Abstract
Attention-deficit/hyperactivity disorder (ADHD) is a common childhood mental disorder with undetermined pathophysiological mechanisms. The gut microbiota and immunological dysfunction may influence brain functions and social behaviours. In the current study, we aimed to explore the correlation of gut microbiome imbalance and inflammation in the pathophysiology of ADHD. Forty-one children with ADHD and thirty-nine healthy-control (HC) individuals were recruited. Faecal samples from all participants were collected and submitted for 16 S rRNA V3–V4 amplicon microbiome sequencing analysis. The plasma levels of 10 cytokines, including TNF-α, IL-6, IL-1β, IL-2, IL-10, IL-13, IL-17A, IFN-α2, IFN-γ, and MCP-1, were determined using a custom-made sandwich enzyme-linked immunosorbent assay (ELISA) developed by Luminex Flowmetrix. There was no significant difference between the ADHD and HC groups in species diversity in the faeces, as determined with α-diversity and β-diversity analysis. In the ADHD group, three differentially abundant taxonomic clades at the genus level were observed, namely Agathobacter, Anaerostipes, and Lachnospiraceae. Top differentially abundant bacteria and representative biological pathways were identified in children with ADHD using linear discriminant analysis (LDA) effect size (LEfSe), and the phylogenetic investigation of communities by reconstruction of unobserved states (PICRUSt) analysis, respectively. The plasma levels of TNF-α were significantly lower in children with ADHD than in HCs. Within the ADHD group, the levels of TNF-α were negatively correlated with ADHD symptoms and diversity of the gut microbiome. Our study provides new insights into the association between gut microbiome dysbiosis and immune dysregulation, which may contribute to the pathophysiology of ADHD.
Similar content being viewed by others
Introduction
Children with attention-deficit/hyperactivity disorder (ADHD) suffer from failures in academic performance and interpersonal relationships, and from multiple psychiatric comorbidities, which, without proper interventions, may persist into adulthood [1]. Although multiple parameters may be involved in ADHD pathophysiology, a major contributing factor is the dysfunctionality of the ‘gut–brain axis’—which refers to the link between intestinal function, gut microbiota and the central nervous system—during the course of ADHD development, due to gut microbial dysbiosis [2,3,4]. However, a systematic review that investigated the role of dysbiosis in neurodevelopmental disorders was inconclusive due to the marked heterogeneity between different studies [5]. Therefore, a thorough investigation of the gut microbiota profiles in children with ADHD may shed new light on its underlying pathophysiology [6,7,8,9,10,11].
To analyse the phylogeny and taxonomy of samples with complex microbiomes or from complex environments, 16 S ribosomal RNA (rRNA) sequencing is commonly used for amplicon sequencing [12]. Although microbiota analyses with case-control design are exploratory, they establish the basis of longitudinal studies to clarify the pathogenesis of ADHD [13]. One previous study found significantly decreased microbial diversity (α-diversity) in patients with ADHD compared with controls [14]. A study using the classical gene-set enrichment analysis approach found an association between ADHD and the genus Desulfovibrio, order Clostridiales [15]. Our research group previously found that the abundance of Bacteroides and Sutterella was different between children with ADHD and HCs [16]. Studies on the potential significance of Bifidobacterium, which is involved in the dopamine neural reward system, suggested that Bifidobacterium abundance may serve as a biomarker for ADHD [17, 18]. Moreover, Faecalibacterium, a genus of the Ruminococcaceae family, Anaerotaenia and Gracilibacter at the genus levels have been associated with attention deficit, which indicates that dysbiosis in gut microbiota is potentially involved in the pathophysiology of ADHD [19,20,21]. Based on these findings, we hypothesised that the gut microbiota imbalance may cause changes in the gut–brain axis and further affect neurotransmitter levels, which contribute to the clinical presentation of ADHD [16]. However, published studies have reported inconclusive results.
The intestinal microbiota may influence the development of multiple organs, including the brain, lungs and the immune system and overall body growth [22, 23]. In an animal study, microbiota composition changes were associated with activation of the neuroimmune response [24]. Children’s brains may be susceptible to pathological insult in combination with instability and immaturity of the gut microbiota [25]. The microbiome and the released factors in the blood may stimulate peripheral immune cells, thus influencing the blood–brain barrier (BBB) and other factors in the neurovascular system [26]. Peripheral immune-cell composition is modulated by the microbiome, which may be also associated with brain development and alterations in neurotransmitter systems [27,28,29].
Imbalanced cytokine levels may represent an important linking mechanism between ADHD and allergies [30, 31]. For example, concentrations of interleukin (IL)-16, IL-13, tumour necrosis factor (TNF)-α, and interferon (IFN)-γ were associated with clinical symptoms and the performance of children with ADHD in the Conner’s Continuous Performance Test (CPT) [32]. In addition, the levels of IFN-γ and IL-13 were slightly higher in children with ADHD than in healthy controls (HCs) [33]. Evidence has also indicated that cytokines affect brain development, particularly the prefrontal cortex and anterior cingulate cortex [34, 35]. Furthermore, TNF-α has been found to influence synaptic development in the hippocampus, whereas IL-1β, IL-6, and IL-10 affect memory function [36]. Serum IL-6 and IL-10 levels were relatively higher in children with ADHD, compared with HCs [37, 38]. A case–control study indicated oxidative stress (glutathione) and immune abnormalities (a trend for lower plasma IL-5 levels) in untreated patients with ADHD [39]. A recent study found a significant correlation between IL-6, TNF-α and hyperactivity/impulsivity symptoms in young people with obesity [40].
We hypothesised that dysbiosis of gut microbiota may be partly associated with the pathogenesis of ADHD. In addition, we proposed that immunological dysregulation is involved in alterations of the gut microbiome. Therefore, our study aimed to explore the correlation between gut microbiome imbalance and inflammatory cytokine levels in the pathophysiology of ADHD.
Materials and methods
Study participants
Our study protocol was approved by the Institutional Review Board (IRB) at Chang Gung Memorial Hospital in Taiwan (IRB No. 201702019A3). Children with ADHD were recruited from Chang Gung Children’s Hospital in Taiwan; HCs were recruited from the community close to the hospital. Study protocols were explained to the participants and their parents or guardians prior to their entry into this study, and informed consents were signed by both the participants and their caregivers upon their agreement.
The following inclusion criteria for patients with ADHD were applied: (a) children diagnosed with ADHD, as confirmed by a senior child psychiatrist through structured interviews using the Kiddie-Schedule for Affective Disorder and Schizophrenia-Epidemiological Version (K-SADS-E) [41, 42]; (b) children aged between 6 and 16 years; and (c) children with no prior history of any medical treatment for ADHD (methylphenidate, atomoxetine or clonidine). The following exclusion criteria of this study were applied: (a) children with a history of major neuropsychiatric diseases, including autism-spectrum disorder (ASD), intellectual disabilities, major depressive disorders (MDD), bipolar disorders, psychotic disorders or substance-use disorders (SUD); (b) children with any major physical illnesses, such as epilepsy, neuroendocrine or gastrointestinal disorders; (c) children on a vegetarian diet or those who were administered any anti-inflammatory drugs, antibiotics or probiotics within one month.
The HC group consisted of children without ADHD diagnosis (aged between 6 and 16 years) within the same catchment area. HCs were also interviewed using the K-SADS-E and were not diagnosed with the aforementioned major neuropsychiatric diseases (ASD, intellectual disabilities, MDD, bipolar disorders, psychotic disorders or SUD). Individuals on a vegetarian diet, suffering with major physical illnesses, or receiving any anti-inflammatory drugs, antibiotics or probiotics during recruitment were also excluded.
Sample collection
Both, children with ADHD and HCs, were asked to collect their faecal samples by scooping a pea-sized piece of faeces in a sterile plastic tube. Samples were stored at 4 °C after collection and transferred to a −80 °C refrigerator in our laboratory within 24 h. Total genomic DNA (gDNA) was extracted from each sample using the QIAamp PowerFecal DNA Kit from Qiagen. The concentration of gDNA was determined with Qubit fluorometric quantitation prior to polymerase-chain reaction (PCR).
16S library preparation and paired-end sequencing
The 16S V3–V4 region was amplified by a specific primer pair (319 F: 5′ CCTACGGGNGGCWGCAG 3′, 806 R: 5′ GACTACHVGGGTATCTAATCC 3′). First, faecal gDNA (12.5 ng) was used for V3–V4 region amplification. The KAPA HiFi HotStart Ready Mix (Roche) was used under the following PCR conditions: 95 °C for 3 min; 25 cycles of 95 °C for 30 sec, 55 °C for 30 sec and 72 °C for 30 sec; 72 °C for 5 min. The length of the PCR product was approximately 500 bp. The main fragments were then purified with AMPure XP beads, followed by a second PCR with dual indices and sequencing adapters (Nextera XT Index Kit, Illumina). Finally, equal amounts of the indexed PCR product were denatured and pooled, and then the 16S library was sequenced on an Illumina MiSeq sequencer (paired-end 2 ×300 bp reads).
Immunological profile
Blood samples were obtained between 7:00 and 8:00 AM. A multiplex bead-based assay was developed using the Luminex Flowmetrix system (Luminex, Austin, TX) to investigate the cytokine levels in the plasma. Each capture antibody for an individual cytokine was coupled to a different bead set (Upstate Biotechnology Beads, NY; Luminex). Recombinant cytokines pre-diluted in pooled blank plasma from HCs were used as standards; test sera from patients or controls were examined with multiplex assays as previously reported [33, 34, 43,44,45] for several cytokines, including TNF-α, IL-6, IL-1β, IL-2, IL-10, IL-13, IL-17A, IFN-α2, IFN-γ and MCP-1. First, beads were incubated with 50-μl diluted standards or plasma and then with detection antibodies for 2 h at room temperature. Next, mixtures were incubated with biotin as a reporter for 1.5 h, followed by incubation for 30 min with phycoerythrin-conjugated streptavidin. Finally, the concentration of cytokines in the bead array was measured with the fluorescence intensity calculated with the Flowmetrix software.
Clinical assessments
A senior psychiatrist confirmed the clinical diagnosis of ADHD and neuropsychiatric disorders in the enroled children with ADHD and HCs through face-to-face interviews [41]. K-SADS-E is designed to assess psychiatric disorders in children and adolescents according to The Diagnostic and Statistical Manual of Mental Disorders, Fourth Edition (DSM-IV) criteria using semi-structured interviews [41]. The Chinese version of K-SADS-E has been established as a reliable and valid assessment tool in Taiwan [42]. Furthermore, the Wechsler Intelligence Scale for Children—Fourth Edition (WISC-IV) [46] was conducted by a child psychologist to assess participants’ cognitive function. The psychologist also performed the Conners’ CPT [47] to examine participants’ attention. Participants’ parents and teachers were requested to complete the parent and teacher forms, respectively, of the Swanson, Nolan, and Pelham Version IV Scale (SNAP-IV), to measure ADHD core symptoms at home and school [48,49,50].
Statistical and bioinformatics analysis
The sample size was estimated using the software package G-Power 3.1; based on the settings of 80% power, p = 0.05. The Statistical Package for the Social Sciences Version 16.0 (SPSS Inc., Chicago, IL, USA) was used for data analysis. Mean ± standard deviation or frequency were presented as variables. The chi-square test was used to analyse the sex distribution between children with ADHD and HCs. The independent t-test was used to determine the differences in demographics, clinical symptoms and neuropsychological function between patients and controls. Two-tailed p < 0.05 was regarded as statistically significant.
For the analysis of 16 S rRNA sequencing data, the main workflow was performed using QIIME (v1.9.1) [51], which contains multiple programmes. All raw data (*.fastq) in the data folder were analysed with FastQC (v0.11.5) followed by MultiQC (v0.9). Trimming of primer sequences was performed using Cutadapt. Then, R1 and R2 of paired-end reads were merged using FLASH v1.2.11, and only merged reads were used in further analysis. Low-quality reads (Q < 20) were removed using the QIIME pipeline. Chimera sequences were identified using UCHIME v4.2 (with the reference Gold database) [52] to obtain the effective tags. Operational taxonomic unit (OTU) picking was performed using the UPARSE [53] function in the USEARCH pipeline (v.7) [54] with 97% identity [55]. Ideally, the sequences in the same OTUs were from the same group, known as a hypothetical classification unit [56]. The RDP classifier (v2.2) algorithm [57] was adapted to annotate taxonomy classifications based on the SILVA database (SILVA v132 rep_set_16 S_only/99/silva_132_99_16S.fna, rep_set_aligned/99/99_alignment.fna) [56, 58] for each representative sequence, which was established with an 80% minimum confidence threshold to identify an assignment. Species annotation was visualised with KRONA (v2.7) [59]. Moreover, sequencing results were further validated with the DADA2 package followed by standard default settings [60]. Differences of bacterial taxonomy between children with ADHD and HCs were analysed using analysis of covariance (ANCOVA) following adjustment with age and sex. Bonferroni-corrected p-values were calculated as 0.05 (alpha error) divided by the number of parameters in each figure.
Results
Demographic data
Faecal samples from 41 children with ADHD (mean age: 8.0 years old, 73.2% male) and 39 HC individuals (mean age: 10.0 years old, 56.4% male) were collected. The characteristics of the ADHD and HC groups in our study are summarised in Table 1. In our cohort, higher male-to-female ratios were found in ADHD, which is consistent with the known ADHD prevalence ratio ranging from 2:1 to 10:1 [61].
Gut microbiota analyses
The 16S V3–V4 library from faecal samples was constructed and sequenced in the Illumina sequencer platform according to standard procedures. We used a rarefaction curve to estimate gut microbiome species diversity. Rarefied OTU tables were generated and α-diversity metrics were established for each rarefied OTU table. For rarefaction curve generation, a certain amount of sequencing data was randomly selected from the samples to represent the number of species. When the curve became flat, a reasonable sequencing depth was acquired, indicating that more sequencing depth was likely to yield only a few additional species. Based on the chart, both the ADHD and HC samples had sufficient sequencing depth for species identification. The X axis illustrates the number of sequences (per sample), and the Y axis demonstrates the number of observed species (Supplementary Fig. 1). Rarefaction curves showed that a plateau of species richness (up to 200 OTUs) was achieved in approximately 40,000 reads per sample (Supplementary Fig. 1), indicating that the sequencing depth included considerable information regarding total species richness.
We further analysed the gut microbiota composition in children with ADHD. The ACE richness estimator, Shannon index and Simpson index were analysed to estimate species richness. Our data showed that no significant difference was observed in the aforementioned indices between the ADHD and HC groups (Fig. 1A). Weighted PCoA and unweighted PCoA plots were performed based on the OTU level to evaluate the variation in gut microbiota composition in both groups (Fig. 1B, C). We also considered sex predilection with respect to gut microbiota. We found that the microbiome community in either male or female participants was similar between ADHD and HC groups (Supplementary Fig. 2). Taken together, α-diversity and β-diversity analyses indicate that the gut microbiome was similar between children in ADHD and HC groups.
A The ACE richness estimator, Shannon and Simpson index were analysed to determine species richness, and it was found that there was no significant difference between the ADHD and healthy-control groups. B The weighted PCoA and C unweighted PCoA plot were performed based on the OTU level to evaluate the variation in gut microbiota composition among the groups. β-diversity analysis indicates the extent of similarity between microbial communities.
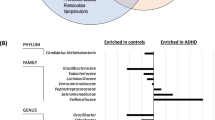
Next, the gut microbiome at different phylogenetic levels from phylum to species was comprehensively determined in children with ADHD and HCs. Figure 2 shows the relative representation of the top-5 bacteria at the phylum level in the ADHD and HC groups. Our data showed that the main phyla in the gut were the Bacteroidotes, Firmicutes, Proteobacteria, Actinobacteriota and Fusobacteriota (Fig. 2A, B). The distribution of the main top-5 phyla was similar in children with ADHD and HCs. The Firmicutes-to-Bacteroidetes ratio (F/B ratio) showed a slightly increasing trend in children with ADHD compared with HCs (Fig. 2C). Supplementary Fig. 3 illustrates the bacterial taxonomic distributions from class to genus levels in the ADHD and HC groups. The most abundant classes, orders, families and genera were Bacteroidia, Bacteroidales, Bacteroidaceae and Bacteroides, respectively. Compared with the HC group, children with ADHD had increased representation of three bacterial genera, including Agathobacter, Anaerostipes and Lachnospiraceae UCG-010 (Supplementary Fig. 4). The p-value was determined using ANCOVA following adjustment for age and sex. Supplementary Fig. 5 shows the bacterial taxonomic distributions at the species level in the ADHD and HC groups.
A The relative abundance of the most prevalent bacteria in phylum level in individual ADHD and healthy was shown. B The main phylum were Bacteroidota, Firmicutes, Proteobacteria, Actinobacteriota and Fusobacteriota in gut microbiome. C The Firmicutes-to-Bacteroidetes ratio (F/B ratio) in ADHD and HCs. The p-value was determined using analysis of covariance (ANCOVA) following adjustment with age and sex.
In addition, we applied the LDA effect-size (LEfSe) algorithm to identify metagenomic biomarkers in children with ADHD. LEfSe analysis revealed differentially abundant bacterial taxa between the ADHD and HC groups (Fig. 3). Our results identified the genera Roseburia, Ruminococcaceae and Agathobacter in the ADHD group, which showed a higher LDA score based on LEfSe, reflecting a significant increase in frequency in the ADHD group compared with HC participants (Fig. 3A, B). Figure 4 demonstrates functional predictions of all samples using the phylogenetic investigation of communities by reconstruction of unobserved states (PICRUSt). Based on the heatmap of the microbiota identified in the HC and ADHD groups, the most representative pathways were proximal tubule bicarbonate reclamation followed by glutamatergic synapses. The nervous and immune systems were also present in the functional prediction of the microbiota. Collectively, the population of some bacteria was enriched in children with ADHD. Altered bacteria composition in ADHD may affect the gut-to-brain axis, thus our PICRUst analysis revealed the most abundant nervous-related pathway in these children. Interestingly, immune-related pathways were also enriched in the functional predictions in our dataset. We then profiled the inflammatory-related cytokines in our cohort.
Immunological profile
The plasma levels of TNF-α were significantly lower in the ADHD group than in the HC group (Fig. 5A). Except for TNF-α, the plasma levels of IL-6, IL-1β, IL-2, IL-10, IL-13, IL-17A, IFN-α2, IFN-γ and MCP-1 were not significantly different between the two groups (Fig. 5A). We further identified the correlation of the cytokine profile with the gut microbiome composition or clinical symptoms in ADHD. In the ADHD group, the plasma levels of TNF-α showed an inverse correlation with the observed OTUs (r = −0.348, p = 0.028) and Shannon index (r = −0.326, p = 0.04) (Fig. 5B). The plasma levels of IL-10 also showed an inverse correlation with the observed OTUs (r = −0.460, p = 0.003) and Shannon index (r = −0.438, p = 0.007). Moreover, TNF-α levels in children with ADHD were negatively correlated with attention deficit (r = −0.436, p = 0.004) and hyperactivity/impulsivity symptoms (r = −0.376, p = 0.015).
A The plasma levels of TNF-α were significantly lower in the ADHD group, compared with the HC group. B In the ADHD group, the plasma levels of IL-10 levels were negatively correlated with observed OTU (r = -0.460, p = 0.003) and Shannon index (r = -0.438, p = 0.007). The plasma levels of TNF-α levels were negatively correlated with observed OTU (r = −0.348, p = 0.028) and Shannon index (r = −0.326, p = 0.04).
Discussion
In this study, the 16 S rRNA sequencing platform was implemented for the screening of bacterial communities from faecal samples of children with ADHD and HC participants. Species richness in the faeces, which was determined by α-diversity and β-diversity analysis, was not significantly different between the participants of ADHD and HC groups. Our results are in line with a previous case–control study [17] that also used the same metrics (α-diversity or relative abundance of Fusobacterium). Nonetheless, our data showed that a higher abundance of three genera Agathobacter, Anaerostipes and Lachnospiraceae UCG-010 was found in the ADHD group compared with the HC group. Furthermore, our study revealed a slightly higher F/B ratio in children with ADHD than in HCs. The F/B ratio has been widely investigated in previous studies regarding gut microbiota in both humans and mice and has been associated with obesity and other diseases [62].
A previous animal study showed that administration of an anti-mouse IL-6 receptor antibody (MR16-1) significantly improved the decreased F/B ratio and normalised the gut microbiota composition in susceptible mice under social-defeat stress [63]. Another study indicated that the composition of the human gut microbiota, particularly the F/B ratio, varies significantly between different age groups [64]. The relative abundance of the components of the F/B ratio is highly variable because a number of lifestyle-associated factors (i.e., diet, antibiotic consumption and physical activity) potentially influence the microbiota composition in the gastrointestinal tract [62]. Therefore, the mechanism underlying the association between the F/B ratio and ADHD requires further exploration.
In children with ADHD, a total of three differentially abundant taxonomic clades were observed, namely Agathobacter, Anaerostipes and Lachnospiraceae UCG-010. Agathobacter is a bacterial genus from the family Lachnospiraceae. The relative abundance of Agathobacter has been associated with physician-diagnosed inhalant allergies in school-age children [65] and was associated with ASD and sleep problems [66]. Anaerostipes is a Gram-positive anaerobic bacterial genus from the family Lachnospiraceae. Products of bacterial metabolism by Anaerostipes and Lachnospiraceae have been related to the aetiology of depression [67]. Nevertheless, the association between ADHD and these three taxonomic clades has not been reported. Our pathway analyses implied that the nervous and immune systems were involved in the functional prediction of the microbiota in children with ADHD. In our studies, we identified that the amount of Bacteroides uniformis was increased in ADHD (Fig. 3A). Nonetheless, it is challenging to use short-read 16 S rDNA-based sequencing technologies for bacterial taxonomy at the species level. In the future, we will expand the cohort and use species-specific primers by quantitative PCR for more precise detection of bacteria at the species level in patients with ADHD.
We found that plasma TNF-α levels were lower in the ADHD group than in the HC group, which is consistent with a previous study [68]. However, another study indicated that TNF-α was neither significantly associated with ADHD diagnosis nor with ADHD symptoms, as assessed by parents [69]. Our findings revealed that the levels of TNF-α were negatively correlated with ADHD symptoms and the diversity of the gut microbiome. Nonetheless, a positive significant correlation between TNF-α levels and ADHD rating scores was observed in another study [40]. In an animal study, anti-TNF-α therapy potentially regulated the gut microbiota and intestinal barrier function, and further ameliorated proteoglycan-induced arthritis [70]. Another study demonstrated that gut microbial dysbiosis is involved in the pathogenesis of Parkinson’s disease (PD) [71]. Mice with PD benefited from faecal microbiota transplantation through reduction of TLR4/TNF-α signalling and suppression of neuroinflammation [71]. Overall, we propose that there may be a link between the gut microbiome, immunological functions and the pathophysiology of ADHD.
Moreover, we did not find an association between ADHD and cytokine levels, other than TNF-α. Many cytokines, including IFN-α, IFN-γ, IL-2, IL-10, IL-1, IL-6 and TNF-α, can cross the BBB and affect nerve development or the function of the hypothalamic–pituitary–adrenal axis [40, 45]. Specifically, an increased level of IL-6 in patients with ADHD was reported in previous studies [37, 68, 69]. The inconsistencies in the findings between our study and previous studies may be related to the small sample size and differences in patient characteristics (environment and allergens). Further evidence is required to clarify whether production of various cytokines may play a role in the pathogenesis of ADHD.
Our study is limited due to the following points. First, our study design was cross-sectional rather than longitudinal; therefore, early life events (mode of birth delivery, history of breastfeeding, previous antibiotic use and dietary patterns) that may impact the initial colonisation and development of microbiota were not available in this study [72, 73]. Second, although we found that TNF-α was significantly correlated with some bacterial taxa in children in the ADHD group, we measured TNF-α only in the plasma, which is not representative of the whole spectrum of immunological functions. The underlying mechanism of interaction between gut microbiota and the immune system still requires further investigation. Third, the age and sex of the recruited children and the HC individuals were not matched. It is possible that the differential abundance of the gut microbiota was confounded by the differences in demographic characteristics. Fourth, faecal samples were not collected at a certain/fixed time during the day. It remains unclear whether gut microbiome taxa are influenced by the various timepoints of the collection [74]. Finally, the gut microbiota composition may vary across different ethnic groups or socioenvironmental circumstances. Caution should be used when comparing out results to those of previous studies. There is heterogeneity between previous studies regarding the sample size, study design, drug-naive status and technology applied for microbiome sequencing and cytokine analysis [3, 75]. Further research is required to determine whether our findings can be extrapolated to other populations.
In summary, a total of three differentially abundant taxonomic clades at the genus level (Agathobacter, Anaerostipes and Lachnospiraceae UCG-010) were observed in children with ADHD. The plasma levels of TNF-α were lower in the ADHD group than in the HC group. Our results indicate that the association between gut microbiome dysbiosis and immune dysregulation may contribute to the pathophysiology of ADHD.
Data availability
The data are available within the paper from the paper corresponding author on reasonable request.
References
Spencer TJ, Biederman J, Mick E. Attention-deficit/hyperactivity disorder: diagnosis, lifespan, comorbidities, and neurobiology. J Pediatr Psychol 2007;32:631–42.
Stilling RM, Dinan TG, Cryan JF. Microbial genes, brain & behaviour - epigenetic regulation of the gut-brain axis. Genes Brain Behav. 2014;13:69–86.
Sukmajaya AC, Lusida MI, Soetjipto SY. Systematic review of gut microbiota and attention-deficit hyperactivity disorder (ADHD). Ann Gen Psychiatry 2021;20:12.
Martins-Silva T, Salatino-Oliveira A, Genro JP, Meyer FDT, Li Y, Rohde LA, et al. Host genetics influences the relationship between the gut microbiome and psychiatric disorders. Prog Neuropsychopharmacol Biol Psychiatry. 2021;106:110153.
Jurek L, Sevil M, Jay A, Schroder C, Baghdadli A, Hery-Arnaud G et al. Is there a dysbiosis in individuals with a neurodevelopmental disorder compared to controls over the course of development? A systematic review. Eur Child Adolesc Psychiatry. 2020.
Sandgren AM, Brummer RJM. ADHD-originating in the gut? The emergence of a new explanatory model. Med Hypotheses. 2018;120:135–45.
Dam SA, Mostert JC, Szopinska-Tokov JW, Bloemendaal M, Amato M, Arias-Vasquez A. The role of the gut-brain axis in attention-deficit/hyperactivity disorder. Gastroenterol Clin North Am 2019;48:407–31.
Boonchooduang N, Louthrenoo O, Chattipakorn N, Chattipakorn SC. Possible links between gut-microbiota and attention-deficit/hyperactivity disorders in children and adolescents. Eur J Nutr 2020;59:3391–403.
Checa-Ros A, Jerez-Calero A, Molina-Carballo A, Campoy C, Munoz-Hoyos A. Current evidence on the role of the gut microbiome in ADHD pathophysiology and therapeutic implications. Nutrients. 2021;13:249.
Bundgaard-Nielsen C, Knudsen J, Leutscher PDC, Lauritsen MB, Nyegaard M, Hagstrom S, et al. Gut microbiota profiles of autism spectrum disorder and attention deficit/hyperactivity disorder: A systematic literature review. Gut Microbes 2020;11:1172–87.
Ligezka AN, Sonmez AI, Corral-Frias MP, Golebiowski R, Lynch B, Croarkin PE et al. A systematic review of microbiome changes and impact of probiotic supplementation in children and adolescents with neuropsychiatric disorders. Prog Neuropsychopharmacol Biol Psychiatry. 2021;108:110187.
Janda JM, Abbott SL. 16S rRNA gene sequencing for bacterial identification in the diagnostic laboratory: pluses, perils, and pitfalls. J Clin Microbiol 2007;45:2761–4.
Oulas A, Pavloudi C, Polymenakou P, Pavlopoulos GA, Papanikolaou N, Kotoulas G, et al. Metagenomics: tools and insights for analyzing next-generation sequencing data derived from biodiversity studies. Bioinform Biol Insights. 2015;9:75–88.
Prehn-Kristensen A, Zimmermann A, Tittmann L, Lieb W, Schreiber S, Baving L, et al. Reduced microbiome alpha diversity in young patients with ADHD. PLoS ONE. 2018;13:e0200728.
Cheng S, Han B, Ding M, Wen Y, Ma M, Zhang L, et al. Identifying psychiatric disorder-associated gut microbiota using microbiota-related gene set enrichment analysis. Brief Bioinform. 2020;21:1016–22.
Wang LJ, Yang CY, Chou WJ, Lee MJ, Chou MC, Kuo HC, et al. Gut microbiota and dietary patterns in children with attention-deficit/hyperactivity disorder. Eur Child Adolesc Psychiatry 2020;29:287–97.
Aarts E, Ederveen THA, Naaijen J, Zwiers MP, Boekhorst J, Timmerman HM, et al. Gut microbiome in ADHD and its relation to neural reward anticipation. PLoS ONE. 2017;12:e0183509.
Bull-Larsen S, Mohajeri MH. The potential influence of the bacterial microbiome on the development and progression of ADHD. Nutrients. 2019;11:2805.
Jiang HY, Zhou YY, Zhou GL, Li YC, Yuan J, Li XH, et al. Gut microbiota profiles in treatment-naive children with attention deficit hyperactivity disorder. Behav Brain Res 2018;347:408–13.
Szopinska-Tokov J, Dam S, Naaijen J, Konstanti P, Rommelse N, Belzer C et al. Investigating the gut microbiota composition of individuals with attention-deficit/hyperactivity disorder and association with symptoms. Microorganisms. 2020;8:406.
Richarte V, Sanchez-Mora C, Corrales M, Fadeuilhe C, Vilar-Ribo L, Arribas L, et al. Gut microbiota signature in treatment-naive attention-deficit/hyperactivity disorder. Transl Psychiatry. 2021;11:382.
Petra AI, Panagiotidou S, Hatziagelaki E, Stewart JM, Conti P, Theoharides TC. Gut-Microbiota-Brain axis and its effect on neuropsychiatric disorders with suspected immune dysregulation. Clin Ther 2015;37:984–95.
Ronan V, Yeasin R, Claud EC. Childhood development and the microbiome: the intestinal microbiota in maintenance of health and development of disease during childhood development. Gastroenterology 2021;160:495–506.
Amini-Khoei H, Haghani-Samani E, Beigi M, Soltani A, Mobini GR, Balali-Dehkordi S, et al. On the role of corticosterone in behavioral disorders, microbiota composition alteration and neuroimmune response in adult male mice subjected to maternal separation stress. Int Immunopharmacol. 2019;66:242–50.
Hsiao EY, McBride SW, Hsien S, Sharon G, Hyde ER, McCue T, et al. Microbiota modulate behavioral and physiological abnormalities associated with neurodevelopmental disorders. Cell 2013;155:1451–63.
Logsdon AF, Erickson MA, Rhea EM, Salameh TS, Banks WA. Gut reactions: How the blood-brain barrier connects the microbiome and the brain. Exp Biol Med. 2018;243:159–65.
Wang Y, Kasper LH. The role of microbiome in central nervous system disorders. Brain Behav Immun. 2014;38:1–12.
Dunn GA, Nigg JT, Sullivan EL. Neuroinflammation as a risk factor for attention deficit hyperactivity disorder. Pharm Biochem Behav. 2019;182:22–34.
Corominas-Roso M, Armario A, Palomar G, Corrales M, Carrasco J, Richarte V, et al. IL-6 and TNF-alpha in unmedicated adults with ADHD: Relationship to cortisol awakening response. Psychoneuroendocrinology 2017;79:67–73.
Anand D, Colpo GD, Zeni G, Zeni CP, Teixeira AL. Attention-Deficit/Hyperactivity Disorder And Inflammation: What Does Current Knowledge Tell Us? A Systematic Review. Front Psychiatry. 2017;8:228.
Pelsser LM, Buitelaar JK, Savelkoul HF. ADHD as a (non) allergic hypersensitivity disorder: a hypothesis. Pediatr Allergy Immunol. 2009;20:107–12.
Oades RD, Myint AM, Dauvermann MR, Schimmelmann BG, Schwarz MJ. Attention-deficit hyperactivity disorder (ADHD) and glial integrity: an exploration of associations of cytokines and kynurenine metabolites with symptoms and attention. Behav Brain Funct. 2010;6:32.
Oades RD, Dauvermann MR, Schimmelmann BG, Schwarz MJ, Myint AM. Attention-deficit hyperactivity disorder (ADHD) and glial integrity: S100B, cytokines and kynurenine metabolism-effects of medication. Behav Brain Funct. 2010;6:29.
Buske-Kirschbaum A, Schmitt J, Plessow F, Romanos M, Weidinger S, Roessner V. Psychoendocrine and psychoneuroimmunological mechanisms in the comorbidity of atopic eczema and attention deficit/hyperactivity disorder. Psychoneuroendocrinology 2013;38:12–23.
Singhal G, Jaehne EJ, Corrigan F, Toben C, Baune BT. Inflammasomes in neuroinflammation and changes in brain function: a focused review. Front Neurosci. 2014;8:315.
Yirmiya R, Goshen I. Immune modulation of learning, memory, neural plasticity and neurogenesis. Brain Behav Immun. 2011;25:181–213.
Darwish AH, Elgohary TM, Nosair NA. Serum Interleukin-6 level in children with attention-deficit hyperactivity disorder (ADHD). J Child Neurol 2019;34:61–7.
Donfrancesco R, Nativio P, Borrelli E, Giua E, Andriola E, Villa MP, et al. Serum cytokines in paediatric neuropsychiatric syndromes: focus on attention deficit hyperactivity disorder. Minerva Pediatr. 2021;73:398–404.
Verlaet AAJ, Breynaert A, Ceulemans B, De Bruyne T, Fransen E, Pieters L, et al. Oxidative stress and immune aberrancies in attention-deficit/hyperactivity disorder (ADHD): a case-control comparison. Eur Child Adolesc Psychiatry. 2019;28:719–29.
Cortese S, Angriman M, Comencini E, Vincenzi B, Maffeis C. Association between inflammatory cytokines and ADHD symptoms in children and adolescents with obesity: A pilot study. Psychiatry Res. 2019;278:7–11.
Kaufman J, Birmaher B, Brent D, Rao U, Flynn C, Moreci P, et al. Schedule for Affective Disorders and Schizophrenia for School-Age Children-Present and Lifetime Version (K-SADS-PL): initial reliability and validity data. J Am Acad Child Adolesc Psychiatry 1997;36:980–8.
Gau SF, Soong WT. Psychiatric comorbidity of adolescents with sleep terrors or sleepwalking: a case-control study. Aust N. Z J Psychiatry 1999;33:734–9.
Kuo HC, Wang CL, Liang CD, Yu HR, Huang CF, Wang L, et al. Association of lower eosinophil-related T helper 2 (Th2) cytokines with coronary artery lesions in Kawasaki disease. Pediatr Allergy Immunol. 2009;20:266–72.
Liang CD, Kuo HC, Yang KD, Wang CL, Ko SF. Coronary artery fistula associated with Kawasaki disease. Am Heart J 2009;157:584–8.
Verlaet AA, Noriega DB, Hermans N, Savelkoul HF. Nutrition, immunological mechanisms and dietary immunomodulation in ADHD. Eur Child Adolesc Psychiatry. 2014;23:519–29.
Baron IS. Test review: Wechsler Intelligence Scale for Children-Fourth Edition (WISC-IV). Child Neuropsychol. 2005;11:471–5.
Conners CK. The computerized continuous performance test. Psychopharmacol Bull. 1985;21:891–2.
Bussing R, Fernandez M, Harwood M, Wei H, Garvan CW, Eyberg SM, et al. Parent and teacher SNAP-IV ratings of attention deficit hyperactivity disorder symptoms: psychometric properties and normative ratings from a school district sample. Assessment 2008;15:317–28.
Gau SS, Lin CH, Hu FC, Shang CY, Swanson JM, Liu YC, et al. Psychometric properties of the Chinese version of the Swanson, Nolan, and Pelham, Version IV Scale-Teacher Form. J Pediatr Psychol 2009;34:850–61.
Gau SS, Shang CY, Liu SK, Lin CH, Swanson JM, Liu YC, et al. Psychometric properties of the Chinese version of the Swanson, Nolan, and Pelham, version IV scale - parent form. Int J Methods Psychiatr Res 2008;17:35–44.
Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–6.
Haas BJ, Gevers D, Earl AM, Feldgarden M, Ward DV, Giannoukos G, et al. Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res. 2011;21:494–504.
Edgar RC. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods. 2013;10:996–8.
Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010;26:2460–1.
Blaxter M, Mann J, Chapman T, Thomas F, Whitton C, Floyd R, et al. Defining operational taxonomic units using DNA barcode data. Philos Trans R Soc Lond B Biol Sci 2005;360:1935–43.
Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013;41:D590–6.
Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 2007;73:5261–7.
Yilmaz P, Parfrey LW, Yarza P, Gerken J, Pruesse E, Quast C, et al. The SILVA and “All-species Living Tree Project (LTP)” taxonomic frameworks. Nucleic Acids Res 2014;42:D643–8.
Ondov BD, Bergman NH, Phillippy AM. Interactive metagenomic visualization in a Web browser. BMC Bioinforma. 2011;12:385.
Yang CY, Li SW, Chin CY, Hsu CW, Lee CC, Yeh YM, et al. Association of exacerbation phenotype with the sputum microbiome in chronic obstructive pulmonary disease patients during the clinically stable state. J Transl Med 2021;19:121.
Willcutt EG. The prevalence of DSM-IV attention-deficit/hyperactivity disorder: a meta-analytic review. Neurotherapeutics 2012;9:490–9.
Magne F, Gotteland M, Gauthier L, Zazueta A, Pesoa S, Navarrete P et al. The Firmicutes/Bacteroidetes Ratio: A relevant marker of gut dysbiosis in obese patients? Nutrients. 2020;12:1474.
Zhang JC, Yao W, Dong C, Yang C, Ren Q, Ma M, et al. Blockade of interleukin-6 receptor in the periphery promotes rapid and sustained antidepressant actions: a possible role of gut-microbiota-brain axis. Transl Psychiatry. 2017;7:e1138.
Vaiserman A, Romanenko M, Piven L, Moseiko V, Lushchak O, Kryzhanovska N, et al. Differences in the gut Firmicutes to Bacteroidetes ratio across age groups in healthy Ukrainian population. BMC Microbiol. 2020;20:221.
Hu C, van Meel ER, Medina-Gomez C, Kraaij R, Barroso M, Kiefte-de Jong J, et al. A population-based study on associations of stool microbiota with atopic diseases in school-age children. J Allergy Clin Immunol 2021;148:612–20.
Hua X, Zhu J, Yang T, Guo M, Li Q, Chen J, et al. The gut microbiota and associated metabolites are altered in sleep disorder of children with autism spectrum disorders. Front Psychiatry. 2020;11:855.
Cheung SG, Goldenthal AR, Uhlemann AC, Mann JJ, Miller JM, Sublette ME. Systematic review of gut microbiota and major depression. Front Psychiatry. 2019;10:34.
Chang JP, Mondelli V, Satyanarayanan SK, Chiang YJ, Chen HT, Su KP, et al. Cortisol, inflammatory biomarkers and neurotrophins in children and adolescents with attention deficit hyperactivity disorder (ADHD) in Taiwan. Brain Behav Immun. 2020;88:105–13.
Elsadek AE, Al-Shokary AH, Abdelghani WE, Kamal NM, Ibrahim AO, El-Shorbagy HH, et al. Serum levels of Interleukin-6 and tumor necrosis factor alpha in children with attention-deficit hyperactivity disorder. J Pediatr Neurosci 2020;15:402–8.
Liu B, Yang L, Cui Z, Zheng J, Huang J, Zhao Q, et al. Anti-TNF-alpha therapy alters the gut microbiota in proteoglycan-induced ankylosing spondylitis in mice. Microbiologyopen 2019;8:e927.
Sun MF, Zhu YL, Zhou ZL, Jia XB, Xu YD, Yang Q, et al. Neuroprotective effects of fecal microbiota transplantation on MPTP-induced Parkinson’s disease mice: Gut microbiota, glial reaction and TLR4/TNF-alpha signaling pathway. Brain Behav Immun. 2018;70:48–60.
Slykerman RF, Coomarasamy C, Wickens K, Thompson JMD, Stanley TV, Barthow C, et al. Exposure to antibiotics in the first 24 months of life and neurocognitive outcomes at 11 years of age. Psychopharmacology. 2019;236:1573–82.
Klugarova J, Janouskova K, Prochazka M, Halek J, Sibravova V, Klugar M. The association between mode of birth delivery and attention-deficit/hyperactivity disorder: a systematic review protocol of epidemiological evidence. Int J Evid Based Healthc 2019;17:S34–S7.
Short MI, Hudson R, Besasie BD, Reveles KR, Shah DP, Nicholson S, et al. Comparison of rectal swab, glove tip, and participant-collected stool techniques for gut microbiome sampling. BMC Microbiol. 2021;21:26.
Hiergeist A, Gessner J, Gessner A. Current limitations for the assessment of the role of the gut microbiome for attention deficit hyperactivity disorder (ADHD). Front Psychiatry. 2020;11:623.
Acknowledgements
This work was supported by grants from the Chang Gung Memorial Hospital (research grant CMRPG8E1441 and CMRPG8K0761) and the Taiwan Ministry of Science and Technology (MOST 108-2628-B-182-001 and MOST 109-2314-B-182-045-MY2). The authors would like to thank Dr. Yu-Lun Kuo at BIOTOOLS Co., Ltd. in Taiwan for kindly supporting the bioinformatics analysis of NGS data, Dr. Ching-Shu Tsai, Wen-Jiun Chou and Min-Jing Lee for assisting to recruit participants, Professor Wei-Tsun Soong for granting us the use of the Chinese version of the K-SADS and Professor Shur-Fen Gau for granting our use of the Chinese version of the SNAP-IV. We also acknowledge the Next Generation Sequencing Core and Bioinformatics core (grant CLRPD1J0013) at Chang Gung University, Taoyuan, Taiwan for technical support. The authors thank the “Molecular Medicine Research Center, Chang Gung University” from The Featured Areas Research Center Programme within the framework of the Higher Education Sprout Project by the Ministry of Education (MOE) in Taiwan (EMRPD1M0281).
Author information
Authors and Affiliations
Contributions
L-JW and S-CL are co-first authors and contribute equally to this paper, they participated in the study design, patient recruitment, review of references and draughting of the paper. S-WL, H-CK and S-YL participated in protocol development. L-HH and C-YC provided technical and material support. C-YY participated in the study design, statistical analysis, interpretation data, and draughted and revised the paper. All authors read and approved the final paper and contributed to the draughting and revision of the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Wang, LJ., Li, SC., Li, SW. et al. Gut microbiota and plasma cytokine levels in patients with attention-deficit/hyperactivity disorder. Transl Psychiatry 12, 76 (2022). https://doi.org/10.1038/s41398-022-01844-x
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41398-022-01844-x
This article is cited by
-
Prenatal and postnatal neuroimmune interactions in neurodevelopmental disorders
Nature Immunology (2024)