Abstract
Despite more effective chemotherapy combined with limb-salvage surgery for the osteosarcoma treatment, survival rates for osteosarcoma patients have stagnated over the past three decades due to the poor prognosis. Osteosarcoma cancer stem cells (OSCs) are responsible for the growth and metastasis of osteosarcoma. The existence of OSCs offers a theoretical explanation for therapeutic failures including tumor recurrence, metastasis, and drug resistance. Understanding the pathways that regulate properties of OSCs may shed light on mechanisms that lead to osteosarcoma and suggest better modes of treatment. In this study, we showed that the expression level of Kruppel-like factor 4 (KLF4) is highly associated with human osteosarcoma cancer stemness. KLF4-overexpressed osteosarcoma cells displayed characteristics of OSCs: increased sphere-forming potential, enhanced levels of stemness-associated genes, great chemoresistance to adriamycin and CDDP, as well as more metastasis potential. Inversely, KLF4 knockdown could reduce colony formation in vitro and inhibit tumorigenesis in vivo, supporting an oncogenic role for KLF4 in osteosarcoma pathogenesis. Furthermore, KLF4 was shown to activate the p38 MAPK signaling pathway to promote cancer stemness. Altogether, our studies uncover an essential role for KLF4 in regulation of OSCs and identify KLF4–p38 MAPK axis as a potential therapeutic target for osteosarcoma treatment.
Similar content being viewed by others
Introduction
Osteosarcoma is a malignant mesenchymal tumor in which the cancerous cells produce osteoid, the organic extracellular matrix of bone [1]. Osteosarcoma is the most common primary tumor of the bone and is an especially aggressive and predominantly pediatric cancer. Despite advances in diagnosis and treatment, the prognosis of osteosarcoma is still poor, and the long-term cure rate for osteosarcoma patients has reached a plateau over the last 30 years. Recent advances in stem cell biology and associated technologies have enabled the identification of minor components of tumorigenic cells, known as cancer stem cells (CSCs), in cancers such as osteosarcoma. Osteosarcoma cancer stem cells (OSCs) share the characteristics of self-renewal, multipotency, tumorigenicity, multiple drug resistance, and metastasis. The presence of OSCs is responsible for therapeutic failure, including recurrence, metastasis, and resistance to chemotherapy [2, 3]. It has been confirmed that OSCs express high levels of CD133, ABCG2, CXCR4, CD117, and ALDH1A1, which have been considered to be specific markers [4,5,6]. Additionally, several studies have suggested that the TGF-β, Notch, Hedgehog, and p42/44 MAPK signaling pathways could influence the OSC phenotype. Increasing evidence has emerged to substantiate that other factors, including BMP-2, TNF-α, SOX2, OCT3/4, might be involved in regulating the maintenance of OSCs [3]. Despite numerous efforts to characterize OSCs, novel therapeutic approaches for the eradication of CSCs are yet to be established due to our limited understanding of OSC biology. Thus additional in-depth study of the OSC characteristics, CSC regulatory mechanism, and related signal pathways could aid in identifying a novel molecular target for therapeutic intervention in osteosarcoma.
Kruppel-like factor 4 (KLF4), a member of the Spl/Kruppel-like zinc-finger-containing transcription factors, plays a critical role in regulating a diverse array of cellular processes, including proliferation, differentiation, apoptosis, cellular migration, embryonic development, and maintenance of tissue homeostasis [7]. Similar to TGF-β, KLF4 can also act as either a tumor suppressor or an oncogene depending on differing cellular contexts and cancer types [8]. Decreased KLF4 expression in colorectal cancer, gastric cancer, and lung cancer could contribute to cellular hyperproliferation and malignant transformation, which support a putative role for KLF4 as a tumor suppressor with its function in cell-cycle arrest and pro-apoptosis. In addition, KLF4 is upregulated in primary breast ductal carcinoma, oral squamous carcinoma, and head and neck cancer and subsequently promotes the survival and progression of tumors [9]. In addition, KLF4 has been reported as one of the four factors used to reprogram adult fibroblasts into induced pluripotent stem cells, implying its possible function in regulating cancer stemness. Recent data indicate that KLF4 is required for maintenance of cancer stem cells in the breast and prostate [10, 11]. However, KLF4 also exhibits the opposite role in suppressing stemness in neuroblastoma and pancreatic ductal adenocarcinoma [12, 13]. Although the mechanism(s) underlying these discrepancies remains undefined, evidence suggests an important role for KLF4 in cancer and highlights the need for further characterization of the specific role of KLF4 in CSCs [7].
A previous study showed that high expression of KLF4 is correlated with poor overall survival and worse metastasis-free survival of osteosarcoma patients [14]. Recently, we reported that KLF4 played an important role in regulating Adriamycin (ADR)-induced cancer stemness and metastasis [15]. However, the functional roles of KLF4 in osteosarcoma genesis and development, particularly in OSC biology, are still open questions. Thus, this study investigated the new role of KLF4 in modulating the osteosarcoma stemness. Data presented in this work show a potential protumorigenic function of KLF4 in regulating the OSCs phenotype associated with metastasis and drug resistance and identify the KLF4–p38 MAPK axis as a potential therapeutic target for osteosarcoma treatment.
Materials and methods
Drugs and reagents
ADR, cisplatin (CDDP), and SB203580 were sourced from Selleck Chemicals (USA). Recombinant human EGF and FGF-basic were purchased from Peprotech (USA). Dimethyl sulfoxide, sulforhodamine B (SRB), trichloroacetic acid (TCA), Tris base, and propidium iodide (PI) were obtained from Sigma-Aldrich (MO, USA).
Cell culture
Primary osteosarcoma cells (MDOS-15, MDOS-20, and MDOS-21) were established from tumor lesions collected from surgical biopsies and grown in Dulbecco's modified Eagle's medium (DMEM)/F12 (Invitrogen, USA), as previously described [16]. KHOS/NP cells were kindly provided by Dr. Ling-tao Wu (University of Southern California, Los Angeles, USA) and were cultured in DMEM (HyClone, USA). U2OS and MG63 cells were purchased from the Cell Bank of the China Science Academy (Shanghai, China) and were cultured in RPMI 1640 (HyClone, USA). In addition, 293FT cells were grown in conditioned medium composed of DMEM, 1 mmol/L l-glutamine (Gibco Laboratories, USA), 1% penicillin–streptomycin (Invitrogen, USA), and 1% MEM-nonessential amino acids (Gibco Laboratories, USA). All media were supplemented with 10% fetal bovine serum (FBS) (Gibco Laboratories, USA). All cells were maintained at 37 °C in a humidified 5% CO2 atmosphere.
Western blotting
Western blot analysis was conducted as described previously [17]. Forty micrograms of the cell lysate were resolved by 8% sodium dodecyl sulfate polyacrylamide gel electrophoresis (and transferred to a poly-vinylidene fluoride membrane (Millipore Corporation, Billerica, MA). Primary antibodies against KLF4, GAPDH, and p38 MAPK were purchased from Santa Cruz; the CXCR4 antibody was obtained from Abcam, and the p-p38 MAPK (Thr180/Tyr182) antibody was obtained from Cell Signaling. Optical densities were analyzed by Quantity One.
Lentivirus transduction
The pCCL-c-MNDU3c-X2-PGK enhanced green fluorescent protein (GFP) plasmid was kindly provided by Dr. D.B. Kohn, Children’s Hospital Los Angeles, University of Southern California. The pCCL-KLF4 plasmid was constructed according to the pMXs-hKLF4 Retroviral Vector (Cell Biolabs, USA) directions. All lentiviral vectors including expression constructs (pCCL-KLF4 or empty pCCL vectors), pRD8.9 packaging plasmids, and pMD.G envelope plasmids were transfected into 293FT cells for packaging. Viral supernatants were collected after 48 h and used in transfection, as previously described [17].
cDNA microarray
Microarray analysis was performed as described previously [4]. The RNA samples were hybridized using a GeneChip PrimeView Human Gene Expression Array from Gene Tech. Microarray data were deposited in the NCBI GEO under accession number GSE96892.
Quantitative real-time PCR
Total RNA was prepared and detected as previously reported [17]. The primers used in quantitative PCR are as follows: CD133, forward: GTCATCCATGACAACTTTGG; reverse: TTGATCCGGGTTCTTACCTG; SOX2, forward: TACAGCATGTCCTACTCGCAG; reverse: GAGGAAGAGGTAACCACAGGG; OCT3/4, forward: CTGGAGAAGGAGAAGCTGGA; reverse: CAAATTGCTCGAGTTCTTTCTG; NANOG, forward: AAAGAATCTTCA CCTATGCC; reverse: GAAGGAAGAGGAGAGACAGT; KLF4, forward: ACCAGGCACT ACCGTAAACACA; reverse: GGTCCGACCTGGAAAATGCT; ALDH1A1, forward: CACC AGGGCCAGTGTTGTAT; reverse: AACACTGTGGGCTGGACAAA; ABCG2, forward: CACCTTATTGGCCTCAGGAA; reverse: CCTGCTTGGAAGGCTCTATG; CD24, forward: CAATATTAAATCTGCTGGAGTTTCATG; reverse: TCCATATTTCTCAAGCCACATTCA; WNT3, forward: ACTTCGGCGTGTTAGTGTCC; reverse: CATTTGAGGTGCATGTGGTC; ZEB2, forward: GCGGCATATGGTGACACACAA; reverse: CATTTGAACTTGCGATTAC CTGC; DNER, forward: CTCCATTTCTGCATGGGTCT; reverse: GAGGAAACCTTGCCAAAACA; JAGGED1, forward: GTCCATGCAGAACGTGAACG; reverse: GCGGGACTG ATACTCCTTGA; ID1, forward: CTCTACGACATGAACGGCTGTT; reverse: TGCTCAC CTTGCGGTTCTG; ID2, forward: CTGTCCTTGCAGGCTTCTGAATTC; reverse: CATGA ACACCGCTTATTCAGCCAC; BMP6, forward: AGCATAACATGGGGCTTCAG; reverse: CACGTGCACCTCACTCACTT; ABCB1, forward: CAGGGAAAGTGCTGCTTGATG; reverse: TCGATGAAGGCATGTATGTTGG; ABCC1, forward: ATGTCACGTGGA ATACCAGC; reverse: GAAGACTGAACTCCCTTCCT; CXCR-4, forward: GGTGGTCTAT GTTGGCGTCT; reverse: TGGAGTGTGACAGCTTGGAG; DDIT3, forward: CCAGAACC AGCAGAGGTCAC; reverse: CTTCATGCGCTGCTTTCCAG; STAT1, forward: GGAGGC GAACCTGACTTCCA; reverse: TCTGGTGCTTCCTTTGGCCT; RAC1, forward: AGACG GAGCTGTAGGTAAAA; reverse: GCAGGACTCACAAGGGA; GAPDH, forward: GTCAT CCATGACAACTTTGG; reverse: GAGCTTGACAAAGTGGTCGT. The results were normalized to those of the housekeeping gene glyceraldehyde-3-phosphate dehydrogenase (GAPDH).
Small interfering RNA (siRNA) transfection
The siRNA duplexes were synthesized by Genepharma, Co. (Shanghai, China). The sequences of the siRNAs used are as follows: KLF4, 5-UGAGAUGGGAACUCUUUGUGUAGGU-3; p38 MAPK, 5-GAACUGCGGUUACUUAAAC-3. The transfection was performed using the oligofectamine reagent (Invitrogen) according to the manufacturer’s instructions.
Cytotoxicity assay
The sensitivities of the osteosarcoma cells to chemotherapeutic drugs were measured via SRB assay. In brief, cells were plated in 96-well plates at a density of 4000 cells per well. After exposure to various concentrations of ADR and CDDP for 72 h, the plates were subjected to the SRB colorimetric assay [18].
Cell proliferation assay
Cell proliferation was determined by cell count using a standard hemocytometer, as previously described [19].
Flow cytometry
Osteosarcoma cells were harvested, washed with phosphate-buffered saline (PBS), and fixed with precooled 70% ethanol at 4 °C. The cells were subsequently suspended in PBS containing RNase A (40 μg/mL) and propidium iodide (10 μg/mL) in the dark for 30 min. The percentage of apoptotic cells (sub-G0) and cell cycle distribution were detected by flow cytometry using CellQuest software (Becton Dickinson).
Metastasis assay
A wound-healing assay was conducted as follows. A scratch was made on a uniform layer of cells using a sterile micropipette tip, and cells were washed to remove debris. Photographs of the same area of the wound were taken after 0 and 24 h. For the transwell migration assay, cells were resuspended in serum-free medium and seeded into the insert well of a 24-well plate (8-μm pores; BD Biosciences) for 24 h. Culture medium containing 10% FBS was used as a chemoattractant and placed in the bottom chamber.
Soft agar assay
Each well of a six-well plate was coated with 1 mL of 10% FBS medium with 1% low-melting-point agarose. After 20 min of incubation at 37 °C, cell suspensions of an equal number (5000) of cancer cells were added in 1 mL of medium with 0.35% agarose. Cells were incubated for 14 days under standard conditions (37 °C, 5% CO2) with the addition of 500 μL of KHOS/NP culture medium to hydrate the exposed agarose. At the end of the incubation period, the wells were examined under a light microscope at ×50 magnification, and the number of colonies larger than 50 μm per well was counted.
Mammosphere formation assay
As previously described, single cells were plated in ultra-low-attachment six-well plates (Corning) at different densities per well and grown in DMEM/F12 (serum free) with 20 ng/mL EGF, 20 ng/mL bFGF, and N2 supplement. The mammosphere (diameter >50 μm) number was counted at 7 days, and pictures were taken [15].
Xenotransplantation
All animal experiments were conducted in accordance with the Institutional Animal Use and Care Committee. For the tumorigenicity assays, 5 × 105 osteosarcoma cells (MDOS-15, MDOS-20, MDOS-21, and KHOS/NP) were injected subcutaneously into 5-week-old BALB/c (nu/nu) mice (National Rodent Laboratory Animal Resource, Shanghai, China). Tumor growth was monitored bi-weekly, and tumor volume (V) was measured and calculated using the following formula: V = (length × width2)/2.
Statistical analysis
Data are presented as the mean ± SD unless otherwise specified. Student’s t-test was performed using Excel, and data were considered statistically significant when P < 0.05.
Results
The expression levels of KLF4 protein are highly correlated with osteosarcoma cancer stemness
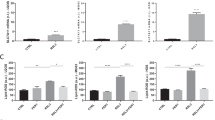
The KLF4 expression pattern and correlation with histopathologic status and the clinical outcome are highly variable among tumors, suggesting distinct roles of KLF4 in individual tumors [7]. To elucidate the function of KLF4 in osteosarcoma, the abundance of KLF4 protein was compared with its tumorigenicity capacity. We observed that the expression levels of KLF4 in primary osteosarcoma MDOS-21 cells was higher than that of other primary osteosarcoma cells, including MDOS-15 and MDOS-20 cells (Fig. 1a), which are positively correlated with its tumorigenicity (Fig. 1b). In addition, we also compared the protein expression of KLF4 in three osteosarcoma cell lines. Western blotting results revealed a higher level of expression of KLF4 in KHOS/NP cells and lower levels in U2OS and MG63 cells (Fig. 1g). Additionally, KHOS/NP cells possess a greater ability to form tumors in vivo than U2OS and MG63 cells, as we previously reported [4]. Therefore, our results implied that the KLF4 expression was positively correlated with tumorigenicity in human osteosarcoma cells.
The expression levels of KLF4 protein are highly correlated with osteosarcoma cancer stemness. a Western blotting analysis of KLF4 expression in primary osteosarcoma cells. b, c The tumor-forming ability of primary osteosarcoma cells was measured in NOD/SCID mice. d Osteospheres showed stem-like properties with increased expression of stem-cell-related genes, including CD133, SOX2, OCT3/4, and NANOG, according to qRT-PCR. e, f KLF4 expression was determined in adherent cells and osteospheres of KHOS/NP and U2OS cells via qRT-PCR and western blotting. GAPDH was used as an internal control. g Western blot analysis of KLF4 expression in osteosarcoma cell lines. Data represent the mean ± SD, n = 3. *P < 0.05; **P < 0.01
Because tumorigenicity is a distinctive characteristic of cancer stem cells, we postulated that this phenomenon could be ascribed to KLF4’s influences on osteosarcoma cancer stemness [20]. OSCs are capable of forming spheres in a serum-free anchorage-independent environment [2, 21]. We thus examined the sphere-forming ability among these primary osteosarcoma cells. Similar phenomena were observed in that MDOS-21 cells exhibited the greatest potential to form osteospheres (Fig. 1c). To further address this possibility, sphere-forming culture was undertaken to enrich OSCs. A panel of stem-cell-related genes, including CD133, SOX2, OCT3/4, and NANOG, were upregulated in sphere-cultured cells compared with monolayer-cultured cells (Fig. 1d). We further determined the expression levels of KLF4 in suspension cultures using quantitative reverse transcriptase polymerase chain reaction (qRT-PCR) and western blotting. The results showed that osteosphere cells had higher expression levels of KLF4 than adherent cells at both the mRNA and protein levels (Fig. 1e, f). Collectively, these results indicate that a high level of KLF4 expression is associated with CSC-enriched populations in osteosarcoma cells.
KLF4 overexpression enhances the proportion of cancer stemness in osteosarcoma
To understand the functional role of KLF4 in CSCs, we first overexpressed KLF4 in osteosarcoma cells using lentivirus. We found that osteosarcoma cells expressing green fluorescent protein were successfully transduced with lentivirus. Moreover, KLF4 overexpression led to a strong increase in osteosphere formation and dimension (Fig. 2a, f). Additionally, overexpression of KLF4 could significantly increase the transcriptional levels of stemness-related genes, including CD133, ALDH1A1, and ABCG2, in KHOS/NP, U2OS, and MDOS-20 cell lines (Fig. 2b). These data suggested that activation of KLF4 expression is sufficient to facilitate the enrichment of stem cell-like osteosarcoma cells.
KLF4 overexpression promotes osteosarcoma cancer stemness. a Left, photomicrographs depict the osteosphere. b Overexpression of KLF4 upregulated the transcription level of osteosarcoma stem cell-related markers. Transduction of osteosarcoma cells including KHOS/NP, U2OS, and MDOS-20 cells with KLF4 or pCCL (lentivirus vector), RNAs from KLF4 cells and pCCL cells were extracted after 72 h transduction. qRT-PCR detected the mRNA level of osteosarcoma stem cell markers, including CD133, ALDH1A1, and ABCG2. c Serial transplantation upregulated signaling molecules of the Wnt, Notch, TGF-β, and Hedgehog signaling pathways. The color bar represents the expression ratio on a log-2 scale (KHOS/NP-KLF4 vs. KHOS/NP-pCCL). d Validation of microarray data by qRT-PCR. Selected genes from different functional groups in the array analysis (Fig. 2c) were examined by qRT-PCR. e Microarray analysis of the transactivation of stem cell marker genes in KHOS/NP-KLF4 cells compared with that in KHOS/NP-pCCL cells. The fold change represents the expression ratio of KHOS/NP-KLF4 vs. KHOS/NP-pCCL. f Transduction of osteosarcoma cells, including KHOS/NP, U2OS, and MDOS-20 cells, with KLF4 or pCCL (lentivirus vector). Cell lysates were prepared for western blotting analysis of KLF4 expression after 72 h of transduction. For (a) and b, the data represent the mean ± SD, n = 3. Scale bar, 50 μm. *P < 0.05; **P < 0.01; ***P < 0.001
We further compared the gene expression profile between control and KLF4-overexpressed KHOS/NP cells. Notably, our data revealed that KLF4 overexpression could upregulate stem-cell-related marker genes and activate several signaling pathways that maintain self-renewal capacity, including Wnt, Notch, TGF-β, and Hedgehog pathways (Fig. 2c). qRT-PCR was used to further confirm the upregulation of several critical genes (Fig. 2d). In addition, it is of particular note that overexpression of KLF4 could also enhance the transcriptional level of stem cell markers, including CD24, CD47, CD52, CD55, CD58, CD59, CD73, CD81, CD109, CD164, and CD276 (Fig. 2e).
KLF4 confers resistance to chemotherapy in osteosarcoma cells
One particularly intriguing property of CSCs is that they are highly resistant to drugs and toxins via the expression of several ATP-binding cassette (ABC) transporters [22]. To further investigate the effect of KLF4 on OSCs, we determined whether KLF4 regulates the sensitivity of osteosarcoma cells to first-line chemotherapeutic drugs, specifically ADR and CDDP. Cell proliferation assay results showed that overexpression of KLF4 could lead to resistance of osteosarcoma cells to drug treatment (Fig. 3a). To further validate this chemoprotective effect of KLF4 on tumor cells, chemotherapy-induced apoptosis was further assessed. As illustrated in Fig. 3b, overexpression of KLF4 could also resist the apoptosis induced by ADR or CDDP in osteosarcoma cells. In an attempt to elucidate the molecular basis for KLF4-induced drug resistance, we detected the changes of the classical ABC drug transporters (ABCB1 and ABCC1). Surprisingly, we found that overexpression of KLF4 does not affect the mRNA levels of any of these genes, implying that neither ABCB1 nor ABCC1 is a relevant mediator of KLF4-induced stemness activity in our model (Fig. 3c). Our study reveals that osteosarcoma cells with KLF4 overexpression are indeed more resistant to chemotherapy than blank cells.
KLF4 inhibits the sensitivity of osteosarcoma cells to chemotherapy drugs. a After transduction of KLF4 or pCCL (lentivirus vector) for 72 h, osteosarcoma cells, including KHOS/NP, U2OS, and MDOS-20 cells, were cultured with various concentrations of the chemotherapy drugs ADR and CDDP for 72 h. Cell proliferation was measured by SRB assay. b After transduction of KLF4 or pCCL (lentivirus vector) for 72 h, osteosarcoma cells, including KHOS/NP, U2OS, and MDOS-20 cells, were cultured with the indicated concentrations of chemotherapy drugs for 48 h. PI staining, followed by flow cytometry to detect apoptosis. c Overexpression of KLF4 had no effect on the transcriptional levels of transporter genes in osteosarcoma cells. After transduction of KLF4 or pCCL (lentivirus vector) for 72 h, osteosarcoma cells, including KHOS/NP, U2OS, and MDOS-20 cells, and the mRNA levels of ABCB1 and ABCC1 genes were examined by qRT-PCR. Data represent mean ± SD, n = 3. *P < 0.05; **P < 0.01
KLF4 promotes migration of osteosarcoma cells
CSCs exhibit selected features of distal metastasis [23]. To gain further insight into the potential role of KLF4 in metastasis, we investigated the impact of KLF4 on metastasis-associated marker CXCR-4 [24]. As displayed in Fig. 4a, b, overexpression of KLF4 could obviously increase the expression of CXCR-4 at both the mRNA and protein levels in osteosarcoma cells. Consistent with these findings, functional analyses further demonstrated that KLF4 overexpression could effectively enhance the cell scratch repair ability as well as the migration capability of osteosarcoma cells (Fig. 4d, e). At the same time, no significant influence on cell viability was observed (Fig. 4c). Overall, these results clarify that KLF4 promotes the migration capacity of osteosarcoma cells, which is an important property of CSCs.
Overexpression of KLF4 enhances migration of osteosarcoma cells. a The mRNA levels of CXCR-4 and GAPDH were detected by qRT-PCR in KHOS/NP-pCCL and KHOS/NP-KLF4 cells. b The protein expression levels of CXCR-4 and GAPDH were detected by western blotting in KHOS/NP-pCCL and KHOS/NP-KLF4 cells. c The SRB assay was performed to assess viability. d Two osteosarcoma cell lines (KHOS/NP, U2OS) and primary osteosarcoma MDOS-20 cells infected with either KLF4 or control pCCL were cultured in media. A scratch wound was created across the subconfluent monolayer of cells. Brightfield images of the exact field as referenced by a mark made on the plate (asterisk) were taken at 0 and 24 h to observe the migration of the cells across the wound. e The transwell migration assay was used to assess migration of osteosarcoma cells. Migration through the transwell inserts was assessed at 24 h after inoculation. Representative images of migrated cells are shown on the left, and the results are summarized on the right. Data are shown as the mean ± SD, n = 3. **P < 0.01, ***P < 0.001
KLF4 knockdown impairs cancer stemness and metastasis of osteosarcoma cells
To test the importance of KLF4 for the maintenance of CSCs, we generated KLF4 knockdown cells by transduction with siRNA. As expected, in contrast with overexpression (Fig. 3), the ablation of KLF4 expression significantly increases cancer cell sensitivity to chemotherapeutic drugs (Fig. 5a). Of note, the transcriptional levels of ABCB1 and ABCC1 were not significantly altered upon KLF4 silencing, whereas the expression of ABCG2 was remarkably downregulated by KLF4 depletion (Fig. 5b). Additionally, KLF4 silencing could inhibit the scratch repair ability and migration potential of KHOS/NP cells and had no influence on cell viability (Fig. 5c, d). As shown in Fig. 5e, KLF4 depletion also led to a significant reduction of CXCR-4 at both the mRNA and protein levels. One reliable measurement of the tumorigenic nature of cells in vitro is the ability to form colonies in soft agar. As illustrated in Fig. 5f, KLF4 knockdown dramatically reduced both the number and size of colonies compared with mock KHOS/NP cells.
Knockdown of KLF4 suppresses cancer stemness of osteosarcoma cells. a KHOS/NP-Mock and KHOS/NP-siKLF4 cells were treated with various concentrations of ADR or CDDP for 72 h, and the cytotoxicities of the treatments were subsequently determined by SRB assay. b The mRNA levels of ABCG2, ABCB1, and ABCC1 were detected by qRT-PCR in KHOS/NP-Mock and KHOS/NP-siKLF4 cells. c KHOS/NP cells infected with either Mock (control siRNA) or siKLF4 were cultured in media. A scratch wound was created across the subconfluent monolayer of cells. Brightfield images of the exact field as referenced by a mark made on the plate (asterisk) were taken at 0 and 24 h to observe the migration of cells across the wound. The SRB assay was performed to assess viability. d The transwell migration assay was used to assess migration of KHOS/NP-Mock and KHOS/NP-siKLF4 cells. Migration through the transwell inserts was assessed at 24 h after inoculation. Representative images of migrated cells are shown on the left, and the results are summarized on the right. e mRNA levels of CXCR-4 and GAPDH were detected by qRT-PCR in KHOS/NP-Mock and KHOS/NP-siKLF4 cells. Protein expression levels of CXCR-4 and GAPDH were detected by western blotting in KHOS/NP-Mock and KHOS/NP-siKLF4 cells. f Quantification of colony formation by cells expressing KLF4 siRNA. Western blotting showing KLF4 levels in KHOS/NP-Mock and KHOS/NP-siKLF4 cells. Scale bar, 50 μm. g Tumor growth curves were plotted for nude mice injected with KHOS/NP-Mock and KHOS/NP-siKLF4 cells. The data are presented as the mean ± SEM of tumors in five mice per cell line. h Upper, representative tumors are shown. Lower, weights of tumors formed by KHOS/NP cells with the KLF4 silencing vs. mock-infected KHOS/NP cells. Data represent mean ± SD, n = 3. *P < 0.05; ***P < 0.001
To investigate whether the stem cell phenotype regulated by KLF4 could extend to in vivo conditions, we further tested the tumorigenicity of mock or si-KLF4 transfected KHOS/NP cells. As shown in Fig. 5g, h, the average tumor volume and weight in the KLF4 silencing group were much smaller than those in the scrambled group, supporting the importance of KLF4 in maintaining CSCs. Taken together, these results indicate that KLF4 is required to maintain the cancer stemness of osteosarcoma cells.
The p38 MAPK signaling pathway is required for KLF4-mediated osteosarcoma stemness
To understand the mechanism of KLF4-induced osteosarcoma cancer stemness, microarray analysis was subsequently performed to identify possible downstream genes. As shown in Fig. 6a, microarray analysis demonstrated that overexpression of KLF4 upregulated serial genes correlated with the p38 MAPK signaling pathway, including DDIT3, RPS6KAS, STAT1, MAX, MEF2A, and RAC1. The upregulation of DDIT3, STAT1, and RAC1 was further confirmed by qRT-PCR (Fig. 6b). Because p38 MAPK is activated through phosphorylation, we next monitored the phosphorylation status of p38 MAPK. Consistent with the microarray data, overexpression of KLF4 could elevate intracellular p38 MAPK phosphorylation (Fig. 6c), while silencing of KLF4 reduced the expression level of p-p38 MAPK (Fig. 6d). These results indicate that activation of p38 MAPK is involved in KLF4-regulated cancer stemness.
p38 MAPK signaling is involved in KLF4-induced osteosarcoma stemness. a Relative expression of p38 MAPK signaling-related genes as determined by microarray analysis (KLF4 vs. pCCL). b qRT-PCR confirmation of genes that are a component of the p38 MAPK pathway from the microarray analysis described in a. c After transduction with pCCL and KLF4, the protein expression levels of p-p38 MAPK and p38 MAPK were detected by western blotting in osteosarcoma cells including KHOS/NP, U2OS, and MDOS-20 cells. d The protein expression levels of p-p38 MAPK and p38 MAPK were detected by western blotting in KHOS/NP-Mock and KHOS/NP-siKLF4 cells. e KHOS/NP-pCCL and KHOS/NP-KLF4 cells were transfected with siRNAs specifically targeting p38 MAPK or control siRNAs for 48 h. Proteins were detected via western blotting analysis using specific antibodies against p-p38 MAPK and GAPDH. f p38 MAPK knockdown with siRNA in KHOS/NP-pCCL and KHOS/NP-KLF4 cells showed a reduction in sphere-forming ability. Left, representative images of the spheres. Right, quantification of the assay. Scale bar, 100 μm. g KHOS/NP-pCCL and KHOS/NP-KLF4 cells were incubated with 20 μmol/L SB203580. Proteins were detected via western blotting analysis using specific antibodies against p-p38 MAPK and GAPDH. h Sphere-forming assay of KHOS/NP-pCCL and KHOS/NP-KLF4 cells treated with or without DMSO and SB203580 with indicated concentrations. Representative images of spheres (left) and quantification of the assays (right). Scale bar, 100 μm. i The transwell migration assay was used to assess migration of KHOS/NP-pCCL and KHOS/NP-KLF4 cells treated with DMSO or SB203580 (20 μmol/L). Migration through the transwell inserts was assessed at 24 h after inoculation. Data represent the mean ± SD, n = 3. *P < 0.05; **P < 0.01; #P < 0.05
Because the p38 MAPK signaling pathway plays an important role in regulating diverse cellular functions, we speculated that p38 MAPK might be a critical downstream target of KLF4 that mediates the OSC-related function. Therefore, we examined whether p38 MAPK is involved in KLF4-mediated cancer stemness in osteosarcoma cells. We ablated the expression of p38 MAPK using specific siRNA in KHOS/NP cells (Fig. 6e). Our study showed that ablation of p38 MAPK significantly decreased the numbers of spheres induced by KLF4 overexpression (Fig. 6f). Similarly, SB203580 (a specific p38 MAPK inhibitor) phenocopied the inhibitory effects on the sphere formation ability of p38 MAPK siRNA (Fig. 6g, h). Moreover, pretreatment of KHOS/NP cells with SB203580 also greatly abolished KLF4-induced cell migration (Fig. 6i). Taken together, all of these findings indicate that the p38 MAPK signaling pathway participates in KLF4-induced osteosarcoma cancer stemness.
Discussion
Advances in the outcome of osteosarcoma patients over the recent decades have plateaued despite great improvements in therapeutic strategies. According to the CSC hypothesis, most current therapies are directed to the majority of the tumor mass, which consists of fast-growing differentiated cells, but not to the relatively slow-growing migrating and resistant OSCs. The existence of OSCs with progenitor properties is responsible for therapeutic failures such as tumor relapses and metastasis. Much attention has been focused on characterization of OSCs due to the potential for discovery and development of OSC-related therapies and the identification of key molecules involved in controlling the unique properties of OSC populations [3]. However, this research is in its infancy, and the underlying molecular mechanisms and genetic drivers controlling the osteosarcoma stemness phenotypes have remained largely undefined. In this study, we reveal a previously unrecognized role for KLF4 as an oncogene in the regulation of osteosarcoma cancer stemness associated with drug resistance and metastasis. Furthermore, we also demonstrated that the p38 MAPK signaling pathway is involved in KLF4-mediated osteosarcoma stemness.
It is well recognized that KLF4 is one of four key factors required for inducing pluripotent stem cells and is intimately implicated in the maintenance of the self-renewal capacity of embryonic stem cells. Recent studies have focused on implications in CSCs, whereas the functional role of KLF4 in modulating cancer stemness remains elusive. It has been shown that paracrine PGI signaling induces the self-renewal and tumorigenic potentials of GSCs through induction of KLF4 in human gliomas [25]. In addition, KLF4 is reported to maintain the stemness of breast cancer cells, and KLF4 ablation attenuated tumor formation capacity and cell meatstasis [10]. Our previous study suggests that ADR treatment induced a stem-like phenotype and promotes metastasis via KLF4 upregulation. KLF4 deletion not only rescued ADR-promoted stem cell phenotype but also impaired the enrichment of CD133+ cells and expression of stem-cell-related markers (CD133, ABCG2, and ALDH1A1) in osteosarcoma cells without ADR treatment [15]. In this work, our studies strengthen the emerging notion that KLF4 plays an oncogenic role in modulating osteosarcoma cancer stemness and further demonstrate that the p38 MAPK signaling pathway is involved.
The mechanism(s) by which KLF4 regulates stemness in osteosarcoma cells is probably complex, but likely results mainly from its function as a direct transcriptional regulator. Our preliminary analysis of the gene expression profiles of parental and KLF4-expressing KHOS/NP cells shows that a number of genes correlated with Wnt, Notch, and Hedgehog are upregulated in KLF4-expressing cells. Moreover, we also found that genes associated with the p38 MAPK signaling pathway were upregulated in KLF4-expressing cells. Using cell models with forced KLF4 overexpression or knockdown, we show that KLF4 affects the activity of the p38 MAPK pathway. A report suggested that activation of the p38 MAPK pathway enhances the survival of colorectal cancer stem cells under hypoxia and serum depletion conditions [26]. Additionally, a recent study shows that Interleukin-17 promotes self-renewal of CD133+ cancer stem-like cells in ovarian cancer through activation of the p38 MAPK signaling pathway [27]. In our current study, we found that suppression of the p38 MAPK pathway reduced the discrepancy in sphere-forming capacity and cell migration between KLF4-overexpressed KHOS/NP cells and control cells, which further confirmed that the p38 MAPK signaling pathway is involved in KLF4-promoted cancer stemness.
Several limitations exist in our work. First, we did not determine the exact mechanism through which KLF4 regulates p38 MAPK signaling, and this topic deserves further investigation. Second, our in vivo results were obtained from osteosarcoma cell lines grown as xenografts in nude mice, and the real situation in patients with osteosarcoma might be different. Therefore, we look forward to the results of clinical trials designed to test combinations of chemotherapy drugs with p38 inhibitors in patients with osteosarcoma.
In summary, our findings suggest that KLF4 plays an important role in promoting osteosarcoma cancer stemness, chemotherapy resistance, and metastasis. The p38 MAPK signaling pathway is involved in KLF4-mediated OSC traits. Hence, our study advances the understanding of cancer stem cell properties and suggests new approaches to targeted therapy in patients with osteosarcoma.
References
Lamoureux F, Trichet V, Chipoy C, Blanchard F, Gouin F, Redini F. Recent advances in the management of osteosarcoma and forthcoming therapeutic strategies. Expert Rev Anticancer Ther. 2007;7:169–81.
Gibbs CP, Kukekov VG, Reith JD, Tchigrinova O, Suslov ON, Scott EW, et al. Stem-like cells in bone sarcomas: implications for tumorigenesis. Neoplasia. 2005;7:967–76.
Yan GN, Lv YF, Guo QN. Advances in osteosarcoma stem cell research and opportunities for novel therapeutic targets. Cancer Lett. 2016;370:268–74.
Ying M, Liu G, Shimada H, Ding W, May WA, He Q, et al. Human osteosarcoma CD49f(−)CD133(+) cells: impaired in osteogenic fate while gain of tumorigenicity. Oncogene. 2013;32:4252–63.
Honoki K, Fujii H, Kubo A, Kido A, Mori T, Tanaka Y, et al. Possible involvement of stem-like populations with elevated ALDH1 in sarcomas for chemotherapeutic drug resistance. Oncol Rep. 2010;24:501–5.
Adhikari AS, Agarwal N, Wood BM, Porretta C, Ruiz B, Pochampally RR, et al. CD117 and Stro-1 identify osteosarcoma tumor-initiating cells associated with metastasis and drug resistance. Cancer Res. 2010;70:4602–12.
Ghaleb AM, Yang VW. Kruppel-like factor 4 (KLF4): what we currently know. Gene. 2017;611:27–37.
Rowland BD, Bernards R, Peeper DS. The KLF4 tumour suppressor is a transcriptional repressor of p53 that acts as a context-dependent oncogene. Nat Cell Biol. 2005;7:1074–82.
Wei D, Kanai M, Huang S, Xie K. Emerging role of KLF4 in human gastrointestinal cancer. Carcinogenesis. 2006;27:23–31.
Yu F, Li J, Chen H, Fu J, Ray S, Huang S, et al. Kruppel-like factor 4 (KLF4) is required for maintenance of breast cancer stem cells and for cell migration and invasion. Oncogene. 2011;30:2161–72.
Chang YL, Zhou PJ, Wei L, Li W, Ji Z, Fang YX, et al. MicroRNA-7 inhibits the stemness of prostate cancer stem-like cells and tumorigenesis by repressing KLF4/PI3K/Akt/p21 pathway. Oncotarget. 2015;6:24017–31.
Shum CK, Lau ST, Tsoi LL, Chan LK, Yam JW, Ohira M, et al. Kruppel-like factor 4 (KLF4) suppresses neuroblastoma cell growth and determines non-tumorigenic lineage differentiation. Oncogene. 2013;32:4086–99.
Yan Y, Li Z, Kong X, Jia Z, Zuo X, Gagea M, et al. KLF4-mediated suppression of CD44 signaling negatively impacts pancreatic cancer stemness and metastasis. Cancer Res. 2016;76:2419–31.
Martins-Neves SR, Paiva-Oliveira DI, Wijers-Koster PM, Abrunhosa AJ, Fontes-Ribeiro C, Bovee JV, et al. Chemotherapy induces stemness in osteosarcoma cells through activation of Wnt/beta-catenin signaling. Cancer Lett. 2016;370:286–95.
Li Y, Xian M, Yang B, Ying M, He Q. Inhibition of KLF4 by statins reverses adriamycin-induced metastasis and cancer stemness in osteosarcoma cells. Stem Cell Rep. 2017;8:1617–29.
Zhang L, Zhou Q, Zhang N, Li W, Ying M, Ding W, et al. E2F1 impairs all-trans retinoic acid-induced osteogenic differentiation of osteosarcoma via promoting ubiquitination-mediated degradation of RARalpha. Cell Cycle. 2014;13:1277–87.
Cao J, Wang Y, Dong R, Lin G, Zhang N, Wang J, et al. Hypoxia-induced WSB1 promotes the metastatic potential of osteosarcoma cells. Cancer Res. 2015;75:4839–51.
Vichai V, Kirtikara K. Sulforhodamine B colorimetric assay for cytotoxicity screening. Nat Protoc. 2006;1:1112–6.
Luo P, Wang A, Payne KJ, Peng H, Wang JG, Parrish YK, et al. Intrinsic retinoic acid receptor alpha-cyclin-dependent kinase-activating kinase signaling involves coordination of the restricted proliferation and granulocytic differentiation of human hematopoietic stem cells. Stem Cells. 2007;25:2628–37.
Clarke MF, Dick JE, Dirks PB, Eaves CJ, Jamieson CH, Jones DL, et al. Cancer stem cells–perspectives on current status and future directions: AACR workshop on cancer stem cells. Cancer Res. 2006;66:9339–44.
Fujii H, Honoki K, Tsujiuchi T, Kido A, Yoshitani K, Takakura Y. Sphere-forming stem-like cell populations with drug resistance in human sarcoma cell lines. Int J Oncol. 2009;34:1381–6.
Dean M, Fojo T, Bates S. Tumour stem cells and drug resistance. Nat Rev Cancer. 2005;5:275–84.
Scheel C, Weinberg RA. Cancer stem cells and epithelial-mesenchymal transition: concepts and molecular links. Semin Cancer Biol. 2012;22:396–403.
Perissinotto E, Cavalloni G, Leone F, Fonsato V, Mitola S, Grignani G, et al. Involvement of chemokine receptor 4/stromal cell-derived factor 1 system during osteosarcoma tumor progression. Clin Cancer Res. 2005;11:490–7.
Zhu XY, Wang L, Luan SH, Zhang HS, Huang WT, Wang NH. The PGI-KLF4 pathway regulates self-renewal of glioma stem cells residing in the mesenchymal niches in human gliomas. Neoplasma. 2014;61:401–10.
Lin SP, Lee YT, Wang JY, Miller SA, Chiou SH, Hung MC, et al. Survival of cancer stem cells under hypoxia and serum depletion via decrease in PP2A activity and activation of p38-MAPKAPK2-Hsp27. PLoS ONE. 2012;7:e49605.
Xiang T, Long H, He L, Han X, Lin K, Liang Z, et al. Interleukin-17 produced by tumor microenvironment promotes self-renewal of CD133+ cancer stem-like cells in ovarian cancer. Oncogene. 2015;34:165–76.
Acknowledgements
We would like to acknowledge Dr. D.B. Kohn for providing lentivirus pR Δ 8.9 and pMD.G plasmids. This work was supported by grants from the National Natural Science Foundation of China (No. 81473227) and grants from the International Science & Technology Cooperation Program of China (2014DFE30050), the Fundamental Research Funds for the Central Universities (2017XZZX011-04) and the Zhejiang Provincial Natural Science Foundation of China (No. LY17H310004) (to D.Z.).
Author contributions
M-dY, Q-jH, BY, X-tQ, and Y-lL designed the research project; X-tQ, Y-lL, D-fZ, TX, and BL performed the experiments; X-tQ and Y-lL analyzed the data; Y-qZ, LF, J-qG, and L-sY contributed reagents; and M-dY, X-tQ, and Y-lL wrote the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Qi, Xt., Li, Yl., Zhang, Yq. et al. KLF4 functions as an oncogene in promoting cancer stem cell-like characteristics in osteosarcoma cells. Acta Pharmacol Sin 40, 546–555 (2019). https://doi.org/10.1038/s41401-018-0050-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41401-018-0050-6
Keywords
This article is cited by
-
Transcriptional regulation of cancer stem cell: regulatory factors elucidation and cancer treatment strategies
Journal of Experimental & Clinical Cancer Research (2024)
-
USP40 promotes hepatocellular carcinoma cell proliferation, migration and stemness by deubiquitinating and stabilizing Claudin1
Biology Direct (2024)
-
Cancer cell plasticity, stem cell factors, and therapy resistance: how are they linked?
Cancer and Metastasis Reviews (2024)
-
Regulation and signaling pathways in cancer stem cells: implications for targeted therapy for cancer
Molecular Cancer (2023)
-
DUB3/KLF4 combats tumor growth and chemoresistance in hepatocellular carcinoma
Cell Death Discovery (2022)