Abstract
Reading Disability (RD) is often characterized by difficulties in the phonology of the language. While the molecular mechanisms underlying it are largely undetermined, loci are being revealed by genome-wide association studies (GWAS). In a previous GWAS for word reading (Price, 2020), we observed that top single-nucleotide polymorphisms (SNPs) were located near to or in genes involved in neuronal migration/axon guidance (NM/AG) or loci implicated in autism spectrum disorder (ASD). A prominent theory of RD etiology posits that it involves disturbed neuronal migration, while potential links between RD-ASD have not been extensively investigated. To improve power to identify associated loci, we up-weighted variants involved in NM/AG or ASD, separately, and performed a new Hypothesis-Driven (HD)–GWAS. The approach was applied to a Toronto RD sample and a meta-analysis of the GenLang Consortium. For the Toronto sample (n = 624), no SNPs reached significance; however, by gene-set analysis, the joint contribution of ASD-related genes passed the threshold (p~1.45 × 10–2, threshold = 2.5 × 10–2). For the GenLang Cohort (n = 26,558), SNPs in DOCK7 and CDH4 showed significant association for the NM/AG hypothesis (sFDR q = 1.02 × 10–2). To make the GenLang dataset more similar to Toronto, we repeated the analysis restricting to samples selected for reading/language deficits (n = 4152). In this GenLang selected subset, we found significant association for a locus intergenic between BTG3-C21orf91 for both hypotheses (sFDR q < 9.00 × 10–4). This study contributes candidate loci to the genetics of word reading. Data also suggest that, although different variants may be involved, alleles implicated in ASD risk may be found in the same genes as those implicated in word reading. This finding is limited to the Toronto sample suggesting that ascertainment influences genetic associations.
Similar content being viewed by others
Introduction
Reading Disability (RD), also known as developmental dyslexia, is a neurodevelopmental disorder affecting children globally. In North America alone, it affects 5–7% of individuals [1,2,3,4,5,6]. RD is characterized by difficulties with word reading and spelling, despite typical intelligence and motivation to learn [7]. Affected children often have comorbid neurodevelopmental disorders, including language or speech impairments, or attention-deficit/hyperactivity disorder (ADHD) [8]. These factors increase social difficulties, decrease self-esteem, and hinder academic/occupational success [9,10,11,12]. RD, therefore, represents a major public health concern.
The genetics and underlying mechanisms of RD are not fully known. Twin and family studies initially demonstrated genetic contributions to RD by examining heritability within families [3, 13]. Researchers went on to identify specific chromosomal regions and genes implicated in RD via linkage analysis followed by fine-mapping association studies. These linked regions and genes were supported, to varying degrees, by independent studies [14] and meta-analyses [15,16,17,18,19,20]; however, sample sizes were small by current standards yielding low power and elevated risk of false-positive findings. Moreover, some results could not be replicated [16, 21, 22]. Therefore, researchers called into question the robustness of the genes as candidates for involvement in RD.
Despite these caveats, a number of candidate genes identified from linkage/fine-mapping studies (KIAA0319, KIAA0319L, DCDC2, DNAAF4 (previously called DYX1C1 and EKN1), and ROBO1) were linked to neuronal migration, suggesting a potential molecular mechanism (but for a critical review see [23]). These associations were pertinent as previous postmortem brain studies (n < 10) found heterotopias and cortical dysplasias, signatures of altered migration, in RD-affected individuals [24, 25]. It was theorized that disrupted neuronal migration (DNM) may be involved in RD etiology [26, 27].
The first evidence to implicate allelic variation of these RD candidate genes in DNM came from studies finding that protein motifs/domains encoded by the genes were predicted to function in migration [28,29,30,31,32]. Further evidence emerged from in utero knockdown experiments of the genes (KIAA0319, KIAA0319L, DCDC2, and DNAAF4/DYX1C1) in the developing brains of mice or rats. When gene expression was reduced, neural cells did not migrate as expected from the lower ventricular zone to the higher cortical plate [33,34,35,36,37,38]. Instead, most cells remained in the ventricular or intermediate zones, indicating disrupted migration, albeit with different patterns of disruption for different genes [33,34,35,36,37,38]. Experiments in which the candidate genes were overexpressed also resulted in neural phenotypes, including aberrant neurite outgrowth (Dcdc2) [39], delayed radial migration (Kiaa0319) [40], and altered axon growth and regeneration (Kiaa0319) [41].
There was, however, skepticism regarding the proposed roles of these genes in migration, when independent knockout experiments did not replicate the results of the knockdown studies [23, 40, 42,43,44]. Discrepancies between knockout and knockdown findings may be due to developmental compensation by altered gene expression [45, 46]. For example, data from Dcdc2 knockout mice supports partial redundancy with Dcx, a member of the same gene family, which functionally compensates for the loss of Dcdc2 [43]. Overall, it remains unclear at this time whether the candidate genes in question are indeed implicated in RD and if this involves effects on neuronal migration [23].
To further identify genes implicated in RD, and as the available techniques advanced, the field moved away from linkage analysis in families to genome-wide association studies (GWAS) -- a more powerful approach for complex traits in which effect sizes of individual risk variants are small. In the last few years, GWAS for RD, reading performance, and/or reading-related tasks have begun to yield results with genome-wide statistical significance (SNP-based analysis (p~10–8) [47,48,49,50]; gene-based analysis (p~10–6) [51,52,53]). Across recent significant and previous non-significant GWAS findings, a number of the top genes are thought to be involved in neuronal migration/axon pathfinding, potentially supporting the DNM hypothesis. For example, in a GWAS by Price et al., 2020, using two samples, the Toronto (n = 624) and Philadelphia Neurodevelopmental Cohort (n = 4430), the most significant SNP (p < 5 × 10–7) in the Toronto sample was located in an intron of ARHGAP23, a gene involved in actin cytoskeleton polymerization/reorganization, neuronal development, and other growth cone/axon related processes [54]. Across both samples in that study, when top-ranked SNPs, at a less stringent p-value threshold (p~10–5), were mapped into or near close proximity genes, additional genes were found to have been previously related to neuronal migration/axon guidance (NM-AG) (as well as growth cone formation which is encompassed within this term). For example, ASTN2, CNTN, TUBB3, NRCAM, DSCAM, UNC5D, and GAP43 [53]. Larger GWAS studies also provided weak support for the DNM hypothesis. A meta-analysis of 22 samples (n~34,000) by the GenLang Consortium, identified genome-wide significant variants in DOCK7 associated with word reading [47]. DOCK7 is critical for cortical neurogenesis, axon formation, and neural polarization [55]. In the largest GWAS study to date, analyzing the 23andMe cohort, the authors identified 42 significant loci associated with self-reported dyslexia (ncases~51,000) of which genes had been previously related to NM/AG [48] (Nature Genetics, in press https://doi.org/10.1101/2021.08.20.21262334). However, it should be noted, a systematic targeted gene-set analysis in that sample found significant overrepresentation of genes related to axon guidance but not for those involved in neuronal migration [48].
The Price et al. study also identified, at the less stringent p-value threshold (p~10–5), variants in or near genes implicated in neurodevelopmental disorders, particularly autism spectrum disorder (ASD) [53]. For example, ANKS1B, CNTN4, RBFOX1, CSMD1, and ASTN2. The study of the self-reported dyslexia in 23andme observed top associated SNPs in ASD-related genes. Although there is little evidence to support phenotypic comorbidity between ASD and RD, both are neurodevelopmental disorders and ASD involves deficits in language and communication skills [56, 57]. Further, there is some preliminary evidence of shared genetic risk factors: rare and de novo variants identified in ASD cases for genes that have been independently associated with RD (PRTG, ROBO1, and KIAA0319L) [58, 59], and altered neuronal migration has been suggested as a etiological mechanism in each condition [60,61,62,63,64]. While putative links between RD and DNM have received much attention in prior literature, few investigations to date have explored potential overlaps in neurobiological foundations of RD and ASD.
Given GWAS-based observations of associated SNPs in genes previously implicated in ASD and genes involved in NM/AG, along with investigations of DNM in previous candidate gene and postmortem studies, we wished to leverage this information to improve power to identify significant variants. Most existing GWAS samples are modest in size, with the exception of [47, 48], and relatively few findings reach statistical significance at the genome-wide level. To leverage power within available samples, we used Hypothesis-Driven (HD)–GWAS, which up-weights or prioritizes variants based on previously established genetic or biological hypotheses [65]. Statistical corrections are performed independently on the up-weighted and nonup-weighted groups, reducing the threshold for significance and increasing power. The HD-GWAS was primarily conducted on the Toronto sample [53]. We also wanted to examine the issues in the context of a larger dataset, so secondarily we used the meta-analysis of the GenLang Consortium [47]. A tertiary analysis was conducted using only those GenLang samples that were selected based on reading and/or language difficulties, to make it more comparable to the Toronto samples.
For our HD-GWAS of word reading, we formulated two separate hypotheses based on the results from previous GWAS (“conventional”, hypothesis-free GWAS) [53] and the literature. Specifically, we hypothesized that variants in (1) genes implicated in NM/AG and (2) genes implicated in ASD, would contribute to word reading.
Methods
Study populations and measures
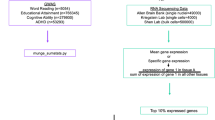
To complete the HD-GWAS analyses, summary statistics from the Toronto conventional GWAS [53] and the meta-analysis of the GenLang Consortium were used [47]. The gene-set analyses made use of the raw genotypes of the Toronto sample and the summary statistics of GenLang. For both samples, quantitative measures of word reading were used as the phenotype.
Toronto sample
The Toronto sample has previously been described and is part of an ongoing genetic study of RD-selected families recruited at the Hospital for Sick Children [53, 56, 66]. At the time of analysis, the sample consisted of children identified with reading difficulties (n = 453) and both their unaffected and affected siblings (n = 171). Children were of European ancestry, as determined by PCA analysis. This choice aimed to reduce variation and create a more homogeneous population; however, it does not fully encompass the complex nature of allelic structure [67]. Children were excluded if there was evidence of other neurodevelopmental disorders, including ASD, apraxia, dyspraxia, central auditory processing disorder, stuttering, and psychiatric disorders, as well as medical conditions that would interfere with reading. Information on psychiatric and neurodevelopmental disorders was obtained using a structured parent interview [68] and a semi-structured teacher interview [69]. Children with ADHD, mild anxiety disorders, and speech/language difficulties were included.
The Toronto sample was measured for word reading using the Wide Range Achievement Test (WRAT) 3 [70]. The mean reading score was 88.4 [53]. Verbal assent and/or written consent was obtained from all children and parents. Procedural approval was given by the Hospital for Sick Children and University Health Network Research Ethics Boards.
The GenLang Consortium
The GenLang Consortium is a large international effort to study genetic contributions to traits related to speech, language, and reading (https://www.genlang.org). It does so through meta-analyses of these traits in population-based samples, as well as family-based and case-control cohorts [47]. For the purposes of the current study, the Toronto sample was analyzed separately, because the overlap between RD and neuronal migration/ASD was originally observed in this sample and formed the basis of the hypotheses being tested in this work.
The GenLang meta-analysis dataset used in this study consisted of 17 samples (n = 26,588) with individuals of European ancestry, as determined by PCA analysis (Table S1) [47]. Data quality control and filtering were performed by each individual sample before the meta-analysis [47]. We refer to this sample collectively as “The GenLang Cohort”.
In the GenLang Cohort, word reading was measured using a variety of standardized tests (depending on the individual sample) [47]. These phenotypic data were aligned across samples prior to the meta-analysis, as described in [47]. Consent was obtained from all participants in the GenLang Consortium and each individual sample’s institution-approved data use.
The GenLang selected subset
Five samples of the GenLang Cohort, selected for reading or language difficulties, were also examined (n = 4152, Table S1) as a subset. With exception of the SLIC cohort, all samples were selected for reading difficulties and participants were between the ages of 7 to 18. We refer to this collectively as the “GenLang selected subset”. In the family-based samples, phenotypic data was available both from probands and their siblings, regardless of affection status; therefore, these samples included a range of quantitative variation.
Data processing and GWAS of Toronto
Genotyping and quality control of the Toronto sample were previously described [53]. Briefly, DNA from each participant was genotyped on the OmniExpress platform and unobserved variants were imputed using the Michigan imputation server [71]. Quality control was conducted over numerous steps. Sex was checked using the heterozygosity of markers on the X chromosome. Sibling relationships were confirmed genetically and individuals with cryptic relationship were removed. In addition, variants with low imputation quality (R2 < 0.3), variants out of Hardy-Weinberg equilibrium (p < 0.0001), and variants with minor allele frequencies less than 5% were removed as well as samples with >2% missing genotypes and call rates <98%. After this filtering, 5.3 million SNPs were included in the analysis.
Because the Toronto sample included sibling pairs, the GWAS analysis was conducted using a linear mixed model in the R package ‘nlme’ to include a random effects term for family relationship [53]. Covariates for population structure (principal components) were also included as fixed effects. Only self-reported European Caucasian individuals were included in the analysis; verified by genotype. Principal components were generated in the program KING [72] and a Tracy-Widom statistic (EIGENSOFT Program) was used to determine significance [53].
Meta-analysis of the GenLang Cohort
Genotyping and quality control of the GenLang Cohort samples were previously described [47]. Individual cohort association analyses were performed with different tools, including SNPTEST [73], GEMMA [74], and PLINK [75] and included covariates for population structure (principal components) and family relationship.
A meta-analysis was performed on the samples using the program METAL [76], again using only individuals of European ancestry as determined by principal components. Effect size estimates were weighted with the inverse of the standard errors and genomic control on [47]. The GenLang selected subset underwent the same meta-analysis process. Summary statistics for the GenLang Cohorts were provided for use in this study after review and approval of the project by the coordinating board of the GenLang Consortium.
The Manhattan and quantile-quantile plots were generated using FUMA [77]. The regional association plot was examined in LocusFocus (https://locusfocus.research.sickkids.ca/) [78].
For both the GenLang Cohort and GenLang selected sample, only SNPs with a MAF ≥ 5%, and variants present in ≥50% of the total sample were used.
HD-GWAS
HD-GWAS serves as a powerful approach to GWAS by incorporating genetic or biological hypotheses based on the previously conducted research. This technique was developed and then tested by [65, 79,80,81]. Variants are divided into two strata: a stratum where it is hypothesized that variants are associated with the trait and a stratum where they are not. An estimated false discovery rate (FDR) is then calculated separately on each stratum. This prioritization leads to a less stringent type 1 error cut-off and increases the power to detect associated SNPs.
Although the Toronto sample did not originally meet SNP-based significance in the conventional GWAS, it did produce SNP level p-values of 10–7 and significant evidence by gene-based analysis. Thus, FDR correction was appropriate to increase power. The GenLang sample was a powerful sample with significant findings by conventional GWAS.
HD-GWAS was run using the stratified False Discovery Rate (sFDR) framework and program (http://www.utstat.toronto.edu/sun/Software/SFDR/index.html) [65, 79, 80]. As input, the program requires a SNP identifier, the p-value, and the weighting status (1- not up-weighted (not prioritized), 2- up-weighted (prioritized)). R (https://www.r-project.org/) and the command “merge ()” were then used to incorporate up-weighting information with summary statistics. Variants that were not in the up-weighted group formed the control stratum. The sFDR commands “-assoc” and “-SFDR” were used. The output included the FDR q-value and a sFDR q-value.
To test the first hypothesis, we up-weighted variants in genes implicated in NM/AG as well as growth cone formation. The gene list was made with AmiGO (http://amigo.geneontology.org), which uses the Gene Ontology (GO) database to annotate genes. The following search terms were used GO:0001764 neuron migration, GO:0007411 axon guidance, and GO:0030426 growth cone.
We broadened neuronal migration to include axon guidance and growth cone formation because many axon guidance molecules are pleiotropic, with diverse functions in multiple tissues and in the brain, including neuronal migration in the developing brain [82, 83] and growth cones are at the tips of the leading processes of migrating neurons and elongating axons [82, 83]. Genes in all three pathways have been implicated in disorders of neuronal migration, including periventricular nodular heterotopia [84], a neuronal migration disorder in which cortical development is compromised, leading to epilepsy and RD [85]. A total of ~115,000 variants in 351 genes were tested (Table S2). Within this gene list, we included the original RD-linked candidate genes that have been implicated in neuronal migration (KIAA0319, DCDC2, DYX1C1, and ROBO1).
To test the second hypothesis, we up-weighted variants in genes implicated in ASD using the gene list from the Simons Foundation Autism Research Initiative (SFARI) database (https://gene.sfari.org/database/human-gene/). More than half of these genes have been implicated in ASD through rare de novo mutations, or copy number variants (CNVs, syndromic or functional), but we also included those identified via genome-wide association studies. All categories were included, which at the time of the analysis consisted of 990 genes (Table S3) (~370,000 variants).
Gene-set analysis
The joint contribution of the genes annotated to each hypothesis was tested using gene-set analysis in MAGMA [86]. For the NM/AG and ASD hypotheses, the AMIGO and the SFARI gene lists, respectively, were used.
Gene-set analysis involves three steps, which were completed in MAGMA (https://ctg.cncr.nl/software/magma). For the first step genes were annotated to SNPs. Input for this step was raw genotype data for the Toronto sample and the reference data of the 1000 genomes project for the GenLang [87]. Second, individual gene analysis was completed to determine the association between each gene and the phenotype. For both samples, this step was performed using linear regression to compute a p-value for each gene. The input data were as follows. For the Toronto sample, the predictor variables were gene annotations from the previous step and principal components for population structure. The outcome variable was the word reading phenotype. For the GenLang datasets, the predictor variables were the gene annotations from the previous step and the outcome was the summary statistic p-values. Lastly, gene annotations were aggregated to their set and tested as a unit to see if they affected the trait. MAGMA took into consideration gene size, gene density, and allele count. The null hypothesis was that the genes tested as a group showed no greater association than a random set of genes.
Threshold for significance
The threshold for significance for the HD-GWAS and gene-set analyses was set at 2.5 × 10–2 (critical threshold 0.05/2 hypotheses). The Toronto sample formed the basis of our hypotheses and was corrected separately from the GenLang meta-analyses. The Toronto sample had no participant overlap with the GenLang Cohort.
Results
Conventional GWAS
The conventional GWAS for the Toronto and the GenLang Cohort have previously been published [47, 53]. The HD-GWAS analyses utilized the GenLang Cohort without the Toronto sample because the potential overlap between reading and neuronal migration/ASD was originally observed in that sample [53].
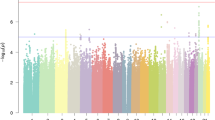
The GWAS of the GenLang selected subset, including only those GenLang samples that were recruited via probands with RD or language disorder, is a novel analysis that has not previously been described. The characteristics of cohorts in the selected subset are available within Supplemental Table 1 of Eising et al (2022). The Manhattan plot and quantile-quantile (Q-Q) plot for the selected subset are shown in Figs. S1–2. The p values depart from the expected line in the Q-Q plot at a p value of 10−8. The λ statistic was a value of 1.02.
For the conventional GWAS of the GenLang selected subset (no hypothesis tested), the top associated locus was on chromosome 21q21.1 in the intergenic region between genes BTG3 and C21orf91. The most significant SNP was rs4818369 (Table 1, p = 2.37 × 10–10, threshold p ≤ 5 × 10–8; results at p < 10–6 shown in Table S4). The regional association plot is depicted in Fig. S3. Rs4818369 was not found to correlate with altered splicing or eQTLs. SNPs (p~10–7) in linkage disequilibrium (LD) (r2 > 0.3) with rs4818369 are eQTLs for the genes BTG3 and C21orf91 (GTEx v8, Table S5). For the conventional GWAS of the GenLang Cohort, this SNP did not show genome-wide significant association (p = 3.00 × 10–3).
HD-GWAS for the Toronto sample
For the HD-GWAS of the Toronto sample, no SNPs passed the threshold for significance when up-weighted based on the two hypotheses (threshold sFDR q ≤ 2.5 × 10–2; top 10 results in Tables S6–S7). However, when we tested the joint contribution of all genes in each of the individual hypotheses, we found the ASD-related gene-set was statistically significant (Table 2, p = 1.45 × 10–2, threshold p ≤ 2.5 × 10–2).
HD-GWAS for the GenLang Cohort
For the HD-GWAS of the GenLang Cohort, two loci on chromosome 1p31.3 and 20q13.33 in DOCK7 and CDH4, respectively, passed the threshold for significance for the NM/AG hypothesis. The most significant SNP was rs1168041 for chromosome 1 and rs6089259 for chromosome 20 (Table 3, rs1168041 p = 6.61 × 10–7, rs6089259 p = 7.03 × 10–6, both sFDR q = 1.02 × 10–2, threshold sFDR q ≤ 2.5 × 10–2; results with q < 0.05 in Tables S8, S10. These SNPs were the top ranked SNPs by sFDR and in the prioritized group. Rs1168041 is an eQTL and splice quantitative trait locus (sQTL) for DOCK7, as are SNPs in LD (r2 > .03) with this marker (GTEx v8, Table S9. This SNP is in LD (r2 = .33) with the top SNP (rs11208009) in the original GenLang meta-analysis (22 samples, including the Toronto sample) located ~45 kb from DOCK7 [47]. Rs11208009 is also an eQTL and sQTL for DOCK7 (Eising et al. (2022) Supplementary) [47]. Rs6089259, intronic to CDH4, is not correlated with altered splicing or eQTLs according to available data.
For the gene-set analysis of the GenLang, no gene-sets passed the threshold for significance (threshold p ≤ 2.5 × 10–2, Table S11).
HD-GWAS for the GenLang selected subset
For the HD-GWAS of the GenLang selected subset, the same locus and SNP (21q21.1, rs4818369) as the conventional GWAS passed the threshold for significance for both hypotheses (Table 4, p = 2.37 × 10–10, sFDR q < 9.00 × 10–4, 8.00 × 10−4, threshold sFDR q ≤ 2.5 × 10–2; results with q < 0.05 Tables S12–S13). This SNP was the top-ranked SNP by sFDR and not in the prioritized group, reflecting the robustness of the HD-GWAS [79]. Within the HD-GWAS and conventional GWAS of the GenLang selected subset, 14 and 18 SNPs, respectively, were previously identified with p < 10–6 in a prior GWAS analysis of word reading by Gialluisi et al. [50] (Tables S14–S15). The GenLang selected subset includes four cohorts which were included in the earlier Gialluisi et al. study [50], although at the time of that study the aforementioned cohorts were smaller in sample size than presently. The GenLang selected subset included one extra cohort (SLIC). The GenLang selected subset comprised 4152 individuals while Gialluisi included 3468 individuals.
For the gene-set analysis of the GenLang selected subset, no gene-sets passed the threshold for significance (threshold p ≤ 2.5 × 10–2, Table S16).
Discussion
Until recently, GWAS investigations of reading skills have identified few associated loci that passed the threshold for genome-wide significance. However, with the organization of large-scale collaborations, such loci are beginning to be found [47,48,49,50]. The number and size of cohorts characterized for reading-related traits has been a limiting factor. Therefore, methods that improve power to capitalize on existing samples are necessary to move the field forward in the short term. Overlap with top loci and genes known to be related to NM/AG and ASD susceptibility were observed in previous GWAS findings [47, 48, 53]. To test these observations, we used HD-GWAS, prioritizing variants in genes implicated in NM/AG or ASD susceptibility. We also tested the joint contribution of the genes and therefore the prioritization hypotheses themselves.
For the hypothesis testing the relationship to ASD, we did not identify significant SNPs by HD-GWAS. However, gene-set analysis determined that the hypothesis itself tested as the joint contribution of ASD-related genes was significant in the Toronto sample. There was no relationship in the GenLang meta-analysis. Previous GWAS studies that examined the relationship between ASD and RD [47, 48, 53] did not find genetic correlations using polygenic risk scores or LD Score Regression (LDSC). This may be because the GWAS for ASD to date are relatively small or because the cohorts were composed of diverse neurodevelopmental phenotypes as previously suggested [48]. Further, the PRS/LDSC analyses depend on summary statistics from GWAS analysis, which use only common variants. The SFARI dataset contains genes implicated in ASD by rare variants or CNV analyses, as well as genes identified from association studies of common variation. Thus, ASD-reading trait overlap may not be detectable via PRS methods because rare variants were not imputed in the GWAS analyses. Another possibility is that there may indeed be shared genes involved, but that the specific risk alleles are different for ASD and reading and are not in LD. HD-GWAS prioritizes genes irrespective of the contributing genetic variants and allows us to more formally quantify word reading–ASD associations in the Toronto sample, which was previously an observation [53].
For the Toronto sample, we excluded children with known or suspected ASD, or with a first-degree relative with ASD, or other global/intellectual disabilities. Overlap between reading- and ASD-associated genes likely stemmed from shared genetic risk for neurodevelopmental disorders, particularly those contributing to language-related difficulties, as opposed to global delays [56, 57].
For the NM/AG hypothesis, HD-GWAS using the GenLang Consortium data identified two associated loci with the top SNPs located in DOCK7 and CDH4. DOCK7, Dedicator of Cytokinesis 7, is involved in axon formation and neuronal polarization [55]. The top marker, rs1168041, is an eQTL and sQTL for DOCK7. SNPs distal to DOCK7 were previously identified as significantly associated with word reading in the GenLang Consortium meta-analysis with the top SNPs also eQTLs and sQTLs for DOCK7 [47]. CDH4, Cadherin 4, encodes a cell-cell adhesion glycoprotein thought to play a role in cortical development and neuronal outgrowth [88]. CDH4 has not been implicated in RD traits in previous studies.
Our HD-GWAS analyses using the selected GenLang Consortium data identified a significant locus associated with word reading for both hypotheses, indicating that the result is robust and found even though this locus is not within the upweighted SNP sets. The top associated markers (rs4818369) are located between the genes BTG3 and C21orf91, and LD SNPs are eQTLs for both genes which are credible candidates for involvement in RD. BTG3, BTG Anti-Proliferation Factor 3, is implicated in neurogenesis [89, 90], and C21orf91, also known as EURL (Early Undifferentiated Retina and Lens), is implicated in oligodendroglia differentiation, influencing the cell’s capacity to mature and to myelinate axons [91].
The results of our HD-GWAS and gene-set analyses, although statistically significant in the separate samples, were not replicated across the Toronto and GenLang datasets. Thus far, few associated loci have been found to overlap between samples of self-reported dyslexia, quantitative measures of reading in population-based, and RD-selected cohorts [50, 53]. Nonetheless, these same studies yield evidence of considerable genetic overlap between quantitative measures of reading and self-report dyslexia, as shown by genetic correlation analyses using the GWAS data (rg = −0.71). The lack of overlap for individual loci may simply be a function of power, which could change with larger sample sizes, but a role for differences in ascertainment cannot be ruled out. Clinically ascertained GWAS samples may be enriched for participants with comorbid disorders as individuals with multiple conditions are more likely to come to clinical attention (Berkson’s Bias) [92], increasing the identification of genes related to those disorders. Alternatively, clinical studies may screen for, and exclude participants with, comorbid or medical conditions or environmental factors that would interfere with reading acquisition. These exclusions could alter the composition of the sample compared to population-based samples and possibly influence gene findings.
In summary, through an HD-GWAS framework, we identified significant associations with reading skills. We also found that genes related to ASD risk contribute to RD in the Toronto sample. Our findings support two core features of the HD-GWAS framework. First, this framework is robust to stratifying misspecification of up-weighted variants (i.e., less than ideal hypotheses [65]). We demonstrated this feature when using HD-GWAS we identified the same chromosomal 21 SNPs from conventional GWAS, even though they were not in the up-weighted group for the GenLang selected subset. Second, this framework increases power to identify genes within hypothesized pathways/mechanisms compared to unstratified approaches. We illustrated this feature when we found that the ASD-related gene-set contributed to reading and identified loci upweighted in the NM/AG hypothesis. Future studies involving larger GWAS samples ascertained through reading and language disorders may help to elucidate shared genetic mechanisms between RD and ASD.
Data availability
Summary statistics for the Toronto sample and the GenLang sample used in this study are available upon application to the GenLang Consortium (http://www.genlang.org) and review of the proposal. To download summary statistics for the entire GenLang (not the sample specific to this study), use http://www.genlang.org or the public GWAS catalogue.
References
Katusic SK, Colligan RC, Barbaresi WJ, Schaid DJ, Jacobsen SJ. Incidence of reading disability in a population-based birth cohort, 1976–1982, Rochester, Minn. Mayo Clin Proc. 2001;76:1081–92.
Snowling MJ, Melby-Lervag M. Oral language deficits in familial dyslexia: A meta-analysis and review. Psychological Bull. 2016;142:498–545.
Shaywitz SE, Shaywitz BA. Dyslexia (specific reading disability). Biol Psychiatry. 2005;57:1301–9.
Wagner RK, Zirps FA, Edwards AA, Wood SG, Joyner RE, Becker BJ, et al. The Prevalence of Dyslexia: A New Approach to Its Estimation. J Learn disabilities. 2020;53:354–65 .
Peterson RL, Pennington BF. Developmental dyslexia. Lancet 2012;379:1997–2007.
Di Folco C, Guez A, Peyre H, Ramus F. Epidemiology of reading disability: A comparison of DSM-5 and ICD-11 criteria. Sci Studies Reading. 2021;26:1–19.
Lyon GR. Part I Defining Dyslexia, Comorbidity, Teachers’ Knowledge of Language and Reading. Ann dyslexia. 2003;53:1–14.
Hendren RL, Haft SL, Black JM, White NC, Hoeft F. Recognizing Psychiatric Comorbidity With Reading Disorders. Front psychiatry. 2018;9:101.
Daniel SS, Walsh AK, Goldston DB, Arnold EM, Reboussin BA, Wood FB. Suicidality, school dropout, and reading problems among adolescents. J Learn disabilities. 2006;39:507–14.
Smart D, Youssef GJ, Sanson A, Prior M, Toumbourou JW, Olsson CA. Consequences of childhood reading difficulties and behaviour problems for educational achievement and employment in early adulthood. Br J Educ Psychol. 2017;87:288–308.
Morgan PL, Farkas G, Wu Q. Do Poor Readers Feel Angry, Sad, and Unpopular? Sci Stud Read. 2012;16:360–81.
Hossain B, Bent S, Hendren R. The association between anxiety and academic performance in children with reading disorder: A longitudinal cohort study. Dyslexia 2021;27:342–54.
Fisher SE, DeFries JC. Developmental dyslexia: genetic dissection of a complex cognitive trait. Nat Rev Neurosci. 2002;3:767–80.
Bates TC, Luciano M, Castles A, Coltheart M, Wright MJ, Martin NG. Replication of reported linkages for dyslexia and spelling and suggestive evidence for novel regions on chromosomes 4 and 17. Eur J Hum Genet: EJHG. 2007;15:194–203.
Deng K-G, Zhao H, Zuo P-X. Association between KIAA0319 SNPs and risk of dyslexia: a meta-analysis. J Genet. 2019;98:62.
Zou L, Chen W, Shao S, Sun Z, Zhong R, Shi J, et al. Genetic variant in KIAA0319, but not in DYX1C1, is associated with risk of dyslexia: an integrated meta-analysis. Am J Med Genet Part B, Neuropsychiatr Genet. 2012;159B:970–6.
Shao S, Niu Y, Zhang X, Kong R, Wang J, Liu L, et al. Opposite Associations between Individual KIAA0319 Polymorphisms and Developmental Dyslexia Risk across Populations: A Stratified Meta-Analysis by the Study Population. Sci Rep. 2016;6:30454.
Zhong R, Yang B, Tang H, Zou L, Song R, Zhu LQ, et al. Meta-analysis of the association between DCDC2 polymorphisms and risk of dyslexia. Mol Neurobiol. 2013;47:435–42.
Muller B, Wilcke A, Czepezauer I, Ahnert P, Boltze J, Kirsten H, et al. Association, characterisation and meta-analysis of SNPs linked to general reading ability in a German dyslexia case-control cohort. Sci Rep. 2016;6:27901.
Schumacher J, Hoffmann P, Schmal C, Schulte-Korne G, Nothen MM. Genetics of dyslexia: the evolving landscape. J Med Genet. 2007;44:289–97.
Bellini G, Bravaccio C, Calamoneri F, Donatella Cocuzza M, Fiorillo P, Gagliano A, et al. No evidence for association between dyslexia and DYX1C1 functional variants in a group of children and adolescents from Southern Italy. J Mol Neurosci. 2005;27:311–4.
Scerri TS, Macpherson E, Martinelli A, Wa WC, Monaco AP, Stein J, et al. The DCDC2 deletion is not a risk factor for dyslexia. Transl psychiatry. 2017;7:e1182.
Guidi LG, Velayos-Baeza A, Martinez-Garay I, Monaco AP, Paracchini S, Bishop DVM, et al. The neuronal migration hypothesis of dyslexia: A critical evaluation 30 years on. Eur J Neurosci. 2018;48:3212–33.
Galaburda AM, Sherman GF, Rosen GD, Aboitiz F, Geschwind N. Developmental dyslexia: four consecutive patients with cortical anomalies. Ann Neurol. 1985;18:222–33.
Humphreys P, Kaufmann WE, Galaburda AM. Developmental dyslexia in women: neuropathological findings in three patients. Ann Neurol. 1990;28:727–38.
Ramus F. Neurobiology of dyslexia: a reinterpretation of the data. Trends Neurosci. 2004;27:720–6.
Galaburda AM, LoTurco J, Ramus F, Fitch RH, Rosen GD. From genes to behavior in developmental dyslexia. Nat Neurosci. 2006;9:1213–7.
Taipale M, Kaminen N, Nopola-Hemmi J, Haltia T, Myllyluoma B, Lyytinen H, et al. A candidate gene for developmental dyslexia encodes a nuclear tetratricopeptide repeat domain protein dynamically regulated in brain. Proc Natl Acad Sci USA. 2003;100:11553–8.
Raskind WH, Peter B, Richards T, Eckert MM, Berninger VW. The genetics of reading disabilities: from phenotypes to candidate genes. Front Psychol. 2012;3:601.
Poelmans G, Buitelaar JK, Pauls DL, Franke B. A theoretical molecular network for dyslexia: integrating available genetic findings. Mol psychiatry. 2011;16:365–82.
Velayos-Baeza A, Toma C, Paracchini S, Monaco AP. The dyslexia-associated gene KIAA0319 encodes highly N- and O-glycosylated plasma membrane and secreted isoforms. Hum Mol Genet. 2008;17:859–71.
Kidd T, Brose K, Mitchell KJ, Fetter RD, Tessier-Lavigne M, Goodman CS, et al. Roundabout controls axon crossing of the CNS midline and defines a novel subfamily of evolutionarily conserved guidance receptors. Cell 1998;92:205–15.
Meng H, Smith SD, Hager K, Held M, Liu J, Olson RK, et al. DCDC2 is associated with reading disability and modulates neuronal development in the brain. Proc Natl Acad Sci USA. 2005;102:17053–8.
Wang Y, Paramasivam M, Thomas A, Bai J, Kaminen-Ahola N, Kere J, et al. DYX1C1 functions in neuronal migration in developing neocortex. Neuroscience 2006;143:515–22.
Platt MP, Adler WT, Mehlhorn AJ, Johnson GC, Wright KA, Choi RT, et al. Embryonic disruption of the candidate dyslexia susceptibility gene homolog Kiaa0319-like results in neuronal migration disorders. Neuroscience. 2013;248:585–93.
Paracchini S, Thomas A, Castro S, Lai C, Paramasivam M, Wang Y, et al. The chromosome 6p22 haplotype associated with dyslexia reduces the expression of KIAA0319, a novel gene involved in neuronal migration. Hum Mol Genet. 2006;15:1659–66.
Burbridge TJ, Wang Y, Volz AJ, Peschansky VJ, Lisann L, Galaburda AM, et al. Postnatal analysis of the effect of embryonic knockdown and overexpression of candidate dyslexia susceptibility gene homolog Dcdc2 in the rat. Neuroscience 2008;152:723–33.
Gonda Y, Andrews WD, Tabata H, Namba T, Parnavelas JG, Nakajima K, et al. Robo1 regulates the migration and laminar distribution of upper-layer pyramidal neurons of the cerebral cortex. Cereb cortex. 2013;23:1495–508.
Massinen S, Hokkanen ME, Matsson H, Tammimies K, Tapia-Paez I, Dahlstrom-Heuser V, et al. Increased expression of the dyslexia candidate gene DCDC2 affects length and signaling of primary cilia in neurons. PloS one. 2011;6:e20580.
Martinez-Garay I, Guidi LG, Holloway ZG, Bailey MA, Lyngholm D, Schneider T, et al. Normal radial migration and lamination are maintained in dyslexia-susceptibility candidate gene homolog Kiaa0319 knockout mice. Brain Struct Funct. 2017;222:1367–84.
Franquinho F, Nogueira-Rodrigues J, Duarte JM, Esteves SS, Carter-Su C, Monaco AP, et al. The Dyslexia-susceptibility Protein KIAA0319 Inhibits Axon Growth Through Smad2 Signaling. Cereb cortex. 2017;27:1732–47.
Rendall AR, Tarkar A, Contreras-Mora HM, LoTurco JJ, Fitch RH Deficits in learning and memory in mice with a mutation of the candidate dyslexia susceptibility gene Dyx1c1. Brain and language. 2015.
Wang Y, Yin X, Rosen G, Gabel L, Guadiana SM, Sarkisian MR, et al. Dcdc2 knockout mice display exacerbated developmental disruptions following knockdown of doublecortin. Neuroscience. 2011;190:398–408.
Guidi LG, Mattley J, Martinez-Garay I, Monaco AP, Linden JF, Velayos-Baeza A, et al. Knockout Mice for Dyslexia Susceptibility Gene Homologs KIAA0319 and KIAA0319L have Unaffected Neuronal Migration but Display Abnormal Auditory Processing. Cereb cortex. 2017;27:5831–45.
Hamada N, Ito H, Nishijo T, Iwamoto I, Morishita R, Tabata H, et al. Essential role of the nuclear isoform of RBFOX1, a candidate gene for autism spectrum disorders, in the brain development. Sci Rep. 2016;6:30805.
Rossi A, Kontarakis Z, Gerri C, Nolte H, Holper S, Kruger M, et al. Genetic compensation induced by deleterious mutations but not gene knockdowns. Nature. 2015;524:230–3.
Eising E, Mirza-Schreiber N, de Zeeuw EL, Wang CA, Truong DT, Allegrini AG, et al. Genome-wide analyses of individual differences in quantitatively assessed reading- and language-related skills in up to 34,000 people. Proc Natl Acad Sci USA. 2022;119:e2202764119.
Doust C, Fontanillas P, Eising E, Gordon SD, Wang Z, Alagoez G, et al. Discovery Of 42 Genome-Wide Significant Loci Associated With Dyslexia. Nat. Genet. in press. 2022.
Truong DT, Adams AK, Paniagua S, Frijters JC, Boada R, Hill DE, et al. Multivariate genome-wide association study of rapid automatised naming and rapid alternating stimulus in Hispanic American and African-American youth. J Med Genet. 2019.
Gialluisi A, Andlauer TFM, Mirza-Schreiber N, Moll K, Becker J, Hoffmann P, et al. Genome-wide association scan identifies new variants associated with a cognitive predictor of dyslexia. Transl Psychiatry. 2019;9:77.
Gialluisi A, Andlauer TFM, Mirza-Schreiber N, Moll K, Becker J, Hoffmann P, et al. Genome-wide association study reveals new insights into the heritability and genetic correlates of developmental dyslexia. Molecular psychiatry. 2020.
Luciano M, Evans DM, Hansell NK, Medland SE, Montgomery GW, Martin NG, et al. A genome-wide association study for reading and language abilities in two population cohorts. Genes, Brain, Behav. 2013;12:645–52.
Price KM, Wigg KG, Feng Y, Blokland K, Wilkinson M, He G, et al. Genome-wide association study of word reading: Overlap with risk genes for neurodevelopmental disorders. Genes, brain, and behavior. 2020:e12648.
Martin-Vilchez S, Whitmore L, Asmussen H, Zareno J, Horwitz R, Newell-Litwa K. RhoGTPase Regulators Orchestrate Distinct Stages of Synaptic Development. PloS One. 2017;12:e0170464.
Watabe-Uchida M, John KA, Janas JA, Newey SE, Van Aelst L. The Rac activator DOCK7 regulates neuronal polarity through local phosphorylation of stathmin/Op18. Neuron 2006;51:727–39.
Price KM, Wigg KG, Misener VL, Clarke A, Yeung N, Blokland K, et al. Language Difficulties in School-Aged Children With Developmental Dyslexia. Journal of learning disabilities. 2021:222194211006207.
Eicher JD, Gruen JR. Language impairment and dyslexia genes influence language skills in children with autism spectrum disorders. Autism Res: Off J Int Soc Autism Res. 2015;8:229–34.
Iossifov I, Ronemus M, Levy D, Wang Z, Hakker I, Rosenbaum J, et al. De novo gene disruptions in children on the autistic spectrum. Neuron. 2012;74:285–99.
Wang HZ, Qin HD, Guo W, Samuels J, Shugart YY. New insights into the genetic mechanism of IQ in autism spectrum disorders. Front Genet. 2013;4:195.
Marchetto MC, Belinson H, Tian Y, Freitas BC, Fu C, Vadodaria KC, et al. Altered proliferation and networks in neural cells derived from idiopathic autistic individuals. Mol psychiatry. 2017;22:820–35.
Pinto D, Delaby E, Merico D, Barbosa M, Merikangas A, Klei L, et al. Convergence of genes and cellular pathways dysregulated in autism spectrum disorders. Am J Hum Genet. 2014;94:677–94.
Gilbert J, Man HY. Fundamental Elements in Autism: From Neurogenesis and Neurite Growth to Synaptic Plasticity. Front Cell Neurosci. 2017;11:359.
Stoner R, Chow ML, Boyle MP, Sunkin SM, Mouton PR, Roy S, et al. Patches of disorganization in the neocortex of children with autism. The New England journal of medicine. 2014;370:1209–19.
Reiner O, Karzbrun E, Kshirsagar A, Kaibuchi K. Regulation of neuronal migration, an emerging topic in autism spectrum disorders. J neurochemistry. 2016;136:440–56.
Sun L, Craiu RV, Paterson AD, Bull SB. Stratified false discovery control for large-scale hypothesis testing with application to genome-wide association studies. Genet Epidemiol. 2006;30:519–30.
Tran C, Wigg KG, Zhang K, Cate-Carter TD, Kerr E, Field LL, et al. Association of the ROBO1 gene with reading disabilities in a family-based analysis. Genes, brain, Behav. 2014;13:430–8.
Birney E, Inouye M, Raff J, Rutherford A, Scally A. The language of race, ethnicity, and ancestry in human genetic research. arXiv. 2021.
Weller EB, Weller RA, Fristad MA, Rooney MT, Schecter J. Children’s Interview for Psychiatric Syndromes (ChIPS). J Am Acad Child Adolesc Psychiatry. 2000;39:76–84.
Tannock R, Hum M, Masellis M, Humphries T, Schachar R Teacher Telephone Interview for Children’s Academic Performance, Attention, Behavior and Learning: DSM-IV Version (TTI-IV). Toronto, Canada.: The Hospital for Sick Children, Unpublished Document; 2002.
Wilkinson GS Wide Range Achievement Test 3-Revision 3. Wilmington, DE: Jastak Associates; 1993.
Das S, Forer L, Schonherr S, Sidore C, Locke AE, Kwong A, et al. Next-generation genotype imputation service and methods. Nat Genet. 2016;48:1284–7.
Manichaikul A, Mychaleckyj JC, Rich SS, Daly K, Sale M, Chen WM. Robust relationship inference in genome-wide association studies. Bioinformatics. 2010;26:2867–73.
Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet. 2007;39:906–13.
Zhou X, Stephens M. Genome-wide efficient mixed-model analysis for association studies. Nat Genet. 2012;44:821–4.
Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75.
Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 2010;26:2190–1.
Watanabe K, Taskesen E, van Bochoven A, Posthuma D. Functional mapping and annotation of genetic associations with FUMA. Nat Commun. 2017;8:1826.
Panjwani N, Wang F, Mastromatteo S, Bao A, Wang C, He G, et al. LocusFocus: Web-based colocalization for the annotation and functional follow-up of GWAS. PLoS computational Biol. 2020;16:e1008336.
Sun L, Rommens JM, Corvol H, Li W, Li X, Chiang TA, et al. Multiple apical plasma membrane constituents are associated with susceptibility to meconium ileus in individuals with cystic fibrosis. Nat Genet. 2012;44:562–9.
Yoo YJ, Bull SB, Paterson AD, Waggott D, Sun L. Were genome-wide linkage studies a waste of time? Exploiting candidate regions within genome-wide association studies. Genet Epidemiol. 2010;34:107–18.
Schork AJ, Thompson WK, Pham P, Torkamani A, Roddey JC, Sullivan PF, et al. All SNPs are not created equal: genome-wide association studies reveal a consistent pattern of enrichment among functionally annotated SNPs. PLoS Genet. 2013;9:e1003449.
Accogli A, Addour-Boudrahem N, Srour M. Neurogenesis, neuronal migration, and axon guidance. Handb Clin Neurol. 2020;173:25–42.
Gonda Y, Namba T, Hanashima C. Beyond Axon Guidance: Roles of Slit-Robo Signaling in Neocortical Formation. Front cell developmental Biol. 2020;8:607415.
Vriend I, Oegema R. Genetic causes underlying grey matter heterotopia. Eur J Paediatr Neurol: EJPN. 2021;35:82–92.
Felker MV, Walker LM, Sokol DK, Edwards-Brown M, Chang BS. Early cognitive and behavioral problems in children with nodular heterotopia. Epilepsy Behav: EB. 2011;22:523–6.
de Leeuw CA, Mooij JM, Heskes T, Posthuma D. MAGMA: generalized gene-set analysis of GWAS data. PLoS computational Biol. 2015;11:e1004219.
Clarke L, Fairley S, Zheng-Bradley X, Streeter I, Perry E, Lowy E, et al. The international Genome sample resource (IGSR): A worldwide collection of genome variation incorporating the 1000 Genomes Project data. Nucleic acids Res. 2016;45:D854–D9.
Wiszniewski W, Gawlinski P, Gambin T, Bekiesinska-Figatowska M, Obersztyn E, Antczak-Marach D, et al. Comprehensive genomic analysis of patients with disorders of cerebral cortical development. Eur J Hum Genet: EJHG. 2018;26:1121–31.
Guehenneux F, Duret L, Callanan MB, Bouhas R, Hayette S, Berthet C, et al. Cloning of the mouse BTG3 gene and definition of a new gene family (the BTG family) involved in the negative control of the cell cycle. Leukemia 1997;11:370–5.
Yoshida Y, Matsuda S, Ikematsu N, Kawamura-Tsuzuku J, Inazawa J, Umemori H, et al. ANA, a novel member of Tob/BTG1 family, is expressed in the ventricular zone of the developing central nervous system. Oncogene 1998;16:2687–93.
Reiche L, Gottle P, Lane L, Duek P, Park M, Azim K, et al. C21orf91 Regulates Oligodendroglial Precursor Cell Fate-A Switch in the Glial Lineage? Front Cell Neurosci. 2021;15:653075.
Smoller JW, Lunetta KL, Robins J. Implications of comorbidity and ascertainment bias for identifying disease genes. Am J Med Genet. 2000;96:817–22.
Acknowledgements
We thank the psychometrists and volunteers that assisted with this project over the years and the participants in this study. We would like to thank also Dr. Lei Sun for her statistical guidance using SFDR. Support for the Toronto project was provided by grants from the Canadian Institutes of Health Research (MOP-133440 and PJT-180419). K.P. was supported by the Hospital for Sick Children Research Training Program. E.E. and S.E.F. are supported by the Max Planck Society. GenLang Consortium Acknowledgements. As stated by Eising et al. (2022): B.M., B.M.-M., B.S.P., C.F., E.E., E.V., G.A., M.v.D., and S.E.F. are supported by the Max Planck Society. A.G. and T.F.M.A. were supported by the Munich Cluster for Systems Neurology (SyNergy), and A.G. was supported by Fondazione Umberto Veronesi. A.T.M. is supported by National Health and Medical Research Council of Australia (NHMRC) Grants 1105008 and 1195955 and Centre of Research Excellence Grant 1116976. A.J.O.W. is supported by NHMRC Grant 1173896. B.S.P. is supported by Simons Foundation Autism Research Initiative Grant 514787. C.Y.S. works in the Medical Research Council Integrative Epidemiology Unit at the University of Bristol (MC_UU_00011/3). D.I.B. acknowledges Royal Netherlands Academy of Science Professor Award PAH/6635. E.E. is supported by NIH Grant R01DC016977. E.G.W. and J.R.G. are supported by National Institute of Child Health and Human Development (NICHD) Grant P50 HD 27802. F.R. is supported by Agence Nationale de la Recherche Grants ANR-06-NEURO-019-01, ANR-17-EURE-0017 IEC, ANR-10-IDEX-0001-02 PSL, and ANR-11-BSV4-014-01 and European Commission Grant LSHM-CT-2005-018696. H.T. is supported by the Netherlands Organization for Scientific Research (NWO) and Netherlands Organisation for Health Research and Development (ZonMW) Grant VICI 016.VICI.170.200. J.C.D. was supported by NICHD Grant P50 HD 27802. J.J.M., J.B.To., and T.K. were supported by NIH Grant R01 DC014489. K.M.P. was supported by the Hospital for Sick Children Research Training Program (Restracomp). K.R. is supported by a Sir Henry Wellcome Postdoctoral Fellowship (213514/Z/18/Z). M.J.S. is supported by Wellcome Trust Grant WT082032MA. S.P. and F.A. are supported by Royal Society Grants UF150663 and RGF\EA\180141. T.B. is supported by Institut Pasteur, the Bettencourt-Schueller Foundation, and Université de Paris. The Adolescent Brain Cognitive Development Study is supported by the NIH and additional federal partners (NIH Grants U01DA041048, U01DA050989, U01DA051016, U01DA041022, U01DA051018, U01DA051037, U01DA050987, U01DA041174, U01DA041106, U01DA041117, U01DA041028, U01DA041134, U01DA050988, U01DA051039, U01DA041156, U01DA041025, U01DA041120, U01DA051038, U01DA041148, U01DA041093, U01DA041089, U24DA041123, and U24DA041147). The Aston Cohort was supported by funding from European Union (EU) Horizon 2020 Programme 641652 and Waterloo Foundation Grant 797/17290. The St. Andrews Bioinformatics Unit is funded by Wellcome Trust Grants 105621/Z/14/Z and 204821/Z/16/Z. ALSPAC is supported by UK Medical Research Council and Wellcome Grant 217065/Z/19/Z and the University of Bristol. A comprehensive list of grant funding is available on the ALSPAC website (http://www.bristol.ac.uk/alspac/external/documents/grant-acknowledgements.pdf). The Basque Center on Cognition, Brain and Language (BCBL) cohort was supported by the Basque Government through the Basic Excellence Research Centre program and the Agencia Estatal de Investigación through BCBL Severo Ochoa excellence accreditation. The Brisbane Adolescent Twin Sample was supported by Australian Research Council Grants A7960034, A79906588, A79801419, DP0212016, and DP0343921, with genotyping funded by the NHMRC Grant 389891. The Colorado Learning Disabilities Research Center cohort was supported by NICHD Grant P50 HD 27802. The Early Language in Victoria Study was supported by NHMRC Grant 436958. The Familial Influences on Literacy Abilities cohort is supported by the University of Amsterdam, the Max Planck Institue Nijmegen, and NWO Grants Rubicon 446-12-005 and VENI 451-15-017. The Iowa study was funded by DC00496 and DC02746 from the National Institute on Deafness and Other Communication Disorders (NIDCD). The GRaD study was funded by the Manton Foundation, NIH Grants P50-HD027802 and K99-HD094902, and the Lambert Family. NeuroDys was funded by an EU Sixth Framework Program grant to the NeuroDys Consortium, Swiss National Science Foundation Grant 32-108130, and Austrian Science Fund Grant 18351-B02. The Netherlands Twin Register is funded by NWO Grants 480-04-004, 481-08-011, 056-32-010, 024.001.003, 480-15-001/674, 184.021.007, 184.033.111, and 56-464-14192; ZonMW Grants 911-09-032 and 912-10-020; the Amsterdam Public Health and Amsterdam Reproduction and Development Research Institutes; European Science Council Grant ERC Advanced 230374; EU Seventh Framework Program (FP7) Grant FP7/2007-2013: 602768; National Institute of Mental Health (NIMH) Grants U24 MH068457-06, R01 MH58799-03, and 1RC2 MH089995; and the Avera Institute for Human Genetics. The Pediatric Imaging, Neurocognition, and Genetics cohort is funded by NIH Grant RC2DA029475, the National Institute on Drug Abuse, and the Eunice Kennedy Shriver NICHD. The Philadelphia Neurodevelopmental Cohort is funded by NIH Grants RC2MH089983 and RC2MH089924, an institutional development award to the Center for Applied Genomics from The Children’s Hospital of Philadelphia, and a donation from Adele and Daniel Kubert and thanks the NIH data repository. The Raine study was supported by long-term funding from NHMRC Grants 572613, 403981, 1059711, 634445, 634509, and 1021105 and Canadian Institutes of Health Research (CIHR) Grant MOP-82893. Funding was also provided by the University of Western Australia, Curtin University, the Women and Infants Research Foundation, the Telethon Kids Institute, Edith Cowan University, Murdoch University, the University of Notre Dame Australia, and the Raine Medical Research Foundation. The Raine study analyses were supported by the Pawsey Supercomputing Centre with funding from the Australian Government and the Government of Western Australia. The Saguenay Youth Study is supported by the CIHR, the Heart and Stroke Foundation of Quebec, and the Canadian Foundation for Innovation. The SLI Consortium was funded by Wellcome Trust Grant 076566 and UK Medical Research Council Grant G1000569. The Twins Early Development Study is supported by UK Medical Research Council Grants MR/V012878/1 and MR/M021475/1, NIH Grant AG046938, and the EU FP7 grant FP7/2007-2013/: 602768. Toronto was supported by CIHR Grant MOP-133440. UK Dyslexia was supported by Wellcome Trust Grants 076566/Z/05/Z and 075491/Z/04, Waterloo Foundation Grant 797–1720, EU Grant 018696, and Royal Society Grant UF100463. The York cohort was funded by Wellcome Trust Grant 082036/B/07/Z.
Author information
Authors and Affiliations
Consortia
Contributions
KMP, LJS, and CLB designed research; KMP, KGW, YF, KB, MW, ENK, SLG, and EE performed research and analyzed data; MWL, ENK, SLG MW, KB and CLB collected the Toronto cohort; The Quantitative Trait Working Group of the GenLang Consortium collected and analysed the data on the cohorts (led by EE and SEF); KMP and CLB wrote the paper with input from all authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Price, K.M., Wigg, K.G., Eising, E. et al. Hypothesis-driven genome-wide association studies provide novel insights into genetics of reading disabilities. Transl Psychiatry 12, 495 (2022). https://doi.org/10.1038/s41398-022-02250-z
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41398-022-02250-z
This article is cited by
-
Genetic architecture of childhood speech disorder: a review
Molecular Psychiatry (2024)
-
Identification of brain cell types underlying genetic association with word reading and correlated traits
Molecular Psychiatry (2023)