mm in diameter and 8 μM pore size; Costar, Corning, NY, USA). The lower chamber contained 600 μL of medium supplemented with 10% FBS. Next, 100 μL of cell suspension (1 × 105 cells per well) with various concentrations of C11 diluted in serum-free medium was added to the upper chamber. After 8 h of incubation, the migrated cells were fixed with 90% ethanol and stained with 0.1% crystal violet in 0.1 M borate and 2% ethanol. The stained cells were photographed, and then, the dye was dissolved with 10% acetic acid. The OD value was determined at 590 nM, and the migration inhibition rate was calculated as [1 − (ODC11 − ODblank) / (ODcontrol − ODblank)] × 100%.
Wound healing assay
Wound healing assays were performed in 24-well plates with automated WoundMaker and Incucyte systems (Essen Bioscience, Michigan, USA). The scratch wounds were made with sterile 10 μL pipette tips. Fresh culture medium containing either DMSO or various concentrations of compounds was added. The Incucyte Scratch Wound software was used to capture and analyze the pictures.
Cell invasion assay
Transwell chamber membranes were coated with 1% gelatin. Medium containing 10% FBS was added to the lower chambers. Cells were suspended in serum-free medium containing various concentrations of compound and added to the upper chambers. Similar to the migration assay, the cells that invaded into the lower chamber were detected and quantitated after 16 h of incubation.
Western blot analysis
Cells (2.0 × 105) were seeded in six-well plates and exposed to C11 at the indicated concentrations. Cells were harvested and subjected to standard western blot analysis using antibodies against phospho-AKT (ser473), AKT, phospho-Erk (Thr202/Tyr204) and Erk (Cell Signaling Technology, Beverly, MA, USA); phospho-FGFR1 (Tyr653/654) and FGFR1 (Santa Cruz Technology, CA, USA); MMP2 and MMP9 (Millipore, MA, USA); and GAPDH (Kangchen, Shanghai, China).
Gelatin zymogram
Cells were incubated in serum-free medium for 24 h with or without C11 treatment. The supernatants were collected and concentrated. Electrophoresis was performed in 7.5% polyacrylamide gels containing 0.1% gelatin. The gels were then washed for 1 h in 2.5% (v/v) Triton X-100 to remove SDS and incubated overnight at 37 °C in 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 0.5 mM ZnCl2, and 10 mM CaCl2 to allow proteolysis of the gelatin substrate. Bands corresponding to activity were visualized by negative staining using 0.5% Coomassie brilliant blue R-250 (Bio-Rad, Hercules, CA, USA).
mRNA extraction and RT-PCR
Cells were lysed with Trizol reagent (Invitrogen), and total RNA was isolated with chloroform and isopropyl alcohol. RNA was subjected to reverse transcription using a DRR037A PrimeScript RT reagent kit (TaKaRa) according to the manufacturer’s instructions. cDNA was amplified using a D334A PCR amplification kit (TaKaRa). The sequences of specific primers were as follows: MMP2, 5ʹ-GTGGATGATGCCTTTGCTCG-3′ (sense) and 5′-CTTCTTGTCGCGGTCGTAGTC-3′ (antisense); MMP9, 5′-CAGATTCCAAACCTTTGAGGGCGA-3′ (sense) and 5′-TCGAAATGGGCGTCTCCCTGAAT-3′ (antisense); and β-actin, 5’-AGCAAGAGAGGCATCCTCACCCTGAAGTACC-3’ (sense) and 5’-CAGATTCTCCTTAATGTCACGCACGATTTCC-3’ (antisense). The PCR products were separated in a 1% agarose gel and visualized by ethidium bromide staining.
Tube formation assay
A 96-well plate was coated with 50 μL of Matrigel (Becton Dickinson Labware, MA, USA), which was allowed to solidify at 37 °C for 1 h. HMEC-1 cells (1.5 × 104 cells/well) were seeded on the Matrigel, cultured in MCDB131 medium containing different concentrations of C11 or solvent control for 8 h, and then photographed under a microscope (Olympus, IX70, Japan). The inhibition rate of tube formation was calculated as [1 − tube numberC11/tube numbercontrol] × 100%.
Chicken chorioallantoic membrane (CAM) assay
Groups of 10 fertilized chicken eggs were incubated in a humidified egg incubator (Lyon, Chula Vista, CA, USA) maintained at 37 °C with 50% humidity and allowed to grow for 7 days. Gentle suction was applied at the hole located at the broad end of the egg to create a false air sac directly over the chicken chorioallantoic membrane, and a 1-cm2 window was immediately removed from the egg shell. A glass coverslip (0.5 × 0.5 cm) saturated with compound or normal saline was placed on an area between preexisting vessels, and the embryos were further incubated for 48 h. The neovascular zones beside the glass coverslips were photographed under a stereomicroscope (Leica, MS5, Heerbrugg, Switzerland), and images of 5 in × 7 in regions were printed. Vessel branches <3 mm long in five random 1 in × 1 in zones per picture were counted, and five eggs were chosen from each group.
Statistical analysis
The significance of differences between means was assessed via Student’s t test, and P < 0.05 was considered statistically significant.
Results
Kinase inhibitory activity of C11 and potential binding mode with FGFR1
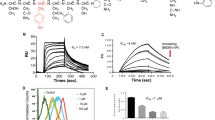
The 1-oxo-1H-phenalene derivatives were recently reported by our group to be a series of potent FGFR1 inhibitors [22]. Compound C11 (Fig. 1a) was identified as one of the most potent in this series of derivatives and was selected for further evaluation. We first determined the kinase suppressive selectivity of C11 against a panel of 20 tyrosine kinases consisting of FGFR family members (FGFRs), VEGFRs, PDGFRs, EGFRs, c-kit, RET, c-Src, Abl, EphA2, EphB2, c-Met, RON and IGF1R. According to the results shown in Table 1, C11 exhibited potent inhibition against FGFR1 (IC50 = 19 nM) and 4 times less potent efficacy against FGFR4 (IC50 = 84 nM) but showed no obvious suppression against the other tested tyrosine kinases, including two other FGFR family members, FGFR2 and FGFR3, and kinases highly homologous to FGFRs (such as PDGFRs), even at a much higher concentration (1 μM). Therefore, the results demonstrated that C11 is a potent and highly selective inhibitor of FGFR1.
To further characterize the potential binding mode of C11 with FGFR1 protein, we conducted a molecular docking analysis using the Glide 4.0 module [23, 24]. As shown in Fig. 1b, several interesting features of possible interactions could be derived from the predicted binding pose of C11 with FGFR1 protein: (1) C11 occupied the cleft of the ATP-binding site, and the scaffold (i.e., the planar and hydrophobic aromatic ring fragment) stretched toward the pocket and was exposed to the solution. Extensive hydrophobic interactions and van der Waals (VDW) contacts with hydrophobic residues (Val 492, Val 561, Ala 512, ILE 545 and Leu 630) were present. (2) An essential hydrogen bond interaction was observed in the hinge region of the kinase domain, i.e., the carbonyl oxygen atom of the cyclohexa-2,5-dienone formed a hydrogen bond with the nitrogen atom of residue Ala 564. These results indicated that C11 might inhibit FGFR1 by directly binding with the protein through hydrogen bond and hydrophobic interactions, which is quite similar to the binding mode of other reported FGFR1 inhibitors [27].
C11 suppresses the proliferation of a panel of tumor cells through inactivation of FGFR1 and downstream signal transduction
FGFRs are potential targets for therapeutic cancer intervention based on next-generation sequencing of a broad array of solid tumors, which identified aberrant signaling of FGFRs in 7% of cancers [28]. We studied the in vitro anti-proliferative activity of C11 against 20 human cancer cell lines from various tissues, including human leukemia cell lines (HL 60 and K562), three human hepatocellular carcinoma cell lines (SMMC-7721, BEL-7402, and HepG-2), two human lung adenocarcinoma cell lines (SGC-7901 and A549), one human gastric carcinoma cell line (MKN45), three human breast adenocarcinoma cell lines (MDA-MB-468, MDA-MB-231, and MCF-7), one human cervical carcinoma cell line (HeLa), one human pancreatic carcinoma cell line (BXPC-3), one human ovarian carcinoma cell line (SKOV3), one human colon carcinoma cell line (HCT-116), one human prostate adenocarcinoma cell line (PC-3), one human bladder carcinoma cell line (T24), one human nasopharyngeal carcinoma cell line (KB), one human epidermoid carcinoma cell line (A431) and one human kidney carcinoma cell line (786-O). As shown in Fig. 2a, C11 potently inhibited the growth of most of the cancer cell lines, with an IC50 value of 7.57 ± 3.78 μM on average. For example, the IC50 value was 2.81 ± 0.25 μM against T24 cells and 3.02 ± 1.59 μM against MDA-MB-231 cells. Meanwhile, the antiproliferative activity against SGC-7901 and HeLa cells was relatively poor (IC50 value of 19.76 ± 6.65 μM and 11.66 ± 4.15 μM, respectively).
a C11 inhibited the proliferation of 20 cancer cell lines. b Protein expression of FGFR1 in T24, MDA-MB-231, MCF-7 and SGC-7901 cells. C11 downregulated the FGFR1 signaling pathway in MDA-MB-231 (c) and T24 (d) cells. Cells were starved in serum-free medium for 24 h and then cultured in the presence of varying doses of C11 for 12 h at 37 °C. bFGF (50 ng/mL) was then added. After a 15 min incubation at 37 °C, whole cell lysates were harvested and assayed via Western blotting
FGFR protein expression in several C11-sensitive and insensitive cancer cell lines (T24, MDA-MB-231, MCF-7 and SGC-7901) was tested. As shown in Fig. 2b, FGFR1 is highly expressed in MDA-MB-231, T24 and MCF7 cells in contrast to SGC-7901 cells. We thus speculated that FGFR1 inhibition might be involved in the antiproliferative activity of C11 and examined the inhibitory activity of C11 on FGFR1 activation and signaling transduction in T24 and MDA-MB-231 cells, respectively. As expected, C11 dose-dependently suppressed FGF-induced FGFR1 activation in both cell types (Fig. 2c, d). C11 also reduced phosphorylation of the downstream molecules of FGFR1, including AKT and Erk. The total protein levels of FGFR1, AKT, Erk and GAPDH were rarely affected. Therefore, C11 inhibited the growth of cancer cells through FGFR1 inactivation and inhibition of downstream signaling transduction.
C11 inhibits migration and invasion of MDA-MB-231 cells by suppressing MMP2 and MMP9 expression
Metastatic spread of cancer cells is the main cause of death of breast cancer patients, and elucidation of the molecular mechanisms underlying this process is a major focus in cancer research. FGFR1 plays an important role in both embryonic and postnatal mammary gland development, which prompted investigation of its relevance to breast cancer and its potential as a therapeutic target [29, 30]. We thus evaluated the influence of C11 on the migration and invasion activities of MDA-MD-231 cells, which is reported to be a highly invasive cell line [31], using a Boyden Chamber transwell assay and wound healing assay, respectively.
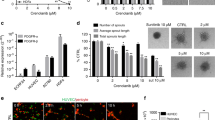
The results of the Boyden Chamber transwell assay (Fig. 3a) showed that the number of migrated cells was dose-dependently reduced after treatment with C11 in comparison with vehicle control. The migration ability was reduced by 80% in the presence of 10 μM C11 for 8 h. Meanwhile, the wound healing assay results (Fig. 3b) showed that C11 exerted significant and dose-dependent inhibition of cell migration compared with the control. Furthermore, the effect of C11 on MDA-MB-231 cell invasion was determined in Matrigel invasion chambers. In contrast to the migration experiment, the transwell chamber membranes were coated with 1% gelatin. C11 treatment potently inhibited cell invasion and suppressed cell invasion by 50% at a concentration of 10 μM (Fig. 3c). These results indicated that C11 dramatically inhibited the migration and invasion of MDA-MB-231 cells.
a C11 inhibited the migration of MDA-MB-231 cells. Migration of MDA-MB-231 cells was detected using transwell Boyden chambers after treatment with vehicle or C11 (1.25, 2.5, 5 and 10 μM) for 8 h. Representative images and summary data show inhibition of cell migration. b Real-time wound healing assay results were analyzed on WoundMaker and Incucyte systems after treatment with vehicle or C11 (1.25, 2.5, 5 and 10 μM). c The inhibitory activity of C11 on MDA-MB-231 cell invasion. Matrigel invasion chambers were used to detect cell invasion. MDA-MB-231 cells were treated with vehicle or C11 (1.25, 2.5, 5 and 10 μM) for 16 h. Representative images and quantification of the inhibition activity are shown. The data shown are the mean ± SD of three independent experiments. *P < 0.05, and **P < 0.01 compared with the vehicle control group
Due to their extracellular matrix degradation capacity, MMP2 and MMP9 have been reported to be associated with the ability of tumor cells to metastasize, and activation of the FGFR signaling pathway likely promotes cell migration and invasion due to enhanced secretion of MMPs [32]. Thus, we validated whether the anti-metastasis potency of C11 in MDA-MB-231 cells is related to down-regulation of MMP2 and MMP9 function, protein level or gene expression. As expected, of gelatin zymography results (Fig. 4a) showed that C11 treatment suppressed the activities of MMP2 and MMP9 in a dose-dependent manner. RT-PCR and Western blotting analyses, as shown in Figs. 4b, c, clearly demonstrated that MMP2 and MMP9 expression was decreased after C11 treatment, consistent with the gelatin zymography results. Meanwhile, C11 exhibited no significant inhibitory activity against the gene expression of other MMPs (MMP1, 3, 11, and 14) or TIMPs (data not shown), indicating that the down-regulation of MMP2 and MMP9 might specifically result from FGFR1 inhibition. Therefore, the results demonstrated that the activity of C11 against migration and invasion of MDA-MB-231 cells might be attributed to inactivation of FGFR1 and subsequent down-regulation of MMP2 and MMP9 secretion.
C11 suppressed MMP2 and MMP9 activity. Cells were seeded overnight and then treated with vehicle or C11 (1.25, 2.5, 5 and 10 μM) in serum-free medium for 24 h. Cell culture supernatants were harvested and subjected to a gelatin zymography assay. Representative image and quantification (a) are shown on the left. RT-PCR (b) and western blotting results (c) also showed inhibition activity of C11 against MMP2 and MMP9. The data shown are the mean ± SD of three independent experiments. *P < 0.05, and **P < 0.01 compared with the vehicle control group
C11 suppresses angiogenesis both in vitro and in vivo
FGFRs have been reported to be responsible for tumor angiogenesis, which is a complex process that includes the proliferation, migration and tube formation of endothelial cells. Therefore, we investigated the antiangiogenic activity of C11 both in vitro and in vivo. First, we investigated the effect of C11 on the in vitro migration of human microvascular endothelial cells (HMEC-1). According to Fig. 5a, treatment with C11 for 8 h reduced the number of migrated cells in a dose-dependent manner. In detail, after exposure to 5 and 10 μM C11, up to 55 and 80% of the migrated HMEC-1 cells were inhibited, respectively. Meanwhile, tube formation occurs in the late stage of angiogenesis, and thus, we investigated the effect of C11 on the formation of functional tubes by plating HMEC-1 cells on Matrigel substratum (Fig. 5b). In the control group, stimulation with 20% serum resulted in a rapid alignment of HMEC-1 cells and the formation of tube-like structures within 8 h. C11 treatment effectively inhibited serum-stimulated tube formation of HMEC-1 cells in a dose-dependent manner. Treatment with 5 and 10 μM C11 induced 70 and 90% inhibition of tube-like structures, respectively.
a C11 inhibited the migration of HMEC-1 cells. Transwell Boyden chambers were used to detect cell migration. HMEC-1 cells were treated with vehicle or C11 (1.25, 2.5, 5 and 10 μM) for 8 h. Representative images and summary data (bar graph) show inhibition of cell migration. b C11 suppressed HMEC-1 cell tube formation on Matrigel. HMEC-1 cells were seeded on Matrigel with vehicle or C11 (1, 5 and 10 μM). Representative images and summary data (bar graph) show inhibition of tube formation after 8 h. c Anti-angiogenic effects of C11 in a chorioallantoic membrane (CAM) model. A glass cover-slip saturated with C11 or normal saline was placed areas between preexisting vessels in the fertilized chick eggs and incubated for 48 h. The glass cover-slip was place was placed on the right side of the field. Representative images and quantification of the inhibitory activity of C11 on angiogenesis in CAM assays. The data shown are the mean ± SD of three independent experiments. *P < 0.05, and **P < 0.01 compared with the vehicle control group
CAM assays have been demonstrated to be a unique in vivo model to investigate the process of new blood vessel formation and the effects of anti-angiogenic agents. Thus, we evaluated the in vivo antiangiogenic effects of C11 using CAM assays. As shown in Fig. 5c, the normal branching pattern of blood vessels formed over 2 days of incubation in the control group, whereas neovascularization of the CAMs was dramatically and dose-dependently decreased by C11 treatment. C11 at 10 ng/egg almost completely blocked angiogenesis in the CAM. These results demonstrated that C11 possesses potent antiangiogenic efficacy both in vitro and in vivo.
Discussion
The FGFR family comprises four highly conserved transmembrane receptor tyrosine kinases (FGFR1–4) and one receptor that lacks the intracellular kinase domain (FGFR5, also named FGFRL1) [1]. FGFRs can be activated by binding with FGFs and initiate a cascade of intracellular events. FGFRs play crucial roles in physiological processes, including tissue and metabolism homeostasis, endocrine functions and wound repair. Dysfunction of the FGF signaling axis has been implicated in oncogenesis, tumor progression and resistance to anticancer therapy in many cancer types. For example, amplification of FGFR1 is found in approximately 17% of squamous non-small-cell lung carcinoma (NSCLC) cases and is an independent adverse prognostic marker in early-stage NSCLC [33,34,35]. FGFR1 amplification is also involved in breast cancer and was reported in nearly 15% of hormone receptor-positive breast cancers and in approximately 5% of the more aggressive triple-negative breast cancers [36,37,38]. FGFRs are now considered promising potent therapeutic targets, which is supported by multiple preclinical and clinical studies. Several first-generation FGFR1 inhibitors have been approved for clinical use. However, their antitumor efficacy is mainly due to their inhibitory activity against other RTKs, such as VEGFRs and PDGFRs, rather than FGFRs. Second-generation of FGFR inhibitors are now emerging and show enhanced RTK selectivity. Currently, none of the selective FGFR inhibitors have been approved for cancer treatment. Therefore, it is vital to develop FGFR inhibitors with a novel scaffold.
We previously designed and synthesized a series of 1-oxo-1H-phenalene derivatives and evaluated their FGFR inhibition activity via cell-free and cell-based assays. C11 is a representative compound in this series. C11 was synthesized via alkylation of naphtho[1,8-ef]isoindole-7,8,10(9 H)-trione with allylic iodide in CH3CN in the presence of K2CO3 at 40 °C under argon. According to the ELISA-based kinase inhibitory assay, C11 dose-dependently inhibited FGFR1 kinase activity, with an IC50 value of 19 nM. The C11 inhibitory activity against another member of the FGFR family (FGFR4) was approximately 4 times less potent than that against FGFR1 (IC50 = 84 nM). In addition, no obvious inhibition was observed of other FGFR family members (FGFR2 and 3) or the other 16 tested RTKs (including VEGFR and PDGFRs). Therefore, C11 is an FGFR1 inhibitor with a new scaffold that can be produced at a high yield through a simple derivation process. Compared with the existing pan-FGFR inhibitors, C11 selectively targets FGFR1 over other RTK family members, such as VEGFR and PDGFRs, and other FGFR family members.
FGFRs are widely implicated in oncogenesis pathways and provide a potential point of therapeutic intervention based on next-generation sequencing of a broad array of solid tumors, which identified aberrant FGFR signaling in 7% of cancers [28]. Therefore, the effects of C11 on the proliferation of human cancer cell lines from 14 different organs were examined. The results demonstrated that C11 efficiently inhibited the in vitro proliferation of most of the tested cells, with an average IC50 of 7.57 ± 3.78 μΜ. The T24 bladder cancer cell line and MDA-MB-231 breast cancer cell line were the most sensitive to C11, with IC50 values of 2.81 ± 0.25 μΜ and 3.02 ± 1.59 μΜ, respectively. FGFR1 expression in T24, MDA-MB-231, MCF-7 and SGC-7901 cells was tested via western blotting, and the protein expression level was generally in accordance with the anti-proliferation activity of C11. Target inhibition results confirmed that C11 dose-dependently inhibited FGFR1 phosphorylation as well as the downstream PI3K/AKT and Erk signaling pathway in T24 and MDA-MB-231 cells.
Breast cancer is the most frequently diagnosed cancer and the second leading cause of cancer death among females and accounts for approximately 23% of the total cancer cases and 14% of cancer deaths [39]. The incidence of breast cancer has increased over the past years, while the associated death rates have decreased, mainly as a result of early detection and improved treatment. Metastatic breast cancer causes the vast majority of the deaths. To date, the US Food and Drug Administration (FDA) have approved targeted therapies for the treatment of breast cancer, including endocrine therapy and anti-HER2 therapies [8]. Other strategies targeting different RTKs are currently under way. FGFR signaling plays a vital role in both embryonic and postnatal mammary gland development, which has prompted scientists to investigate its relevance in breast cancer and its potential as a therapeutic target [30]. The FGF/FGFR signal transduction pathway has been implicated in cancer cell proliferation, metastasis, angiogenesis and survival.
Tumor metastasis is a multistep process, during which cancer cells disseminate from the primary tumor to distant secondary tissues. After acquiring advantageous characteristics, tumor cells escape from the primary tumor, invade through the extracellular matrix (ECM) to intravasate into the surrounding blood supply and extravasate into distant tissue to form metastatic foci. Metastasis is the leading cause of cancer-related death and a major obstacle to cancer therapy. Hase et al found that bFGF and FGFR1 were more highly expressed in high-invasive oral squamous cell carcinoma (OSCC) specimens than in low-invasive OSCC specimens. Moreover, these expression levels were well correlated with patient prognosis [40]. Sato et al reported that FGFR-1 overexpression was correlated with liver metastasis in patients with colorectal carcinoma [41]. In addition, bFGF/FGFR was shown to promote tumor invasion via modulation of the downstream PI3K/AKT and Erk signaling pathway in vestibular schwannoma [42].
We have demonstrated that C11 can inhibit breast cancer MDA-MB-231 cell proliferation by down-regulating FGFR1 and downstream signaling transduction. According to the Boyden Chamber transwell assays and wound healing assays, the selective FGFR1 inhibitor C11 effectively suppressed the migration of MDA-MB-231 cells in a dose-dependent manner. The migration ability of breast cancer cells was reduced by up to 80% after treatment with 10 μΜ C11 for 8 h. C11 also inhibited the invasion of MDA-MB-231 cells based on the Matrigel invasion assay results. C11 dose-dependently inhibited MDA-MB-231 cell invasion.
The tumor microenvironment, particularly the ECM, is emerging as a key factor in influencing cancer progression. ECM degradation is a key tool used by cancer cells to initiate the metastatic cascade program. MMPs are critical molecules involved in degradation of ECM components and thus promote angiogenesis, tumor growth, and metastasis [43]. MMPs are present in nearly all human cancers (e.g., gastric, pancreatic, prostate, and breast cancer) [44]. In breast cancer, MMP-2 expression levels are confirmed to be correlated with poor overall survival [32]. For example, mRNA levels of MMP-2 and MMP-9 are significantly higher in MDA-MB-231 and MCF-7 breast cancer cell lines compared with the normal HS578Bst cell line [45]. Furthermore, low MMP-2 and MMP-9 mRNA levels are associated with better overall survival for breast cancer patients [45]. More than 20 years ago, broad-spectrum inhibitors targeting MMP activity failed in clinical trials, and since then, enthusiasm for the use of MMP inhibition as a therapeutic approach has faded [46]. However, after expanding our knowledge of MMP biology, we realized that MMP inhibition is still a promising treatment strategy via advanced selective targeting of individual MMPs combined with specific delivery of therapeutics to the tumor microenvironment [32, 47]. In our study, C11 inhibited MMP-2 and MMP-9 secretion in MDA-MB-231 cells in a dose-dependent manner based on gelatin zymography. RT-PCR and WB results also revealed the C11-induced suppression of MMP-2 and MMP-9 mRNA and protein levels, in accordance with the gelatin zymography results. The selectivity of C11 toward MMPs was also tested. C11 exhibited no obvious inhibitory activity against the gene expression of other MMPs, namely, MMP1, 3, 11, and 14.
Angiogenesis is the sprouting of novel capillaries from preexisting blood vessels and involves the basement membrane, migration of endothelial cells and the formation of new capillary loops. Angiogenesis plays key roles in tumor pathogenesis, progression and metastasis and is regulated by VEGFR, PDGFR and FGFR tyrosine kinases, which suggests a promising tumor therapeutic strategy with few side effects. FGFs were the first identified angiogenic factors, and their role in endothelial cell proliferation, migration, and other angiogenic processes has been extensively studied [48]. Inhibition of FGFR1 and FGFR2 activity results in suppression of angiogenesis [49]. Therefore, the anti-angiogenesis effect of C11 was tested in this study. Boyden chamber transwell assays confirmed that C11 could reduce the number of migrated HMEC-1 cells by up to 55 and 80% after treatment with 5 and 10 μM C11, respectively. The tube formation inhibition activity of C11 was examined, and the formation of tube-like structures was suppressed by 70 and 90% with 5 and 10 μM C11, respectively. Furthermore, C11 also decreased neovascularization in a CAM model in a dose-dependent manner.
In sum, we have identified C11 as a small-molecule FGFR1-selective inhibitor with a new scaffold that acts as an anti-tumor agent. C11 inhibited the proliferation of various cancer cells and displayed anti-metastasis activity in breast cancer cell MDA-MB-231 cells via FGFR1 and downstream signal transduction inhibition. MMP-2 and MMP-9 were shown to be involved in the anti-metastasis activity of C11 due to their ECM degradation capacity. Furthermore, C11 suppressed angiogenesis both in vitro and in vivo. These data warrant investigation of the PK and in vivo efficacy of C11 in mouse models and support further research on the use of C11 for the treatment of breast cancers.
References
Babina IS, Turner NC. Advances and challenges in targeting FGFR signalling in cancer. Nat Rev Cancer. 2017;17:318–32.
Powers CJ, McLeskey SW, Wellstein A. Fibroblast growth factors, their receptors and signaling. Endocr-Relat Cancer. 2000;7:165–97.
Tomlinson DC, Lamont FR, Shnyder SD, Knowles MA. Fibroblast growth factor receptor 1 promotes proliferation and survival via activation of the mitogen-activated protein kinase pathway in bladder cancer. Cancer Res. 2009;69:4613–20.
Ornitz DM, Itoh N. The fibroblast growth factor signaling pathway. WIREs Dev Biol. 2015;4:215–66.
Acevedo VD, Ittmann M, Spencer DM. Paths of FGFR-driven tumorigenesis. Cell Cycle. 2009;8:580–8.
Itoh N, Ornitz DM. Fibroblast growth factors: from molecular evolution to roles in development, metabolism and disease. J Biochem. 2011;149:121–30.
Tiseo M, Gelsomino F, Alfieri R, Cavazzoni A, Bozzetti C, De Giorgi AM, et al. FGFR as potential target in the treatment of squamous non small cell lung cancer. Cancer Treat Rev. 2015;41:527–39.
Perez-Garcia J, Munoz-Couselo E, Soberino J, Racca F, Cortes J. Targeting FGFR pathway in breast cancer. Breast. 2018;37:126–33.
Li XQ, Guise CP, Taghipouran R, Yosaatmadja Y, Ashoorzadeh A, Paik WK, et al. 2-Oxo-3, 4-dihydropyrimido[4, 5-d]pyrimidinyl derivatives as new irreversible pan fibroblast growth factor receptor (FGFR) inhibitors. Eur J Med Chem. 2017;135:531–43.
Brameld KA, Owens TD, Verner E, Venetsanakos E, Bradshaw JM, Phan VT, et al. Discovery of the irreversible covalent FGFR inhibitor 8-(3-(4-acryloylpiperazin-1-yl)propyl)-6-(2,6-dichloro-3,5-dimethoxyphenyl)-2-(methylamino)pyrido[2,3-d]pyrimidin-7(8H)-one (PRN1371) for the treatment of solid tumors. J Med Chem. 2017;60:6516–27.
Wang Y, Li L, Fan J, Dai Y, Jiang A, Geng M, et al. Discovery of potent irreversible pan-Fibroblast Growth Factor Receptor (FGFR) inhibitors. J Med Chem. 2018. https://doi.org/10.1021/acs.jmedchem.7b01843
Angevin E, Lopez-Martin JA, Lin CC, Gschwend JE, Harzstark A, Castellano D, et al. Phase I study of Dovitinib (TKI258), an oral FGFR, VEGFR, and PDGFR inhibitor, in advanced or metastatic renal cell carcinoma. Clin Cancer Res. 2013;19:1257–68.
Bello E, Colella G, Scarlato V, Oliva P, Berndt A, Valbusa G, et al. E-3810 is a potent dual inhibitor of VEGFR and FGFR that exerts antitumor activity in multiple preclinical models. Cancer Res. 2011;71:1396–405.
Yeung KT, Cohen EEW. Lenvatinib in advanced, radioactive Iodine-refractory, differentiated thyroid carcinoma. Clin Cancer Res. 2015;21:5420–6.
Gozgit JM, Wong MJ, Moran L, Wardwell S, Mohemmad QK, Narasimhan NI, et al. Ponatinib (AP24534), a multitargeted pan-FGFR inhibitor with activity in multiple FGFR-amplified or mutated cancer models. Mol Cancer Ther. 2012;11:690–9.
Hilberg F, Roth GJ, Krssak M, Kautschitsch S, Sommergruber W, Tontsch-Grunt U, et al. BIBF 1120: Triple angiokinase inhibitor with sustained receptor blockade and good antitumor efficacy. Cancer Res. 2008;68:4774–82.
Gavine PR, Mooney L, Kilgour E, Thomas AP, Al-Kadhimi K, Beck S, et al. AZD4547: An orally bioavailable, potent, and selective inhibitor of the fibroblast growth factor receptor tyrosine kinase family. Cancer Res. 2012;72:2045–56.
Guagnano V, Kauffmann A, Wohrle S, Stamm C, Ito M, Barys L, et al. FGFR genetic alterations predict for sensitivity to NVP-BGJ398, a selective pan-FGFR inhibitor. Cancer Discov. 2012;2:1118–33.
Perera TPS, Jovcheva E, Mevellec L, Vialard J, De Lange D, Verhulst T, et al. Discovery and pharmacological characterization of JNJ-42756493 (Erdafitinib), a functionally selective small-molecule FGFR family inhibitor. Mol Cancer Ther. 2017;16:1010–20.
Ebiike H, Taka N, Matsushita M, Ohmori M, Takami K, Hyohdoh I, et al. Discovery of [5-amino-1-(2-methyl-3H-benzimidazol-5-yl)pyrazol-4-yl]- (1H-indol-2-yl)methanone (CH5183284/Debio 1347), an orally available and selective Fibroblast Growth Factor Receptor (FGFR) inhibitor. J Med Chem. 2016;59:10586–600.
Zhao GS, Li WY, Chen DH, Henry JR, Li HY, Chen ZG, et al. A novel, selective inhibitor of fibroblast growth factor receptors that shows a potent broad spectrum of antitumor activity in several tumor xenograft models. Mol Cancer Ther. 2011;10:2200–10.
Chen Z, Wang X, Zhu WP, Cao XW, Tong LJ, Li HL, et al. Acenaphtho[1,2-b]pyrrole-based selective fibroblast growth factor receptors 1 (FGFR1) inhibitors: Design, synthesis, and biological activity. J Med Chem. 2011;54:3732–45.
Mohammadi M, Froum S, Hamby JM, Schroeder MC, Panek RL, Lu GH, et al. Crystal structure of an angiogenesis inhibitor bound to the FGF receptor tyrosine kinase domain. EMBO J. 1998;17:5896–904.
Halgren TA, Murphy RB, Friesner RA, Beard HS, Frye LL, Pollard WT, et al. Glide: a new approach for rapid, accurate docking and scoring. 2. Enrichment factors in database screening. J Med Chem. 2004;47:1750–9.
Li MY, Lv YC, Lj Tong, Peng T, Qu R, Zhang T, et al. DW10075, a novel selective and small-molecule inhibitor of VEGFR, exhibits antitumor activities both in vitro and in vivo. Acta Pharmacol Sin. 2017;37:398–407.
Liu YQ, Wang YN, Lu XY, Tong LJ, Li Y, Zhang T, et al. Identification of compound D2923 as a novel anti-tumor agent targeting CSF1R. Acta Pharmacol Sin. 2018. https://doi.org/10.1038/s41401-018-0056-0
Mohammadi M, McMahon G, Sun L, Tang C, Hirth P, Yeh BK, et al. Structures of the tyrosine kinase domain of fibroblast growth factor receptor in complex with inhibitors. Science. 1997;276:955–60.
Helsten T, Elkin S, Arthur E, Tomson BN, Carter J, Kurzrock R. The FGFR landscape in cancer: Analysis of 4,853 tumors by next-generation sequencing. Clin Cancer Res. 2016;22:259–67.
Fantozzi A, Christofori G. Mouse models of breast cancer metastasis. Breast Cancer Res. 2006;8:212 https://doi.org/10.1186/bcr1530
Brady N, Chuntova P, Bade LK, Schwertfeger KL. The FGF/FGFR axis as a therapeutic target in breast cancer. Expert Rev. Endocrinol Metab. 2013;8:391–402.
Marcus N, Christian F, Burkhard K, Frieder B, Seibel MJ. Site-specific human breast cancer (MDA-MB-231) metastases in nude rats: Model characterisation and in vivo effects of ibandronate on tumour growth. Int J Cancer. 2003;107:468–77.
Tauro M, Lynch CC. Cutting to the chase: how matrix metalloproteinase-2 activity controls breast-cancer-to-bone metastasis. Cancers. 2018;10. https://doi.org/10.3390/cancers10060185
Cihoric N, Savic S, Schneider S, Ackermann I, Bichsel-Naef M, Schmid RA, et al. Prognostic role of FGFR1 amplification in early-stage non-small cell lung cancer. Brit J Cancer. 2014;110:2914–22.
Weiss J, Sos ML, Seidel D, Peifer M, Zander T, Heuckmann JM, et al. Frequent and focal FGFR1 amplification associates with therapeutically tractable FGFR1 dependency in squamous cell lung cancer. Sci Transl Med. 2010;2:62ra93.
Yang W, Yao YW, Zeng JL, Liang WJ, Wang L, Bai CQ, et al. Prognostic value of FGFR1 gene copy number in patients with non-small cell lung cancer: a meta-analysis. J Thorac Dis. 2014;6:803–9.
Reis JS, Simpson PT, Turner NC, Lambros MB, Jones C, Mackay A, et al. FGFR1 emerges as a potential therapeutic target for lobular breast carcinomas. Clin Cancer Res. 2006;12:6652–62.
Courjal F, Cuny M, Simony-Lafontaine J, Louason G, Speiser P, Zeillinger R, et al. Mapping of DNA amplifications at 15 chromosomal localizations in 1875 breast tumors: Definition of phenotypic groups. Cancer Res. 1997;57:4360–7.
Lee HJ, Seo AN, Park SY, Kim JY, Park JY, Yu JH, et al. Low prognostic implication of fibroblast growth factor family activation in triple-negative breast cancer subsets. Ann Surg Oncol. 2014;21:1561–8.
Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. Ca-Cancer J Clin. 2011;61:69–90.
Hase T, Kawashiri S, Tanaka A, Nozaki S, Noguchi N, Kato K, et al. Correlation of basic fibroblast growth factor expression with the invasion and the prognosis of oral squamous cell carcinoma. J Oral Pathol Med. 2006;35:136–9.
Sato T, Oshima T, Yoshihara K, Yamamoto N, Yamada R, Nagano Y, et al. Overexpression of the fibroblast growth factor receptor-1 gene correlates with liver metastasis in colorectal cancer. Oncol Rep. 2009;21:211–6.
Blair KJ, Kiang A, Wang-Rodriguez J, Yu MA, Doherty JK. Ongkeko WMEGF and bFGF promote invasion that is modulated by PI3/Akt kinase and Erk in vestibular schwannoma. Otol Neurotol. 2011;32:308–14.
Jezierska A, Motyl T. Matrix Metalloproteinase-2 involvement in breast cancer progression: a mini-review. Med Sci Monit. 2009;15:Ra32–40.
Cathcart J, Pulkoski-Gross A, Cao J. Targeting matrix metalloproteinases in cancer: Bringing new Life to old ideas. Genes Dis. 2015;2:26–34.
Li H, Qiu ZW, Li F, Wang CL. The relationship between MMP-2 and MMP-9 expression levels with breast cancer incidence and prognosis. Oncol Lett. 2017;14:5865–70.
Coussens LM, Fingleton B, Matrisian LM. Cancer therapy - Matrix metalloproteinase inhibitors and cancer: Trials and tribulations. Science. 2002;295:2387–92.
Tauro M, McGuire J, Lynch CC. New approaches to selectively target cancer-associated matrix metalloproteinase activity. Cancer Metast Rev. 2014;33:1043–57.
Presta M, Dell’Era P, Mitola S, Moroni E, Ronca R, Rusnati M. Fibroblast growth factor/fibroblast growth factor receptor system in angiogenesis. Cytokine Growth F R. 2005;16:159–78.
Auguste P, Gursel DB, Lemiere S, Reimers D, Cuevas P, Carceller F, et al. Inhibition of fibroblast growth factor/fibroblast growth factor receptor activity in glioma cells impedes tumor growth by both angiogenesis-dependent and -independent mechanisms. Cancer Res. 2001;61:1717–26.
Acknowledgements
This work was financially supported by the National Natural Science Foundation of China (grants 81173080, 81321092, 21236002 and 21302054), the National Basic Research Program of China (973 Program, 2010CB126100), the National High Technology Research and Development Program of China (863 Program, 2011AA10A207), the National Science & Technology Pillar Program (2009ZX09103–102), the Shanghai Committee of Science and Technology (grant 13ZR1453100) and the Fundamental Research Funds for the Central Universities.
Author contributions
JD, HX, Y-fX, X-hQ and H-lL designed research; ZC, L-jT, B-yT, H-yL, XW, TZ and YC performed research; ZC, XW and X-wC contributed new reagents or analytic tools; HX and ZC analyzed data; HX and ZC wrote the paper.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
We declare that we do not have any commercial or associative interest that represents a conflict of interest in connection with the work submitted.
Rights and permissions
About this article
Cite this article
Chen, Z., Tong, Lj., Tang, By. et al. C11, a novel fibroblast growth factor receptor 1 (FGFR1) inhibitor, suppresses breast cancer metastasis and angiogenesis. Acta Pharmacol Sin 40, 823–832 (2019). https://doi.org/10.1038/s41401-018-0191-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41401-018-0191-7
Keywords
This article is cited by
-
Whole exome and transcriptome analysis revealed the activation of ERK and Akt signaling pathway in canine histiocytic sarcoma
Scientific Reports (2023)
-
TNFAIP3 mediates FGFR1 activation-induced breast cancer angiogenesis by promoting VEGFA expression and secretion
Clinical and Translational Oncology (2022)
-
The 3D in vivo chorioallantoic membrane model and its role in breast cancer research
Journal of Cancer Research and Clinical Oncology (2022)
-
Discovery of a novel third-generation EGFR inhibitor and identification of a potential combination strategy to overcome resistance
Molecular Cancer (2020)