Abstract
Depression is currently the leading cause of disability around the world. We conducted an epigenome-wide association study (EWAS) in a sample of 58 depression score-discordant monozygotic twin pairs, aiming to detect specific epigenetic variants potentially related to depression and further integrate with gene expression profile data. Association between the methylation level of each CpG site and depression score was tested by applying a linear mixed effect model. Weighted gene co-expression network analysis (WGCNA) was performed for gene expression data. The association of DNA methylation levels of 66 CpG sites with depression score reached the level of P < 1 × 10−4. These top CpG sites were located at 34 genes, especially PTPRN2, HES5, GATA2, PRDM7, and KCNIP1. Many ontology enrichments were highlighted, including Notch signaling pathway, Huntington disease, p53 pathway by glucose deprivation, hedgehog signaling pathway, DNA binding, and nucleic acid metabolic process. We detected 19 differentially methylated regions (DMRs), some of which were located at GRIK2, DGKA, and NIPA2. While integrating with gene expression data, HELZ2, PTPRN2, GATA2, and ZNF624 were differentially expressed. In WGCNA, one specific module was positively correlated with depression score (r = 0.62, P = 0.002). Some common genes (including BMP2, PRDM7, KCNIP1, and GRIK2) and enrichment terms (including complement and coagulation cascades pathway, DNA binding, neuron fate specification, glial cell differentiation, and thyroid gland development) were both identified in methylation analysis and WGCNA. Our study identifies specific epigenetic variations which are significantly involved in regions, functional genes, biological function, and pathways that mediate depression disorder.
Similar content being viewed by others
Introduction
Depression is currently the leading cause of disability worldwide [1], and it is predicted to be one of the three leading causes of illness by 2030 [2]. Although the heavy social and economic burden, the potential molecular mechanisms underlying depression remain poorly understood.
The depression risk is influenced by both genetic and environmental factors. It is suggested that epigenetic modification could mediate the lasting increasing depression risk resulting from exposure to adverse life events and provide a mechanistic framework, where genetic and environmental factors were integrated [3, 4]. DNA methylation was one important form of epigenetic modification, and one recent review of 67 studies concluded that there was evidence for the association of DNA methylation variation with depression risk [5]. Additionally, candidate gene studies discovered that BDNF and SLC6A4 hypermethylation were related to depression or major depressive disorder (MDD) [5]. Even currently some significant methylation modifications were found to be associated with depression, however, no consistent results were identified.
Nowadays, using monozygotic twins discordant for a trait or disease has been proved to be a powerful and popular design for EWAS in linking the environmental basis of epigenetic modification variation to disease status while controlling for individual genetic component [6, 7]. This design has been extensively used to explore specific DNA methylation variation associated with diseases, such as cognitive function decline [8], Alzheimer’s disease [9], and rheumatoid arthritis [10]. Since the Chinese population are different from other ethnic populations worldwide concerning genetic constitutions, environmental exposure, a multitude of life styles and occupations, the DNA methylation variation may also differ. However, to our knowledge, yet no EWAS has been performed to explore the DNA methylation variation associated with depression using Chinese monozygotic twin samples.
Accordingly, we aimed to conduct an EWAS to detect DNA CpG sites associated with depression and further integrate with gene expression data in a sample of middle and ole-aged Chinese monozygotic twins.
Material and methods
The primary materials and methods of this study were similar to our previously published studies [8, 11, 12].
Participants
Participants recruitment and collection were described in detail previously [13]. Participants who suffered from cerebrovascular disorders, stroke, traumatic brain injury, central nervous system (CNS) tumor, CNS infections, and alcohol or drug dependence were excluded. Meanwhile, participants who were unconscious, unable, or unwilling to cooperate were also dropped. Finally, a total of 58 complete monozygotic twin pairs with a mean age of 52 years (SD: 7.5) were included in the methylation analysis, and a subsample consisted of 12 twin pairs were included in the gene expression analysis. The median of absolute values of intrapair depression score difference (∆(depression score)) of all twins was 4 (range: 1–15). The number of twin pairs for ∆(depression score) ranging in 1–5, 6–10, and 11–15 was 39, 15, and 4 in the methylation analysis and 9, 3, and 0 in the gene expression analysis, respectively. The median of ratio difference calculated as |∆(depression score)|/max(depression score) was 0.41 (range: 0.14–1.00).
After providing written informed consent, all participants took a standardized questionnaire and underwent a health examination. This study was approved by the Regional Ethics Committee of the Qingdao CDC Institutional Review Boards. And the ethical principles of the Helsinki Declaration were also followed.
Assessment of depression
Depression was assessed by the Geriatric Depression Scale-30 (GDS-30) [14]. The GDS-30 had 30 items, and participants were asked to answer “yes” or “no” to the items based on how they felt over the past 1 week. The higher the total score was, the more severe the participant’s mental condition was.
Reduced representation bisulfite sequencing (RRBS) analysis
The total DNA was extracted from whole blood and sent to one biomarker technology corporation in China for the RRBS experiment. Briefly, genomic DNA was first digested to generate short fragments that contained CpG dinucleotides at the ends. Then the CpG-rich DNA fragments were extracted and bisulfite-converted. The cDNA library was constructed and sequenced to get raw sequencing data, which was then preprocessed and mapped by Bismark [15] and smoothed by R package BiSeq [16]. The methylation β-value was transformed to M-value for statistical modeling. Finally, a total of 551,447 CpG sites were included.
Cell-type composition estimation
Considering total DNA was extracted from whole blood, different methylation profiles of distinct cell types may lead to false discoveries [17]. We used ReFACTor method to control for the cell-type composition effect on DNA methylation in EWAS [18]. ReFACTor is an unsupervised reference-free method that selects methylation sites, which are informative about the cell composition in the data to apply to principal component analysis (PCA) and further uses the top components of PCA to construct surrogates for the underlying cell-type compositions for adjustment in statistical analysis. In our study, the top five components were chosen as covariates to correct the cell-type heterogeneity.
RNA library construction, sequencing, and quality control
Briefly, after total mRNA was extracted from whole blood, the RNA-Seq library was constructed and sequenced to get the sequenced data. The data was then mapped to the human genome by TopHat2 [19]. The gene expression level was estimated by FPKM value through Cufflinks software [20].
Statistical analysis
Methylation analysis
Epigenome-wide association analysis
The association between the methylation level of each CpG site and depression score was tested by a linear mixed effect model, which was equivalent to the regression model as proposed by Tan et al. [6]. The fixed effect variables of age, gender, and cell-type composition as well as random effect variable of twin pairing were adjusted for in the model.
Predicting functions of cis-regulatory regions and ontology enrichments analysis
The identified epigenome CpG sites (P < 0.05) were submitted to the Genomic Regions Enrichment of Annotations Tool (GREAT) online to analyze the functional significance of cis-regulatory regions and ontology enrichments [21]. The default “basal plus extension” association rule was chosen. In this rule, a “basal regulatory region” irrespective of the presence of neighboring genes which extended 5 kb upstream and 1 kb downstream of the transcription start site (TSS) were firstly assigned. Then each gene’s regulatory domain was extended up to the basal regulatory region of the nearest upstream and downstream genes, but no longer than 1 Mb in each direction. FDR < 0.05 was set as statistically significant in ontology enrichments analysis.
Differentially methylated region (DMR) analysis
Based on the bisulfite sequencing data and corresponding EWAS results, the DMRs associated with depression score were detected by using the comb-p approach [22]. Significant enriched DMRs were evaluated by Stouffer-Liptak-Kechris (slk) corrected P-value < 0.05.
Gene expression analysis
Weighted gene co-expression network analysis (WGCNA)
The WGCNA package is a comprehensive collection of R functions for performing various aspects of weighted correlation network analysis [23, 24]. Briefly, we first established a weighted adjacency matrix. Then the topological overlap matrix (TOM) was constructed [25,26,27] and used as input for hierarchical clustering analysis [28]. Gene modules were detected by using a dynamic tree cutting algorithm. The module eigengenes (MEs) were correlated with the trait of depression score. Relationships among modules were illustrated by a hierarchical clustering dendrogram of MEs [29], and a heatmap plot of the corresponding eigengene network. Intramodular hub genes were defined following criteria of depression score based gene significance (GS) > 0.70 and module membership (MM) > 0.90 with a threshold of P-value < 0.01 [30].
For the genes clustered in the module associated with depression score, GO enrichment analysis and BIOCARTA, KEGG, and REACTOME pathway enrichment analysis were conducted by the DAVID tool [31, 32]. The modified fisher exact P-value < 0.05 was considered as enrichment cut-off criterion.
Differentially expressed genes analysis
Five depression cases (depression score > 10) and eight health controls were included. The gene expression levels of 46 genes (including the genes where the top CpG sites and the DMRs were located) between the two groups were compared by the Wilcoxon rank sum test. The P-value < 0.05 was considered as statistically significant.
Results
Methylation analysis
A total of 58 monozygotic twin pairs with a mean age of 52 years (SD: 7.5) were included. The median of depression score was 8 (95% range: 0–21). Most of the clinical indicators were statistically intrapair correlated, indicating that the co-twin design could be beneficial (Table 1).
Epigenome-wide association analysis
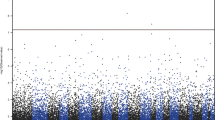
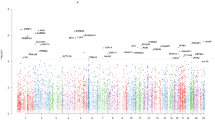
As shown in the Manhattan plot (Fig. 1) and Table 2, the association of DNA methylation levels of 66 top CpG sites with depression score reached the level of P < 1 × 10−4. The numbers of these top CpG sites located at chr1, chr4, chr16, chr19, chr20, chr11, chr7, chr6, chr12, and chr17 were 9, 8, 7, 6, 6, 5, 5, 4, 3, and 3, respectively. The strongest association (β = 0.403, P = 2.951 × 10−8) was detected for the CpG site (chr16: 90,143,728 bp) located near PRDM7. All of the top CpG sites (P < 1 × 10−4) were located at 34 genes, and there were 5, 4, 4, 4, 3, 3, 3, 3, and 3 CpG sites located at/near PTPRN2, HES5, PRDM7, RIOK1, FCGBP, HELZ2, HPF1, LAMA5, and TRIM69, respectively.
Predicting functions of cis-regulatory regions and ontology enrichments analysis
A total of 15,978 genomic cis-regulatory regions were identified to be associated with one or more genes. (Supplementary Fig. 1) Many important pathway terms probably related to depression were significantly enriched, such as Notch signaling pathway, nicotine pharmacodynamics pathway, Huntington disease, p53 pathway by glucose deprivation, Parkinson disease, and hedgehog signaling pathways. Moreover, the GO enriched terms mainly highlighted DNA binding and nucleic acid metabolic process (Table 3).
Differentially methylated region (DMR) analysis
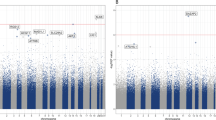
Among the 19 DMRs identified (Fig. 2 and Table 4), the methylation levels of 14 DMRs (1, 2, 3, 5–12, 15, 17, 19) were positively and two DMRs (13, 14) negatively correlated with depression score. But it was difficult to determine the trend of associations between three DMRs (4, 16, 18) and depression score.
The DMRs were located at/near 19 genes, among which DGKA and NIPA2 might play an important roles in regulating depression. Interestingly, several DMRs covered the top CpG sites listed in Table 2. The DMR1 (located at CAGE1), DMR3 (PTPRN2), and DMR9 (PRDM7) covered 4 CpG sites, and the DMR7 (located at FCGBP) covered three CpG sites.
Gene expression analysis
There were 12 twin pairs (including seven male pairs) with a median age of 53 years (95% range: 43–65) and a median depression score of 7.5 (range: 1–27) included in the gene expression analysis.
Weighted gene co-expression network analysis (WGCNA)
As Fig. 3 illustrated, genes clustered in pink module (including 3629 genes) were positively correlated with both depression score (r = 0.62, P = 0.002) and disease status (r = 0.49, P = 0.02). For this module, neuroactive ligand–receptor interaction, nicotine addiction, calcium signaling pathway, glutam4atergic synapse, and nervous system development were significantly enriched. (Table 5) MM and depression score-based GS exhibited a very significant positive correlation in pink module (r = 0.67, P < 0.001) (Supplementary Fig. 2), and 27 hub genes were identified (Supplementary Table 1).
Each row in the table corresponds to a consensus module, and each column to a trait. Numbers in the table report the correlations of the corresponding module eigengenes and trait with the P-values printed below the correlations in parentheses. The table is color coded by correlation according to the color legend.
The common genes and enrichment terms between methylation analysis and WGCNA
The CpG sites (P < 0.05) were annotated to 2808 genes, of which 404 genes were also clustered in the pink module in WGCNA. Among these common genes, DENND5B, KBTBD13, TENM3, and BMP2 were also identified as hub genes following our strict criteria. And genes including PRDM7, KCNIP1, PLEKHH3, GRIK2, PROB1, PAX3 were where the top CpG sites or DMRs were located. (Supplementary Table 2)
Many common enrichment terms were also found, including extra cellular matrix (ECM)-receptor interaction pathway, maturity onset diabetes of the young pathway, complement and coagulation cascades pathway, DNA binding, neuron fate specification, glial cell differentiation, thyroid gland development, and cellular response to hormone stimulus.
Differentially expressed genes related to depression
Genes including HELZ2 (P = 0.013), PTPRN2 (P = 0.040), GATA2 (P = 0.013), ZNF624 (P = 0.019) were found differentially expressed between the two independent groups.
Discussion
In this study based on monozygotic twins, we detected some important epigenetic variants underlying depression by EWAS. A total of 66 interesting CpG sites (P < 1 × 10−4) and 19 DMRs were identified. Moreover, many crucial GREAT ontology enrichments were identified. Genes clustered in the pink module were positively correlated with depression score in WGCNA, and many genes and enrichment terms were overlapped between methylation analysis and WGCNA. Finally, four genes were found to be differentially expressed in depression cases and health controls.
In EWAS, some genes where the top CpG sites and DMRs were located (Tables 2 and 4) may play essential roles in regulating depression status: (1) PTPRN2: DNA methylation variation of PTPRN2 was found to be associated with mood state disturbances across [33]; (2) HES5: HES5 could negatively regulate 5-HT1A receptor gene, which was related to MDD and suicide [34]; (3) GATA2: It was reported that overexpression of human GATA2 interfered with spine formation and produced depressive behavior in rats [35]; (4) DGKA: Blood transcript levels of DGKA differed significantly between participants with MDDs and nondepressed controls [36]; (5) NIPA2: It was suggested that rare copy number variants (CNVs) in NIPA2 could increase the risk of MDD by disrupting regulatory regions [37]; (6) PRDM7: The protein encoded by this gene was involved in lysine degradation pathway, and lysine methylation was a physiological post-translational modification of tau protein which played an important role in aging and Alzheimer’s disease [38]; (7) KCNIP1: The protein encoded by this gene was a member of the family of cytosolic voltage-gated potassium (Kv) channel-interacting proteins (KCNIPs), and could regulate rapidly inactivating (A-type) currents and hence neuronal membrane excitability; (8) GRIK2: GRIK2, as one glutamate-related gene, might be related to risk for mood disorders [39], and the gene polymorphism of GRIK2 was associated with depressive symptoms [40]. The other genes have an unknown function in terms of depression now, whereas they may also be interesting potential candidates to be future researched and validated.
As additional validation, we integrated the methylation data with gene expression data. Genes clustered in the pink module were positively correlated with depression score in WGCNA. And some genes were in common with EWAS findings, like BMP2, PRDM7, KCNIP1, and GRIK2. It was indicated that histone deacetylases could control neurogenesis in embryonic brain by inhibiting BMP2/4 signaling [41]. The other three genes had been discussed above. Additionally, four genes HELZ2, PTPRN2, GATA2, and ZNF624 were differentially expressed between depression cases and health controls. PTPRN2 and GATA2 have been discussed above, whereas the biological of HELZ2 and ZNF624 involved in depression remained to be studied further.
Two strengths can be noticed in our study. Since the case co-twin design using monozygotic twins discordant for a trait or disease was a powerful tool for EWAS [6, 7], our results based on the twin data would be credible. Meanwhile, considering the various genetic constitutions, environmental exposures, and a multitude of life styles in different ethnic populations worldwide, our findings will specifically help elucidate the underlying pathogenesis of depression in the Chinese population.
Nevertheless, some limitations of our study should also be considered. First, the sample size of the present study was relatively limited due to the challenges of recruiting and identifying qualified twin pairs. We’ll further validate the top CpG sites, essential genes, and biological pathways in a community population. And we’ll also evaluate if the physical distribution of top CpG sites at different chromosomes is over-represented in the regulatory domain of one specific biological pathway. Additionally, we’ll conduct a causal effect analysis based on one specific biological pathway by integrating data of genetic variation, epigenetic variation, and environmental factors. Second, the Townsend deprivation index (TDI) was indicated to be associated with depression [42, 43]. However, we couldn’t add this factor as a covariate in the linear mixed effects model, because we didn’t investigate the corresponding information of TDI during the epidemiological survey. We’ll consider the TDI factor in the validation analysis in the future.
In summary, we confirm that epigenetic factors are significant in explaining depression through twin-based analysis. We detected multiple CpG sites, genes, DMRs, and pathways that were potentially associated with depression. The findings provided an important clues to further elucidate the pathogenesis of depression and helped to identify new diagnostic biomarkers and therapeutic targets for this disease.
Data availability
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
Friedrich MJ. Depression is the leading cause of disability around the world. JAMA 2017;317:1517.
Mathers CD, Loncar D. Projections of global mortality and burden of disease from 2002 to 2030. PLoS Med. 2006;3:e442.
Penner-Goeke S, Binder EB. Epigenetics and depression. Dialog Clin Neurosci. 2019;21:397–405.
Peña CJ, Nestler EJ. Progress in epigenetics of depression. Prog Mol Biol Transl Sci. 2018;157:41–66.
Li M, D'Arcy C, Li X, Zhang T, Joober R, Meng X. What do DNA methylation studies tell us about depression? A systematic review. Transl. Psychiatry 2019;9:68.
Tan QH, Christiansen L, Hjelmborg JV, Christensen K. Twin methodology in epigenetic studies. J Exp Biol. 2015;218:134–9.
Li WL, Christiansen L, Hjelmborg J, Baumbach J, Tan QH. On the power of epigenome-wide association studies using a disease-discordant twin design. Bioinformatics 2018;34:4073–8.
Wang W, Li W, Jiang W, Lin H, Wu Y, Wen Y, et al. Genome-wide DNA methylation analysis of cognitive function in middle and old-aged Chinese monozygotic twins. J Psychiatr Res. 2020;136:571–80.
Konki M, Malonzo M, Karlsson IK, Lindgren N, Ghimire B, Smolander J, et al. Peripheral blood DNA methylation differences in twin pairs discordant for Alzheimer’s disease. Clin Epigenet. 2019;11:130.
Webster AP, Plant D, Ecker S, Zufferey F, Bell JT, Feber A, et al. Increased DNA methylation variability in rheumatoid arthritis-discordant monozygotic twins. Genome Med. 2018;10:64.
Li W, Zhang D, Wang W, Wu Y, Mohammadnejad A, Lund J, et al. DNA methylome profiling in identical twin pairs discordant for body mass index. Int J Obes. 2019;43:2491–9.
Wang W, Jiang W, Hou L, Duan H, Wu Y, Xu C, et al. Weighted gene co-expression network analysis of expression data of monozygotic twins identifies specific modules and hub genes related to BMI. BMC Genomics 2017;18:872.
Duan H, Ning F, Zhang D, Wang S, Zhang D, Tan Q, et al. The Qingdao twin registry: a status update. Twin Res Hum Genet. 2013;16:79–85.
Yesavage JA, Brink TL, Rose TL, Lum O, Huang V, Adey M, et al. Development and validation of a geriatric depression screening scale: a preliminary report. J Psychiatr Res. 1982;17:37–49.
Krueger F, Andrews SR. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics 2011;27:1571–2.
Hebestreit K, Dugas M, Klein HU. Detection of significantly differentially methylated regions in targeted bisulfite sequencing data. Bioinformatics 2013;29:1647–53.
Jaffe AE, Irizarry RA. Accounting for cellular heterogeneity is critical in epigenome-wide association studies. Genome Biol. 2014;15:R31.
Rahmani E, Zaitlen N, Baran Y, Eng C, Hu D, Galanter J, et al. Sparse PCA corrects for cell type heterogeneity in epigenome-wide association studies. Nat Methods 2016;13:443–5.
Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36.
Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–5.
McLean CY, Bristor D, Hiller M, Clarke SL, Schaar BT, Lowe CB, et al. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol. 2010;28:495–501.
Pedersen BS, Schwartz DA, Yang IV, Kechris KJ. Comb-p: software for combining, analyzing, grouping and correcting spatially correlated P-values. Bioinformatics 2012;28:2986–8.
Langfelder P, Horvath S. Fast R functions for robust correlations and hierarchical clustering. J Stat Softw. 2012;46:i11.
Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinform. 2008;9:559.
Yip AM, Horvath S. Gene network interconnectedness and the generalized topological overlap measure. BMC Bioinform. 2007;8:22.
Li A, Horvath S. Network neighborhood analysis with the multi-node topological overlap measure. Bioinformatics 2007;23:222–31.
Zhang B, Horvath S. A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol Biol. 2005;4:Article17.
Ravasz E, Somera AL, Mongru DA, Oltvai ZN, Barabasi AL. Hierarchical organization of modularity in metabolic networks. Science 2002;297:1551–5.
Langfelder P, Horvath S. Eigengene networks for studying the relationships between co-expression modules. BMC Syst Biol. 2007;1:54.
Horvath S, Dong J. Geometric interpretation of gene coexpression network analysis. PLoS Comput Biol. 2008;4:e1000117.
Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57.
Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13.
Liang F, Lv K, Wang Y, Yuan Y, Lu L, Feng Q, et al. Personalized epigenome remodeling under biochemical and psychological changes during long-term isolation environment. Front Physiol. 2019;10:932.
Albert PR, Lemonde S. 5-HT1A receptors, gene repression, and depression: guilt by association. Neuroscientist 2004;10:575–93.
Choi M, Wang SE, Ko SY, Kang HJ, Chae SY, Lee SH, et al. Overexpression of human GATA-1 and GATA-2 interferes with spine formation and produces depressive behavior in rats. PLoS ONE 2014;9:e109253.
Redei EE, Andrus BM, Kwasny MJ, Seok J, Cai X, Ho J, et al. Blood transcriptomic biomarkers in adult primary care patients with major depressive disorder undergoing cognitive behavioral therapy. Transl. Psychiatry 2014;4:e442.
Zhang X, Abdellaoui A, Rucker J, de Jong S, Potash JB, Weissman MM, et al. Genome-wide burden of rare short deletions is enriched in major depressive disorder in four cohorts. Biol. Psychiatry 2019;85:1065–73.
Huseby CJ, Hoffman CN, Cooper GL, Cocuron JC, Alonso AP, Thomas SN, et al. Quantification of Tau protein lysine methylation in aging and Alzheimer’s disease. J. Alzheimers Dis. 2019;71:979–91.
de Sousa RT, Loch AA, Carvalho AF, Brunoni AR, Haddad MR, Henter ID, et al. Genetic studies on the tripartite glutamate synapse in the pathophysiology and therapeutics of mood disorders. Neuropsychopharmacology 2017;42:787–800.
Myung W, Song J, Lim SW, Won HH, Kim S, Lee Y, et al. Genetic association study of individual symptoms in depression. Psychiatry Res. 2012;198:400–6.
Shakèd M, Weissmüller K, Svoboda H, Hortschansky P, Nishino N, Wölfl S, et al. Histone deacetylases control neurogenesis in embryonic brain by inhibition of BMP2/4 signaling. PLoS ONE 2008;3:e2668.
Wilson KC, Chen R, Taylor S, McCracken CF, Copeland JR. Socio-economic deprivation and the prevalence and prediction of depression in older community residents. The MRC-ALPHA Study. Br J Psychiatry 1999;175:549–53.
Ye J, Wen Y, Sun X, Chu X, Li P, Cheng B, et al. Socioeconomic deprivation index is associated with psychiatric disorders: an observational and genome-wide gene-by-environment interaction analysis in the UK Biobank Cohort. Biol. Psychiatry 2021;89:888–95.
Acknowledgements
This work was supported by the National Natural Science Foundation of China [grant number 82073641].
Author information
Authors and Affiliations
Contributions
W.J.W. and D.F.Z. designed the work. Y.L.W., H.P.D., and X.C.T. collected samples and phenotypes. Y.L.W., H.P.D., and X.C.T. managed sample data and sequencing data. W.J.W., W.L.L., S.X.L., Q.H.T., and D.F.Z. analyzed the sequencing data and interpreted the analysis results. W.J.W. and W.L.L. drafted the manuscript. S.X.L., Q.H.T., and D.F.Z. substantively revised the manuscript. All authors have read and approved this version of the article to be published. All authors agreed to be responsible for all aspects of the work.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical approval
The authors state that all participants have provided written informed consent for participating in the study, and this study was approved by the local ethics committee at Qingdao CDC, Qingdao, China.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Wang, W., Li, W., Wu, Y. et al. Genome-wide DNA methylation and gene expression analyses in monozygotic twins identify potential biomarkers of depression. Transl Psychiatry 11, 416 (2021). https://doi.org/10.1038/s41398-021-01536-y
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41398-021-01536-y
This article is cited by
-
Mediation role of DNA methylation in association between handgrip strength and cognitive function in monozygotic twins
Journal of Human Genetics (2024)
-
Identification of key DNA methylation changes on fasting plasma glucose: a genome-wide DNA methylation analysis in Chinese monozygotic twins
Diabetology & Metabolic Syndrome (2023)
-
Epigenome-wide association study in Chinese monozygotic twins identifies DNA methylation loci associated with blood pressure
Clinical Epigenetics (2023)
-
Epigenome-wide DNA methylation in obsessive-compulsive disorder
Translational Psychiatry (2022)
-
DNA methylation and waist-to-hip ratio: an epigenome-wide association study in Chinese monozygotic twins
Journal of Endocrinological Investigation (2022)