Abstract
Autophagy, a form of cellular self-digestion by lysosome, is associated with various disease processes including cancers, and modulating autophagy has shown promise in the treatment of various malignancies. A number of natural products display strong antitumor activity, yet their mechanisms of action remain unclear. To gain a better understanding of how traditional Chinese medicine agents exert antitumor effects, we screened 480 natural compounds for their effects on autophagy using a high content screening assay detecting GFP-LC3 puncta in HeLa cells. Tubeimoside-1 (TBMS1), a triterpenoid saponin extracted from Bolbostemma paniculatum (Maxim) Franquet (Cucurbitaceae), was identified as a potent activator of autophagy. The activation of autophagy by TBMS1 was evidenced by increased LC3-II amount and GFP-LC3 dots, observation of autophagosomes under electron microscopy, and enhanced autophagic flux. To explore the mechanisms underlying TBMS1-activated autophagy, we performed cheminformatic analyses and surface plasmon resonance (SPR) binding assay that showed a higher likelihood of the binding between Akt protein and TBMS1. In three human breast cancer cell lines, we demonstrated that Akt–mTOR–eEF-2K pathway was involved in TBMS1-induced activation of autophagy, while Akt-mediated downregulations of Mcl-1, Bcl-xl, and Bcl-2 led to the activation of apoptosis of the breast cancer cells. Inhibition of autophagy enhanced the cytotoxic effect of TBMS1 via promoting apoptosis. Our results demonstrate the role and mechanism of TBMS1 in activating autophagy, suggesting that inhibition of cytoprotective autophagy may act as a therapeutic strategy to reinforce the activity of TBMS1 against cancers.
Similar content being viewed by others
Introduction
Autophagy is a cellular catabolic process by which proteins aggregate and damaged organelles are degraded to generate ATP and various macromolecules as a response to metabolic stress. The process of autophagy consists of three main steps: sequestration of cytoplasmic materials into a double-membrane vesicle, known as the autophagosome; fusion of the autophagosome with the lysosome to form the autolysosome; and lastly, degradation of the sequestered materials for recycling and energy production [1]. Autophagy is associated with pathogenesis of various human diseases, including cancer, cardiac disease, and neurodegeneration [2]. Autophagy suppresses tumor initiation and development by eliminating damaged proteins and organelles and by preventing genomic instability [3,4,5]. Once cancer is established, this catabolic process can provide nutrients and energy to allow cancer cells to survive against a variety of stresses, including nutrient deprivation, hypoxia, chemotherapy, or radiation, thereby contributing to tumor progression and therapeutic resistance [6,7,8]. Targeting autophagic survival has been considered a novel strategy to reinforce the efficacy of anticancer therapy. Autophagy may also promote cell death in tumor cells in response to certain chemotherapeutic agents or under certain circumstances and is thus referred to as type II programmed cell death [9,10,11,12].
Natural compounds are important resources for anticancer drug development. TBMS1 is a triterpenoid saponin extracted from the Chinese medicinal herb Bolbostemma paniculatum (Maxim) Franquet (Cucurbitaceae), which is conventionally used for treatment of snake venoms and inflammation [13,14,15]. TBMS1 has also been proven to possess potent anticancer activity. For example, it has been reported that TBMS1 can induce apoptosis in human prostate cancer cells [16], lung cancer cells [17], liver cancer cells [18, 19], cervical cancer cells [20], and gastric cancer cells [21]. TBMS1 was demonstrated to inhibit cell invasion [22], induce cell cycle arrest [16, 18, 23, 24], and suppress tumor angiogenesis [25]. However, the modulatory effect of TBMS1 on autophagy remains unknown.
The serine/threonine kinase Akt promotes cell growth, proliferation, and survival [26]. Constitutive activation of Akt plays a critical role in cancer development and progression [27,28,29]. Thus Akt is identified as an attractive target for cancer therapy. It is known that the anticancer action of Akt inhibitors results from apoptosis induction via suppression of survival-associated signaling pathways such as those modulated by Bcl-2 family proteins [30,31,32]. In addition, it has been found that autophagy is also activated when Akt is inhibited [33,34,35].
In this study, we demonstrate for the first time that TBMS1 can induce autophagy in cancer cells, and the Akt-mediated signaling pathway is involved in autophagy and apoptosis activation caused by this compound. Inhibition of cytoprotective autophagy can enhance the cytocidal effect of TBMS1 in breast cancer cells by promoting apoptotic cell death.
Materials and methods
Cell culture
Human breast cancer cell lines MCF-7 (Cell Bank of Chinese Academy of Sciences, Beijing, China) and T47D (Cell Bank of Chinese Academy of Sciences, Shanghai, China) were cultured in Dulbecco’s modified Eagle’s medium/high glucose (HyClone) medium supplemented with 10% fetal bovine serum (Gibco) at 37 °C with 5% CO2. MDA-MB-231 (Cell Bank of Chinese Academy of Sciences, Beijing, China) was cultured in L-15 (HyClone) medium supplemented with 10% fetal bovine serum at 37 °C with 100% air.
Chemical reagents and antibodies
TBMS1 was purchased from Pufei De Biotech (Chengdu, China). Chloroquine was purchased from Sigma (USA). NH125 was purchased from Selleck (Shanghai, China). Protease inhibitor and phosphatase inhibitor cocktails A and B were purchased from Selleck (Shanghai, China). Antibodies against light chain 3 (LC3; cat. no. 12741), Mcl-1 (cat. no. 5453), Bcl-xl (cat. no. 2764), Bcl-2 (cat. no. 2870), Akt (cat. no. 4691), p-Akt (Thr308) (cat. no. 13038), p-Akt (Ser473) (cat. no. 4060), p70S6K (cat. no. 2708), p-p70S6K (Thr389) (cat. no. 9234), eukaryotic translation elongation factor 2 (eEF-2; cat. no. 2332), p-eEF-2 (Thr56) (cat. no. 2331), eEF-2K (cat. no. 3692), Beclin1 (cat. no. 4122), poly ADP-ribose polymerase (PARP; cat. no. 9532), and cleaved caspase-3 (cat. no. 9664) were purchased from Cell Signaling Technology (Danvers, MA, USA). Antibody against β-actin was purchased from Proteintech. Recombinant human Akt1 protein (cat. no. 10763-H08B) was purchased from Sino Biological (Beijing, China). The enhanced chemiluminescence (ECL) kit was purchased from Beijing Com Win Biotech Co, Ltd. (Cwbio, China). Cell Counting Kit-8 (CCK-8) was purchased from Bimake (Shanghai, China).
High content screening
High content screening and data analysis were performed as previously described [36]. Briefly, HeLa cells stably expressing pEGFP-LC3 plasmids were seeded in a 96-well plate (clear bottom, black; PerkinElmer) overnight. Cells were then treated with different natural product compounds in triplicates [37]. After 48 h, cells were fixed with 4% paraformaldehyde, and fluorescence images were acquired using an Opera High Content Screening System (PerkinElmer) under a ×40–H2O objective. The green fluorescent protein (GFP)-LC3 spots were quantified accordingly.
Surface plasmon resonance (SPR) assay
To measure the interaction between TBMS1 and Akt, a SPRi instrument was used to monitor the whole procedure in real time. Briefly, a chip with a well-prepared biomolecular microarray was assembled with a plastic flow cell for sample loading. The purified recombinant Akt1 protein samples were dissolved in phosphate-buffered saline (PBS) running buffer (pH = 7.4, 0.1% Tween 20), and 10 mM glycine-HCl buffer (pH = 2.0) was used as the regeneration buffer. A typical binding curve was obtained by flowing the protein sample at 0.5 μL/s for 600 s of association and then the running buffer for 360 s of dissociation, after which the regeneration buffer was flowed at 2 μL/s for 200 s. To determine the binding affinity, three gradient concentrations of Akt1 proteins were prepared and flowed in order. Binding data were collected and analyzed by commercial SPRi analysis software (Plexera SPR Data Analysis Model; Plexera).
Western blot analysis
After treatment with TBMS1, cells were washed with PBS and lysed in RIPA buffer supplemented with protease inhibitor and phosphatase inhibitor cocktails A and B (Selleck) for 30 min on ice. The cell lysates were spun in a centrifuge at 12 000 × g for 15 min. The protein concentration of the supernatant was determined using a BCA assay kit according to the manufacturer’s protocol. Equivalent amounts of protein were loaded into sodium dodecyl sulfate–polyacrylamide gels for electrophoresis and transferred onto a polyvinylidene difluoride membrane. Next, the membranes were blocked with 5% skim milk in Tris-buffered saline with Tween 20 and then probed with primary antibodies and peroxidase-conjugated secondary antibodies. Subsequently, the membranes were visualized with an enhanced chemiluminescent detection kit.
Preliminary identification of the possible target by multitarget structure–activity relationship (SAR) models
Based on the related literature published in recent years, we preliminarily chose eight important targets in the autophagy process: extracellular signal–regulated kinase, Akt, c-Jun N-terminal kinase, p38, AMPK, Raf, mammalian target of rapamycin (mTOR), and eEF-2 kinase (eEF-2K). For these selected targets, we collected their ligands, which are small, drug-like molecules, from the ChEMBL database and Binding database. For each target, activity data were filtered to keep only data that had half-maximum inhibitory concentration (IC50), half-maximum effective concentration (EC50), or Ki values. A compound was considered a positive sample when its activity value was below the median. Otherwise, this compound was considered a negative sample. All the compounds were used to construct SAR classification models based on CATS descriptors and the random forest method. Here the following popular statistical parameters were applied to evaluate the performance of these classification models: sensitivity; specificity; accuracy (ACC); area under receiver operating characteristic curve (AUC); Matthews correlation coefficient; and F1 score. Next, the SAR models were applied to predict the probable relationship between the autophagy-related target and TBMS1. According to the predicted results, the most likely target for TBMS1 could be preliminarily identified for further study.
Plasmid and small interfering RNA (siRNA) transfection
MCF-7 cells were transfected with GFP-LC3 plasmid using Lipofectamine 2000 reagent according to the manufacturer’s instructions. In brief, cells in the exponential phase of growth were plated in 6-well tissue culture plates at 1 × 105 cells per well, grown for 24 h, and transfected with a mixture of GFP-LC3 plasmid and Lipofectamine 2000 (Invitrogen) in OPTI-MEM reduced serum medium. To determine the autophagic response, GFP-LC3 puncta were inspected by fluorescence microscopy. For siRNA targeting eEF-2K (AAGCUCGAACCAGAAUGUCAA) or Beclin1 (GGTCTAAGACGTCCAACAA) (Ribobio) transfection, cells were transfected with a mixture of siRNA and Lipofectamine 2000 (Invitrogen) in OPTI-MEM reduced serum medium.
Apoptosis assay
Cell apoptosis was determined by flow cytometric analysis of Annexin V-fluorescein isothiocyanate/propidium iodide (PI) double staining. After treatment with TBMS1 for 48 h, the cells were harvested and rinsed with cold PBS twice by centrifugation at 1000 × g, resuspended in 100 μL 1× binding buffer (BD cat. no.556547) (1 × 105 cells). Five μL Annexin-V (BD cat. no.556420) and 5 μL PI (BD cat. no.556463) were then added to the solution, and the cells were gently vortexed and incubated for 15 min at room temperature in the dark. At the end of the incubation period, the samples were added to 400 μL 1× binding buffer for flow cytometric (Becton Dickinson) analysis.
Transmission electron microscopy
MDA-MB-231 cells were treated with 4 μM TBMS1 for 24 h. The collected cells were fixed with NaCl/Pi containing 3% glutaraldehyde and then postfixed with NaCl/Pi containing 1% OsO4. The samples were dehydrated in graded alcohol, embedded, and sectioned. Ultrathin sections were stained with uranyl acetate and lead citrate and examined using a JEM-1200 transmission electron microscope (JEOL, Tokyo, Japan).
Cell viability assay
Cell viability was measured using the CCK-8 assay according to the manufacturer’s protocol. Briefly, cells were seeded at 5 × 103 cells per well in 96-well plates. At the end of TBMS1 treatment, cells were incubated with 10 μL CCK-8 reagent for 2 h at 37 °C with 5% CO2. The results were detected at 450 nm wavelength.
Clonogenic assay
Two hundred cells per well were plated in 6-well tissue culture plates and treated with TBMS1 in the presence or absence of 20 μM chloroquine (CQ) for 24 h. The cells were incubated at 37 °C with 5% CO2 for 8 days, and the medium was replaced every 2 days. At the end of incubation, cells were fixed with methanol and stained with 1% crystal violet for 1 h. The cells were then washed with water, and the colonies were counted (a colony contains at least 100 cells).
Statistical analysis
GraphPad Prism 5 was used to perform statistical analysis. Data were presented as the mean ± S.D. or mean ± S.E.M using Student’s t test analysis of variance for analyzing the difference between groups. Levels of P < 0.05 were considered statistically significant.
Results
TBMS1 induces autophagy in breast cancer cells
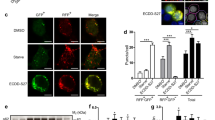
To identify novel autophagic regulators from natural compounds, we performed a high content screening assay to examine the effects of 480 natural compounds on autophagy and found that TBMS1, as shown in plate No. 2C9, increased the number of GFP-LC3 dots (Figure S1). As shown in Figure S2, GFP-LC3 displayed marked accumulation of puncta in cells treated with TBMS1 compared to control cells (hydroxychloroquine (HCQ) was used as a positive control). These results suggest that TBMS1 has a stimulatory effect on autophagy. The chemical structure of TBMS1 is shown in Fig. 1. To validate the role of TBMS1 in activating autophagy in cancer cells, breast cancer cells MCF-7, MDA-MB-231, and T47D were treated with TBMS1, and LC3 levels were measured. As shown in Fig. 2a, b, TBMS1 increased LC3-II levels in a time- and dose-dependent manner. We further measured autophagic flux and found that LC3-II significantly accumulated in the presence of the autophagy inhibitor CQ, indicating that autophagic flux was enhanced by TBMS1 (Fig. 2c). The activating effect of TBMS1 on autophagy was further demonstrated by an increase in the number of GFP-LC3 dots (Fig. 2d) and by autophagosome formation, as observed by transmission electron microscopy (Fig. 2e). These results further support that TBMS1 could induce autophagy in breast cancer cells.
TBMS1 induces autophagy in breast cancer cells. MDA-MB-231, MCF-7, or T47D cells were treated with a series of concentrations of TBMS1 for 24 h (a) or were treated with 4 μM TBMS1 for different periods of time (b), and LC3 levels were measured by Western blot. c MDA-MB-231, MCF-7, or T47D cells were pretreated with 20 μM CQ for 1 h followed by treatment with TBMS1 for 24 h. LC3 levels were measured by Western blot. β-Actin was used as a loading control. d MCF-7 cells were transfected with 4 μg GFP-LC3 plasmid followed by treatment with 8 μM TBMS1 for 12 h, and GFP-LC3 puncta were observed with a fluorescence microscope. e MDA-MB-231 cells were incubated with 4 μM TBMS1 for 24 h. The cellular ultrastructure was examined by transmission electron microscopy. Arrows indicate autophagic vacuoles
The Akt-mediated signaling pathway contributes to the effect of TBMS1 on inducing autophagy
To explore the molecular mechanism by which TBMS1 activates autophagy, we preliminarily identified the most likely targets for TBMS1 by cheminformatic analysis. Eight important proteins in the autophagy process were collected, and the SAR model for each protein was constructed individually. After a series of strict model validation and evaluation, we obtained eight robust classification models. As shown in Fig. 3a, the ACC (= 0.784–0.914) and AUC (= 0.807–0.965) of cross validation are adequate for each SAR model. The statistical parameters indicate that these classification models can reliably predict the relationship between protein and specific compound (Table 1). Based on these models, we predicted the potential autophagy-related proteins that bind to TBMS1. The predictive result for TBMS1 was outputted as a probability value. A higher probability value indicates that there is a higher likelihood of binding between an autophagy-related protein and TBMS1. We found that the most likely target for TBMS1 is Akt (prediction probability is 0.876) (Table 1). We next sought to determine whether there was an interaction between TBMS1 and Akt. First, we performed SPR-based direct binding assays and found that TBMS1 could directly bind to the recombinant human Akt1 protein and that the binding signal became stronger as the concentration increased (Fig. 3b). The kinetic parameters for TBMS1 and Akt1 binding are shown in Table 2. We next measured the effect of TBMS1 on Akt activity. As shown in Fig. 3c, TBMS1 treatment caused decreased Akt activity in a dose-dependent manner as demonstrated by decreased phosphorylation of Akt at both Ser473 and Thr308, whereas Akt expression remained unchanged. These results suggest that TBMS1 can bind to Akt and inhibit its activity and that the Akt-mediated signaling pathway may contribute to TBMS1’s effect on inducing autophagy in breast cancer cells.
TBMS1 inhibits Akt activity. a The prediction statistics (accuracy and AUC) of eight SAR models for eight autophagy-related proteins. b SPRi graph showing the interaction of TBMS1 and Akt1. c MDA-MB-231, MCF-7, or T47D cells were treated with a series of concentrations of TBMS1 for 24 h, and the levels of p-Akt (S473, T308) and Akt were measured by Western blot. β-Actin was used as a loading control
Akt–mTOR–eEF-2K is involved in autophagy activation induced by TBMS1
To further explore the molecular mechanism mediating autophagy activation by inhibiting Akt in TBMS1-treated breast cancer cells, we measured the role of mTOR, an important regulator of autophagy that acts as a downstream effector of Akt. Figure 4a, b show that TBMS1 inhibited mTOR activity in a time- and concentration-dependent manner, as shown by decreased phosphorylation levels of its substrate, p70S6K. eEF-2K, a positive regulator of autophagy under cellular stress [38,39,40], is a downstream effector of mTOR [41], which inhibits eEF-2K activity by phosphorylation at Ser78/Ser366 [42]. We found that eEF-2 phosphorylation at Thr56, the only known substrate of eEF-2K, was increased in a time- and dose-dependent manner in breast cancer cells treated with TBMS1 (Fig. 4a, b), suggesting that eEF-2K was activated by this natural product. We next found that silencing eEF-2K expression led to a decrease in LC3-II (Fig. 4c), indicating that autophagic activity induced by TBMS1 was inhibited by suppression of eEF-2K. A decrease in LC3-II was also observed in breast cancer cells cotreated with NH125, a small molecule inhibitor of eEF-2K (Fig. 4d), providing further support for the key role of eEF-2K in activating autophagy induced by TBMS1. We further found that the decrease in p-p70S6K and increases in p-eEF-2 and LC3-II induced by TBMS1 could be reversed by Akt overexpression (Fig. 4e). These results demonstrate that the Akt–mTOR–eEF-2K pathway mediates autophagy induction triggered by TBMS1.
TBMS1 induces autophagy via the Akt-mTOR-eEF-2K signaling pathway. MDA-MB-231, MCF-7, or T47D cells were treated with 4 μM TBMS1 for different periods of time (a) or were treated with a series of concentrations of TBMS1 for 24 h (b). At the end of treatment, the protein levels were measured by Western blot. c MDA-MB-231 or T47D cells were transfected with nontargeting siRNA or eEF-2K-targeted siRNA followed by treatment with 8 μM TBMS1 for 24 h. The protein levels were measured by Western blot. d MDA-MB-231 or T47D cells were pretreated with 300 nM NH125 for 1 h, followed by treatment with 8 μM TBMS1 for 24 h. The protein levels were measured by Western blot. e MCF-7 cells were transfected with 2 μg Akt plasmid followed by treatment with 8 μM TBMS1 for 24 h, and the protein levels were measured by Western blot
Inhibition of autophagy enhances the cytotoxic effect of TBMS1 against breast cancer cells by promoting apoptosis
Recently, TBMS1 has been reported to trigger apoptosis in several types of cancer cells [16,17,18,19, 21]; however, the effect of TBMS1 on human breast cancer cells has not been studied. We found that TBMS1 induced cell apoptosis in a time- and concentration-dependent manner, as determined by an increase in cleaved PARP and caspase-3 (Fig. 5a, b) and Annexin V staining (Fig. 5c). Fig. 3 shows that TBMS1 targeted and inhibited Akt activity. It is known that Akt inhibits cell apoptosis by influencing Bcl-2 family proteins, including Mcl-1, Bcl-xl, and Bcl-2. Figure 5d, e shows that treatment of breast cancer cells with TBMS1 resulted in a time- and dose-dependent decrease in the levels of Mcl-1, Bcl-xl, and Bcl-2. Downregulation of Mcl-1, Bcl-xl, and Bcl-2 in TBMS1-treated cells was rescued by Akt overexpression (Fig. 5f), indicating that Akt-mediated downregulation of these anti-apoptotic proteins plays a critical role in activating apoptosis triggered by this compound. We next aimed to determine whether there is a relationship between autophagy and apoptosis in TBMS1-treated cells. Fig. 6a shows that inhibition of autophagy by CQ, a chemical inhibitor of autophagy, augmented apoptosis compared to treatment by TBMS1 alone, as determined by cleaved caspase-3 and PARP, indicating that autophagy plays a pro-survival role. Enhancement of TBMS1-induced apoptosis by inhibition of autophagy was also evidenced by an increased in Annexin V staining (Fig. 6b). To further demonstrate the association between autophagy and apoptosis, we suppressed autophagy by silencing Beclin1 and found that the apoptotic activity of TBMS1 was also markedly increased in these cells, as demonstrated by cleaved caspase-3 and PARP (Fig. 6c). We further determined the effects of autophagy on the viability of the breast cancer cells treated with TBMS1 and observed that inhibition of autophagy with CQ or the silencing of Beclin1 expression enhanced the cytotoxicity of this compound against breast cancer cells (Fig. 6d–f). As eEF-2K plays a key role in activating autophagy induced by TBMS1, we also examined cell viability after silencing eEF-2K. Figure 6g shows that breast cancer cells with knockdown of eEF-2K expression were more sensitive to TBMS1 than the cells without knockdown of eEF-2K. Our results suggest that autophagy acts as a survival mechanism, and inhibition of autophagy can sensitize breast cancer cells to TBMS1.
Akt-mediated downregulation of anti-apoptotic Bcl-2 family proteins is involved in TBMS1-induced apoptosis in breast cancer cells. MDA-MB-231 or T47D cells were treated with 4 μM TBMS1 for 24 h or 48 h (a) or were treated with 4 μM or 8 μM TBMS1 for 48 h (b), and the protein levels were measured by Western blot. c MDA-MB-231 cells were treated with 4 or 8 μM TBMS1 for 48 h, and apoptosis was examined by measuring Annexin V staining; one from three identical experiments is shown. *P < 0.05, **P < 0.01, t- test (n = 3, Mean ± S.D.). MDA-MB-231, MCF-7, or T47D cells were treated with 4 μM TBMS1 for different periods of time (d) or were treated with a series of concentrations of TBMS1 for 48 h (e), and the protein levels were measured by Western blot. f MCF-7 cells were transfected with 2 μg Akt plasmid followed by treatment with 8 μM TBMS1 for 24 h, and the protein levels were measured by Western blot
Inhibition of autophagy enhances the cytotoxic effect of TBMS1 against breast cancer cells by promoting apoptosis. a MDA-MB-231 or T47D cells were pretreated with 20 μM CQ for 1 h followed by treatment with TBMS1 for 48 h. The protein levels were measured by Western blot. b MDA-MB-231 cells were incubated with TBMS1 in the presence or absence of CQ (20 μM) for 48 h, and apoptosis was examined by measuring Annexin V staining; one from three identical experiments was shown. *P < 0.05, ##P< 0.01, t- test, *TBMS1 vs. the control group, #TBMS1 vs. TBMS1+CQ (n = 3, Mean ± S.D.). c MDA-MB-231 cells were transfected with nontargeting siRNA or Beclin1-targeted siRNA followed by treatment with 8 μM TBMS1 for 48 h. The protein levels were measured by Western blot. d MDA-MB-231, MCF-7, or T47D cells were treated with a series of concentrations of TBMS1 for 48 h in the absence or presence of 20 μM CQ. At the end of treatment, cell viability was measured by CCK-8 reagent. The results are shown as the mean ± S.D. from one of three identical experiments; *P < 0.05, **P < 0.01, t- test (n = 3, Mean ± S.D.). e MDA-MB-231 cells were pretreated 20 μM CQ for 1 h followed by treatment with 8 μM TBMS1 for 24 h. After treatment, the cells were incubated in normal medium for 8 days, and the cell colonies were fixed and stained for photography. The results are displayed as the mean ± S.D. of three experiments; ** P < 0.01, ##P < 0.01, *TBMS1 vs the control group, #TBMS1 vs TBMS1+CQ. f MDA-MB-231 cells were transfected with Beclin1 siRNA or nontargeting siRNA and then treated with a series of concentrations of TBMS1 for 48 h. g MDA-MB-231 cells were transfected with eEF-2K siRNA or nontargeting siRNA and then treated with a series of concentrations of TBMS1 for 48 h. At the end of treatment, cell viability was measured by CCK-8 reagent. The results are shown as the mean ± S.E.M. of three identical experiments; * P < 0.05, **P < 0.01, t- test
Discussion
In this study, we not only uncovered a previously unknown effect of TBMS1 on autophagy induction in cancer cells but also provided the first evidence that TBMS1 activates apoptosis in breast cancer cells. We found that Akt is a critical regulator of TBMS1-induced tumor cell autophagy and apoptosis (Fig. 7). In addition, we also demonstrated that inhibition of autophagy promoted apoptotic activity and sensitized TBMS1 cytotoxicity in breast cancer cells.
Autophagy can cause either cell death or survival depending on different stimuli and the intracellular environment [5]. A better understanding of the exact roles that autophagy plays in tumor cells subjected to therapeutic stress would be beneficial to explore autophagy as a therapeutic strategy against cancer. Numerous studies have reported that autophagy is activated in tumor cells in response to therapeutic treatments, including chemotherapy, targeted therapy, and radiotherapy, and plays a cytoprotective role [7, 43, 44]. Therefore, inhibition of autophagy-mediated cell survival is now considered a novel strategy to reverse chemoresistance and enhance the effectiveness of anticancer treatments. The autophagy inhibitors CQ or HCQ have been tested in clinical trials and exhibited improving outcomes in several cancer types. In this study, we found that TBMS1 can induce autophagy in breast cancer cells, and autophagy can affect the apoptotic activity induced by this compound. These results suggest that modulating autophagy and apoptosis by a natural compound may be an effective therapeutic intervention against cancer [4].
Akt, an oncoprotein that promotes cell proliferation and survival, is an attractive therapeutic target for cancer treatment. Inhibition of Akt can induce both apoptosis and autophagy. To explore the regulatory mechanisms by which autophagy and apoptosis were modulated in breast cancer cells subjected to TBMS1 treatment, we performed cheminformatic analysis to predict potential autophagy-related proteins that bind to TBMS1 and found that TBMS1 has a high likelihood to interact with Akt, further demonstrating that TBMS1 indeed binds to Akt and inhibits its activity experimentally. As the mTOR signaling pathway is an important downstream effector of Akt [39, 45, 46], we further examined the effect of TBMS1 on mTOR and found that this compound inhibited mTOR activity. eEF-2K is a negative regulator of protein synthesis, and its activity can be inhibited via phosphorylation at Ser78 and Ser366 by the mTOR pathway [42]. We previously demonstrated that eEF-2K participated in autophagy activation in cancer cells under various stresses [38,39,40, 47]. In this study, we found that silencing eEF-2K suppressed autophagic activity triggered by TBMS1, indicating that eEF-2K was involved in autophagy induction activated by TBMS1. Therefore, targeting eEF-2K could be utilized as a strategy to reinforce the anticancer effect of TBMS1. These results are consistent with our previous study, which demonstrated that eEF-2K played an essential role in autophagy induction triggered by Akt inhibition [38]. It is well known that Akt inhibits cell apoptosis by influencing Bcl-2 family proteins. Akt has been reported to promote cell survival by upregulating anti-apoptotic proteins Bcl-2 or Mcl-1 [48,49,50]. Inhibition of Akt triggers apoptosis by downregulating expression of Bcl-2, Mcl-1 or Bcl-xl, thereby leading to tumor suppression in vitro and in vivo [32, 51, 52]. We found that TBMS1 decreased the levels of Mcl-1, Bcl-xl, and Bcl-2 in a time- and dose-dependent manner in breast cancer cells, and overexpression of Akt rescued the downregulation of these anti-apoptotic proteins. A better understanding of the molecular mechanisms and signaling pathways underlying the anticancer effect of TBMS1 may help provide novel strategies and resources for anticancer drug development.
In summary, our study indicates that Akt-mediated downregulation of Mcl-1, Bcl-xl, and Bcl-2 are involved in apoptosis activation induced by TBMS1, and the Akt–mTOR–eEF-2K pathway plays an essential role in autophagy activation and in determining the fate of cancer cells in response to TBMS1 treatment. Inhibition of autophagy may represent an effective means to enhance TBMS1 cytotoxicity by promoting apoptotic activity.
References
Chen Y, Scarcelli V, Legouis R. Approaches for studying autophagy in Caenorhabditis elegans. Cells. 2017;6. https://doi.org/10.3390/cells6030027.
Cheng Y, Ren X, Hait WN, Yang JM. Therapeutic targeting of autophagy in disease: biology and pharmacology. Pharmacol Rev. 2013;65:1162–97.
Gozuacik D, Kimchi A. Autophagy as a cell death and tumor suppressor mechanism. Oncogene. 2004;23:2891–906.
Galluzzi L, Pietrocola F, Bravo-San Pedro JM, Amaravadi RK, Baehrecke EH, Cecconi F, et al. Autophagy in malignant transformation and cancer progression. EMBO J. 2015;34:856–80.
Law BYK, Mok SWF, Chen J, Michelangeli F, Jiang ZH, Han Y, et al. N-desmethyldauricine induces autophagic cell death in apoptosis-defective cells via Ca2+ mobilization. Front Pharmacol. 2017;8:388.
Kimmelman AC, White E. Autophagy and tumor metabolism. Cell Metab. 2017;25:1037–43.
Piya S, Andreeff M, Borthakur G. Targeting autophagy to overcome chemoresistance in acute myleogenous leukemia. Autophagy. 2017;13:214–5.
Chen X, Wang P, Guo F, Wang X, Wang J, Xu J, et al. Autophagy enhanced the radioresistance of non-small cell lung cancer by regulating ROS level under hypoxia condition. Int J Radiat Biol. 2017;93:764–70.
Park MA, Reinehr R, Haussinger D, Voelkel-Johnson C, Ogretmen B, Yacoub A, et al. Sorafenib activates CD95 and promotes autophagy and cell death via Src family kinases in gastrointestinal tumor cells. Mol Cancer Ther. 2010;9:2220–31.
Levine B, Klionsky DJ. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev Cell. 2004;6:463–77.
Cheng Y, Sk UH, Zhang Y, Ren X, Zhang L, Huber-Keener KJ, et al. Rational incorporation of selenium into temozolomide elicits superior antitumor activity associated with both apoptotic and autophagic cell death. PLoS One. 2012;7:e35104.
Wong VKW, Zeng W, Chen J, Yao XJ, Leung ELH, Wang QQ, et al. Tetrandrine, an activator of autophagy, induces autophagic cell death via PKC-alpha inhibition and mTOR-dependent mechanisms. Front Pharmacol. 2017;8:351.
Ma R, Song G, You W, Yu L, Su W, Liao M, et al. Anti-microtubule activity of tubeimoside I and its colchicine binding site of tubulin. Cancer Chemother Pharmacol. 2008;62:559–68.
Wu Q, Sun G, Yuan X, Soromou LW, Chen N, Xiong Y, et al. Tubeimoside-1 attenuates LPS-induced inflammation in RAW 264.7 macrophages and mouse models. Immunopharmacol Immunotoxicol. 2013;35:514–23.
Xu Y, Chiu JF, He QY, Chen F. Tubeimoside-1 exerts cytotoxicity in HeLa cells through mitochondrial dysfunction and endoplasmic reticulum stress pathways. J Proteome Res. 2009;8:1585–93.
Yang JB, Khan M, He YY, Yao M, Li YM, Gao HW, et al. Tubeimoside-1 induces oxidative stress-mediated apoptosis and G0/G1 phase arrest in human prostate carcinoma cells in vitro. Acta Pharmacol Sin. 2016;37:950–62.
Hao W, Wang S, Zhou Z. Tubeimoside-1 (TBMS1) inhibits lung cancer cell growth and induces cells apoptosis through activation of MAPK-JNK pathway. Int J Clin Exp Pathol. 2015;8:12075–83.
Yin Y, Chen W, Tang C, Ding H, Jang J, Weng M, et al. NF-kappaB, JNK and p53 pathways are involved in tubeimoside-1-induced apoptosis in HepG2 cells with oxidative stress and G(2)/M cell cycle arrest. Food Chemical Toxicol. 2011;49:3046–54.
Wang Y, Deng L, Zhong H, Wang Y, Jiang X, Chen J. Natural plant extract tubeimoside I promotes apoptosis-mediated cell death in cultured human hepatoma (HepG2) cells. Biol Pharm Bull. 2011;34:831–8.
Xu Y, Ching YP, Zhou Y, Chiu JF, Chen F, He QY. Multiple pathways were involved in tubeimoside-1-induced cytotoxicity of HeLa cells. J Proteomics. 2011;75:491–501.
Zhang Y, Xu XM, Zhang M, Qu D, Niu HY, Bai X, et al. Effects of tubeimoside-1 on the proliferation and apoptosis of BGC823 gastric cancer cells in vitro. Oncol Lett. 2013;5:801–4.
Bian Q, Liu P, Gu J, Song B. Tubeimoside-1 inhibits the growth and invasion of colorectal cancer cells through the Wnt/beta-catenin signaling pathway. Int J Clin Exp Pathol. 2015;8:12517–24.
Xu Y, Wang G, Chen Q, Lin T, Zeng Z, Luo Q, et al. Intrinsic apoptotic pathway and G2/M cell cycle arrest involved in tubeimoside I-induced EC109 cell death. Chin J Cancer Res. 2013;25:312–21.
Chen WJ, Yu C, Yang Z, He JL, Yin J, Liu HZ, et al. Tubeimoside-1 induces G2/M phase arrest and apoptosis in SKOV-3 cells through increase of intracellular Ca2+ and caspase-dependent signaling pathways. Int J Oncol. 2012;40:535–43.
Gu Y, Korbel C, Scheuer C, Nenicu A, Menger MD, Laschke MW. Tubeimoside-1 suppresses tumor angiogenesis by stimulation of proteasomal VEGFR2 and Tie2 degradation in a non-small cell lung cancer xenograft model. Oncotarget. 2016;7:5258–72.
Altomare DA, Testa JR. Perturbations of the AKT signaling pathway in human cancer. Oncogene. 2005;24:7455–64.
Riemenschneider MJ, Betensky RA, Pasedag SM, Louis DN. AKT activation in human glioblastomas enhances proliferation via TSC2 and S6 kinase signaling. Cancer Res. 2006;66:5618–23.
Manning BD, Cantley LC. AKT/PKB signaling: navigating downstream. Cell. 2007;129:1261–74.
Pal I, Mandal M. PI3K and Akt as molecular targets for cancer therapy: current clinical outcomes. Acta Pharmacol Sin. 2012;33:1441–58.
Garcia-Echeverria C, Sellers WR. Drug discovery approaches targeting the PI3K/Akt pathway in cancer. Oncogene. 2008;27:5511–26.
Qi SM, Lv J, Meng Y, Qi ZL, Ling LF. Effect of sodium aescinate in inducing human breast cancer MCF-7 cells apoptosis by inhibiting AKT, ERK and upstream signal SRC activity. China J Chin Mater Med. 2015;40:3267–72.
Datta SR, Dudek H, Tao X, Masters S, Fu H, Gotoh Y, et al. Akt phosphorylation of BAD couples survival signals to the cell-intrinsic death machinery. Cell. 1997;91:231–41.
Degtyarev M, De Maziere A, Orr C, Lin J, Lee BB, Tien JY, et al. Akt inhibition promotes autophagy and sensitizes PTEN-null tumors to lysosomotropic agents. J Cell Biol. 2008;183:101–16.
Fujiwara K, Iwado E, Mills GB, Sawaya R, Kondo S, Kondo Y. Akt inhibitor shows anticancer and radiosensitizing effects in malignant glioma cells by inducing autophagy. Int J Oncol. 2007;31:753–60.
Xue JF, Shi ZM, Zou J, Li XL. Inhibition of PI3K/AKT/mTOR signaling pathway promotes autophagy of articular chondrocytes and attenuates inflammatory response in rats with osteoarthritis. Biomed Pharmacother. 2017;89:1252–61.
Xu D, Lao Y, Xu N, Hu H, Fu W, Tan H, et al. Identification and characterization of anticancer compounds targeting apoptosis and autophagy from Chinese native Garcinia species. Planta Med. 2015;81:79–89.
Wang J, Shi ZQ, Zhang M, Xin GZ, Pang T, Zhou P, et al. Camptothecin and its analogs reduce amyloid-beta production and amyloid-beta42-induced IL-1beta production. J Alzheimers Dis. 2015;43:465–77.
Cheng Y, Ren X, Zhang Y, Patel R, Sharma A, Wu H, et al. eEF-2 kinase dictates cross-talk between autophagy and apoptosis induced by Akt Inhibition, thereby modulating cytotoxicity of novel Akt inhibitor MK-2206. Cancer Res. 2011;71:2654–63.
Cheng Y, Ren X, Zhang Y, Shan Y, Huber-Keener KJ, Zhang L, et al. Integrated regulation of autophagy and apoptosis by EEF2K controls cellular fate and modulates the efficacy of curcumin and velcade against tumor cells. Autophagy. 2013;9:208–19.
Guan YD, Jiang SL, Yu P, Wen M, Zhang Y, Xiao SS, et al. Suppression of eEF-2K-mediated autophagy enhances the cytotoxicity of raddeanin A against human breast cancer cells in vitro. Acta Pharmacol Sin. 2018;39:642–8.
Cheng Y, Ren X, Yuan Y, Shan Y, Li L, Chen X, et al. eEF-2 kinase is a critical regulator of Warburg effect through controlling PP2A-A synthesis. Oncogene. 2016;35:6293–308.
Kenney JW, Moore CE, Wang X, Proud CG. Eukaryotic elongation factor 2 kinase, an unusual enzyme with multiple roles. Adv Biol Regul. 2014;55:15–27.
Qadir MA, Kwok B, Dragowska WH, To KH, D Le, Bally MB, et al. Macroautophagy inhibition sensitizes tamoxifen-resistant breast cancer cells and enhances mitochondrial depolarization. Breast Cancer Res Treat. 2008;112:389–403.
Amaravadi RK, Yu D, Lum JJ, Bui T, Christophorou MA, Evan GI, et al. Autophagy inhibition enhances therapy-induced apoptosis in a Myc-induced model of lymphoma. J Clin Invest. 2007;117:326–36.
Hay N, Sonenberg N. Upstream and downstream of mTOR. Genes Dev. 2004;18:1926–45.
Hers I, Vincent EE, Tavare JM. Akt signalling in health and disease. Cell Signal. 2011;23:1515–27.
Cheng Y, Li H, Ren X, Niu T, Hait WN, Yang J. Cytoprotective effect of the elongation factor-2 kinase-mediated autophagy in breast cancer cells subjected to growth factor inhibition. PLoS One. 2010;5:e9715.
Pugazhenthi S, Nesterova A, Sable C, Heidenreich KA, Boxer LM, Heasley LE, et al. Akt/protein kinase B up-regulates Bcl-2 expression through cAMP-response element-binding protein. J Biol Chem. 2000;275:10761–6.
Wang JM, Chao JR, Chen W, Kuo ML, Yen JJ, Yang-Yen HF. The antiapoptotic gene mcl-1 is up-regulated by the phosphatidylinositol 3-kinase/Akt signaling pathway through a transcription factor complex containing CREB. Mol Cell Biol. 1999;19:6195–206.
Sheikh AM, Malik M, Wen G, Chauhan A, Chauhan V, Gong CX, et al. BDNF-Akt-Bcl2 antiapoptotic signaling pathway is compromised in the brain of autistic subjects. J Neurosci Res. 2010;88:2641–7.
Yang P, Zhao J, Hou L, Yang L, Wu K, Zhang L. Vitamin E succinate induces apoptosis via the PI3K/AKT signaling pathways in EC109 esophageal cancer cells. Mol Med Rep. 2016;14:1531–7.
Ramalingam M, Kim SJ. Reactive oxygen/nitrogen species and their functional correlations in neurodegenerative diseases. J Neural Transm. 2012;119:891–910.
Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China (Nos. 81422051, 81472593, and 31401208) and the Hunan Natural Science Foundation of China (No. 2016JJ1020).
Author contributions
YC, DSC, and XJX designed, conceived the study, and revised the manuscript. SLJ, YDG, XSC, and PG performed the experiments and analyzed data. SLJ and YDG drafted the manuscript. XLW, YZL, SSX, YZ, and JMY provided technical support in the experiments.
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Jiang, Sl., Guan, Yd., Chen, Xs. et al. Tubeimoside-1, a triterpenoid saponin, induces cytoprotective autophagy in human breast cancer cells in vitro via Akt-mediated pathway. Acta Pharmacol Sin 40, 919–928 (2019). https://doi.org/10.1038/s41401-018-0165-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41401-018-0165-9
Keywords
This article is cited by
-
Combined treatment of mitoxantrone sensitizes breast cancer cells to rapalogs through blocking eEF-2K-mediated activation of Akt and autophagy
Cell Death & Disease (2020)
-
Structural diversity, natural sources, and pharmacological potential of plant-based saponins with special focus on anticancer activity: a review
Medicinal Chemistry Research (2020)
-
Tubeimoside-I sensitizes colorectal cancer cells to chemotherapy by inducing ROS-mediated impaired autophagolysosomes accumulation
Journal of Experimental & Clinical Cancer Research (2019)