Abstract
Suicide is the second leading cause of death globally among young people representing a significant global health burden. Although the molecular correlates of suicide remains poorly understood, it has been hypothesised that epigenomic processes may play a role. The objective of this study was to identify suicide-associated DNA methylation changes in the human brain by utilising previously published and unpublished methylomic datasets. We analysed prefrontal cortex (PFC, n = 211) and cerebellum (CER, n = 114) DNA methylation profiles from suicide completers and non-psychiatric, sudden-death controls, meta-analysing data from independent cohorts for each brain region separately. We report evidence for altered DNA methylation at several genetic loci in suicide cases compared to controls in both brain regions with suicide-associated differentially methylated positions enriched among functional pathways relevant to psychiatric phenotypes and suicidality, including nervous system development (PFC) and regulation of long-term synaptic depression (CER). In addition, we examined the functional consequences of variable DNA methylation within a PFC suicide-associated differentially methylated region (PSORS1C3 DMR) using a dual luciferase assay and examined expression of nearby genes. DNA methylation within this region was associated with decreased expression of firefly luciferase but was not associated with expression of nearby genes, PSORS1C3 and POU5F1. Our data suggest that suicide is associated with DNA methylation, offering novel insights into the molecular pathology associated with suicidality.
Similar content being viewed by others
Introduction
Suicide represents a global public health problem, with approximately 800,000 people dying worldwide from suicide annually and suicide attempts up to 20 times more frequent than completed suicide1. Moreover, suicide is the second leading cause of death among young people worldwide and ranks among the 20th leading causes of death across all ages2. The risk for suicidal acts is multifactorial, and consists of a range of biological, psychiatric, psychosocial, and cultural risk factors3. Despite its economic and social burden, the underlying biological aetiology of suicidal behaviour (SB) remains poorly understood.
To date, large-scale genome-wide association studies (GWAS)4,5,6,7 have failed to identify robust associations suggesting that the risk of SB is highly polygenic in nature and that individual gene variants are likely to account only for a small proportion of the total phenotypic variability8. Other factors, such as the environment, behavioural traits, psychiatric diagnosis, lifestyle, and coping mechanisms, are essential regulators of suicide risk and likely to account for more sizeable effects9. Recently, increased understanding of epigenetic processes that occur in the brain has opened promising avenues in suicide research. The epigenome is potentially malleable—changing with age10, in response to specific environmental11 and psychosocial factors12—providing a mechanism for the interaction between genotype and the environment13. Epigenetic processes, including DNA methylation, have recently been implicated in the aetiology of numerous mental health disorders14,15,16,17,18,19,20,21 and SB22,23.
In the last decade, research aiming to understand the contribution of epigenetic mechanisms to SB has implicated the role for key biological pathways, including hypothalamic pituitary adrenal axis, stress response, polyamine system, neurotrophic signalling, and lipid metabolism8. However, studies examining DNA methylation differences associated with SB have primarily focussed on candidate genes24,25,26,27 and few have examined genome-wide DNA methylation changes in the brains of suicide completers22,28. The availability of brain samples is a major challenge for psychiatric research and many previous studies examining DNA methylation variation in suicide are performed on a limited number of post-mortem brain samples22,29,30. Such small studies have reduced statistical power to detect small changes in DNA methylation. The objective of this study was to identify suicide-associated DNA methylation changes in the human brain by utilising previously published and unpublished methylomic datasets.
Genome-wide DNA methylation profiles were available from post-mortem brain samples of suicide completers and non-psychiatric, sudden-death controls for a total of seven cohorts. Methylomic data available for two different brain regions —prefrontal cortex (PFC) and cerebellum (CER) (PFC: 4 cohorts, n = 211; CER: 3 cohorts, n = 114)—were meta-analysed across the suicide cohorts for each brain region separately. We report evidence for altered DNA methylation in suicide cases compared to non-psychiatric controls in both the PFC and CER and examined the functional implications of a top-ranked PFC suicide-associated differentially methylated region (DMR) on gene expression levels in that region. Finally, gene ontology enrichment analysis was performed in each brain region separately to identify pathways of genes associated with suicide completion.
Materials and methods
Sample collection/data recruitment
For the PFC meta-analysis, we included four independent previously published studies22,31,32,33 aimed at profiling DNA methylation in human PFC in individuals with a diagnosed axis-I psychiatric disorder and healthy non-psychiatric controls. Only data from individuals who died by suicide and non-psychiatric controls were included for the initial meta-analysis. In two of the four studies selected31,32, DNA methylation was profiled from fluorescence-activated nuclei sorted neurones, with the remaining two studies performed in bulk tissue22,33. Raw DNA methylation data for all four studies are deposited in the Gene Expression Omnibus (GEO) database (accession number: GSE89707, GSE88890, GSE98203, GSE41826) and full details of the sample cohort can be obtained from the original studies22,31,32,33. For the CER meta-analysis, three DNA methylation datasets were included two of which are currently unpublished (GSE137222 and GSE137223). Raw DNA methylation data for the CER published EWAS study33 is deposited in GEO database (accession number: GSE89702). The unpublished studies were approved by the University of Exeter Medical School Research Ethics Board (REB). In all three CER cohort’s DNA methylation profiles were derived from bulk brain tissue and cases were individuals who died by suicide (hanging, jumping from height, intentional poisoning, self-harm/bleeding). Cause and manner of death as well as joint presence of psychiatric diagnosis were determined by a forensic pathologist after evaluating autopsy results, circumstances of death, data from extensive toxicological testing, police reports, family interviews, and medical records. Controls were individuals who died suddenly (e.g. cardiac failure, viral infection, or accidents) and did not have evidence of axis-I disorders. See Supplementary Table S1 for a complete description of sample selection, numbers, and demographic characteristics of each cohort.
DNA methylation analysis
DNA methylation was measured using the Illumina HumanMethylation450K BeadChip (‘Illumina 450K array’) or Infinium MethylationEPIC BeadChip (‘Illumina EPIC array’, one CER cohort) platform (Illumina Inc., San Diego, CA, USA). To ensure consistency of the methodological approach, raw DNA methylation data (idat files) were recovered and each cohort was independently reanalysed, applying the same quality control (QC) and pre-processing pipelines. Briefly, QC checks, quantile normalisation, and separate background adjustment of methylated and unmethylated intensities of type I and II probes were employed using the wateRmelon package in R34. Probes on the X- and Y-chromosomes were used to confirm sample sex. Only samples which passed stringent QC measures (>1% of sites with a detection P value (P) >0.01) were included. Probes with a detection P > 0.01 in at least 1% of samples and/or a beadcount <3 in 5% of samples, non-specific probes, potentially cross-reactive probes, or probes near SNPs35,36 were removed across all samples. Only probes common to both the 450k array and EPIC array were included in downstream analyses for the CER. For the annotation of probes, the University of California, Santa Cruz (UCSC) RefGene name from Illumina’s annotation file and enhanced annotation to the UCSC Known Gene were used. All annotations used the human February 2009 (GRCh37/hg19) assembly.
Estimating differential neuronal proportions
The R package (available at www.cran.r-project.org), Cell EpigenoType Specific (CETS) mapper, designed for the quantification and normalisation of differing neuronal proportions in genome-wide DNA methylation datasets was used as previously described32 to estimate brain cellular heterogeneity in each of the four PFC cohorts. Similar estimates could not be obtained for the CER cohorts as the algorithm for the correction of brain cellular heterogeneity bias was developed using post-mortem frontal cortex data and NeuN is not expressed in CER purkinje neurons32.
Data analysis
Statistical analyses were performed using R statistical package (version 3.4.3). The β-value is a ratio between methylated probe intensity and total probe intensities (sum of methylated and unmethylated probe intensities) and ranges from 0 to 1. Linear regression was used to examine differences in DNA methylation scores (reported as change in β-value (∆β)) between suicide cases and controls at each CpG site, controlling for potential confounders. Covariates included in all models were age, sex, and chip. We also included ethnicity or brain bank as covariates in the model for those cohorts where that information was available and represented a potential source of variation. In the PFC cohorts only, we also adjusted for estimated neuronal proportions. For one study22, DNA methylation differences were investigated across individual-matched cortical regions (Brodmann area 11 (BA11), Brodmann area 25 (BA25)) by fitting a linear mixed-effect model (LMM) using the lme4 R package (available at https://cran.r-project.org37). Whereby, brain region and sample ID were included in the model as random effects (‘within participants’ factors) while diagnosis, age, sex, PH, and cellular composition were included in the model as fixed effects.
Meta-analyses
Suicide completers versus non-psychiatric controls
The results obtained from the linear regression were then meta-analysed for each brain region independently. A fixed-effect model, using the ‘metagen’ function in the R package ‘meta’, was applied by providing the regression coefficients and standard errors from each individual cohort to calculate weighted pooled estimates and to test for significance. Experiment-wide significance (P < 1E-07) (threshold estimated from permutation analysis in a larger dataset (N = 675 individuals) generated previously by our group38) was chosen as a multiple testing threshold to determine statistically significant DNA methylation changes.
Suicide completers versus non-suicide psychiatric controls
In order to assess whether the observed suicide-associated DNA methylation changes identified in our original meta-analysis were driven by the psychiatric disorder comorbidity rather than being suicide-specific changes, a second exploratory analysis was performed in additional samples obtained from the CER datasets only, which had additional non-suicide psychiatric samples with DNA methylation data available (not included in the primary analysis report here). In the second meta-analysis, each CER cohort consisted of suicide cases (that were included in the original meta-analysis) and psychiatric controls, where individuals had a diagnosed axis-I disorders (major depressive disorders (MDD), schizophrenia (SZ), bipolar disorder (BD)) but had no documented evidence of SB. In total, the secondary meta-analysis included 130 samples (case group, N = 50; psychiatric control group, N = 80). Results obtained from the linear regression were then meta-analysed using a fixed-effect model as described previously.
Region-based analysis
The results obtained from both the PFC and CER meta-analyses were used to perform a regional-based analysis using the Python module Comb-p39 to identify suicide-associated DMRs. The Comb-p software groups spatially correlated DMPs (seed P < 1E-03, minimum of three probes) at a maximum distance of 500 bp in each brain region. DMR P were corrected for multiple testing using Šidák correction40.
Gene ontology term enrichment analysis
A previously described logistic regression approach41 was used to test if genes (Illumina UCSC gene annotation) annotated to probes in our PFC and CER meta-analyses (DMPs with P ≤ 1E-04) predicted pathway membership, while controlling for the number of probes annotated to each gene. Briefly, pathways were downloaded from the Gene Ontology (GO) website (http://geneontology.org/) and all genes annotated to parent terms were also included. Genes containing at least one Illumina probe and annotated to at least one GO pathway were considered. Pathways were filtered to those containing between 10 and 2000 genes and a list of significant (after correction for multiple testing—Bonferroni correction) pathways were identified as previously described41.
Functional follow-up of significant DNA methylation findings
Tissue (N = 71) from two regions of the cortex, BA11 (N = 38) and BA25 (N = 33), collected from 20 MDD suicide cases and 20 non-psychiatric sudden-death controls was obtained from the Douglas Bell Canada Brain Bank (DBCBB) (http://douglasbrainbank.ca/), further details are available in ref. 22. Previously, our group performed DNA methylation profiling in these samples22 and the results of that study were included in this meta-analysis study. To examine whether our identified suicide-associated DMR (PSORS1C3 DMR; Chr6:31,148,370-31,148,553 (Hg19), 2694 bp downstream the TSS of PSORS1C3 gene) is associated with the expression of nearby genes, we measured expression levels of two nearby genes in these brain tissue samples. We tested for an association with gene expression firstly at the closest transcription start site (TSS) gene—the lncRNA gene, PSORS1C3—and then at the second closest gene, POU5F1 (see Supplementary Fig. S1 for details).
Gene expression analysis
Thirty milligrams of frozen PFC tissue from each brain sample was homogenised with Qiazol Lysis Reagent (Qiagen, Valencia, CA, USA), as per the manufacturer’s instructions, before running it through a QIAshredder (Qiagen, Valencia, CA, USA). Total RNA was extracted using the Qiagen miRNeasy Mini column-purification system and treated with DNase I as outlined by the manufacturer. The Agilent 2100 Bioanalyzer was used to check the quality and concentration of the extracted RNA samples. One microgram of total RNA was reverse transcribed into complementary DNA (cDNA) (20 μL reactions) according to the manufacturer’s instructions using the Invitrogen VILO cDNA synthesis kit (Life Technologies Ltd, Paisley, UK). Three housekeeping genes Ubiquitin Conjugating Enzyme E2 D2 (UBE2D2), Cytochrome C1 (CYC1), and Ribosomal Protein L13 (RPL13) identified previously42 as being among the most stably expressed in the brain were selected to normalise the target gene expression. Next, quantitative RT-PCR was performed in triplicate for each assay using the StepOnePlus Real-Time PCR machine (Applied Biosystems, Foster City, Calif) and pre-optimised Taqman gene expression assays (Applied Biosystems, Foster City, Calif). A full list of the qPCR assays used is given in Supplementary Table S2. PCR cycling conditions were as follows: 50 °C for 2 min, 95 °C for 20 s, and 40 cycles of 95 °C for 10 s, and 60 °C for 20 s. We undertook stringent QC of raw qPCR data, repeating samples where there was high variability between triplicates (Ct > 0.5). The abundance of each test gene was determined by the comparative Ct method43, expressed relative to the geometric mean of the three housekeeping genes. Data were log2-transformed to ensure normal distribution and presented as a fold-difference in expression of suicide cases relative to controls using the 2−ΔΔCT method. To assess whether POU5F1 expression levels were associated with a history of suicide, we used a LMM using the lme4 R package (available at https://cran.r-project.org)37 where ΔCt values of the target gene (POU5F1) was the response variable. Brain region and sample ID were included in the model as random effects (‘within participants’ factors) while diagnosis, age, sex, and neuronal proportion were included in the model as fixed effects. Finally, since 450K array data were available from the same individuals, we examined the correlation between gene expression levels and mean DNA methylation levels at the DMR.
Reporter constructs
The PSORS1C3 DMR sequence was inserted into the pCpGL-basic vector (see ref. 44 for details), which is devoid of CpG sites and was generously provided by the Rehli laboratory44. Briefly, the cleaned PSORS1C3 PCR amplicon was inserted into a digested pCpGL-basic plasmid using T4 ligase and buffer (Invitrogen, California, USA). Ligated plasmids were transformed into One Shot PIR1 E. coli (ThermoFisher Scientific, Massachusetts, USA) to allow for monoclonal amplification of the recombinant plasmids. For the transformation, 50 µl of One Shot PIR1 E. coli was used, including a negative ligation control and a positive transformation control (pUC19) using standard procedures.
Clones were subsequently checked by clonal PCR, restriction digest using BglII and NcoI and Sanger sequencing (see Supplementary Fig. S2) to confirm the DMR had been inserted in the correct orientation. The pCpGL construct was methylated in vitro using M.sssI methyltransferase (New England Biolabs, Massachusetts, USA) in the presence of S-adenosylmethionine (SAM) following the manufacture’s protocol. An empty pCpGL-basic vector was also methylated to act as a control. To confirm successful methylation, the plasmids underwent digestion with the methylation sensitive enzyme HpaII (see Supplementary Fig. S3).
Cell culture and transfections in HEK293 cells were cultured in Dulbecco’s modified Eagle’s medium (þ4.5 g l_1 d-glucose, l-glutamate, pyruvate) (Gibco) with 10% foetal bovine serum (Gibco) at 37 °C and 5% CO2. Briefly, 2 × 105 cells were seeded in six-well plates. The following day media was removed, the cells washed with PBS, and 1.5 ml of fresh growth media was added. Five hundred nanograms of PSORS1C3 or pCpGL plasmid and 100 ng of pGL4.74[hRluc/TK] reporter control vector (Promega, Wisconsin, USA) were diluted in 500 µl of Opti-MEM reduced serum media (Gibco, Massachusetts, USA) in an Eppendorf and left to equilibrate for 5 min at room temperature. In all, 4.5 µl of Lipofectamine LTX Reagent (ThermoFisher Scientific, Massachusetts, USA) was then added to each Eppendorf and incubated for 30 min at room temperature. Following incubation, 500 µl of LTX plasmid mix was added to the HEK293 cells in six-well plates. Cells were then incubated at 37 °C, 5% CO2, for 24 h to allow expression of firefly and Renilla luciferases.
Dual-luciferase assay
Twenty-four hours following transfection a dual-luciferase reporter assay (Promega, Wisconsin, USA) was carried out to measure the expression of firefly luciferase and Renilla luciferase in the transfected cells as per the manufacturers’ instructions. Each experiment contained three technical repeats and the experiment was repeated three times. The injections and light absorbance were carried out automatically using the pherastar plate reader. The average firefly luciferase activity was calculated by averaging absorbance readings between 2 and 10 s. The average Renilla luciferase activity was calculated by averaging absorbance readings between 14 and 22 s. Data analysis was carried out as described in ref. 45. All data are presented as a normalised firefly luciferase activity relative to Renilla luciferase. Fold change expression was calculated by dividing the unmethylated normalised firefly luciferase activity by the respective methylated normalised luciferase activity. A Student’s T-test was used to compare the methylated versus unmethylated vectors.
Results
Suicide-associated DMPs in human cortex and cerebellum
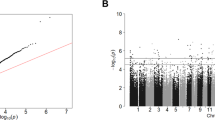
An overview of the methodological approach used in this study is given in Supplementary Fig. S4. We identified one DMP (cg00963169) in the PFC, which reached experiment-wide significance (P = 3.30E-08 (Fig. 1a)). The effect size at this CpG site, located downstream of exon 1 of the neuron-specific protein coding gene, ELAV-like RNA binding protein 4 (ELAVL4), was largely consistent across all cohorts included (see Supplementary Fig. S5), showing hypomethylation in suicide cases relative to controls. Interestingly, the 20 most significant (P < 5E-05) suicide-associated differentially methylated loci identified in the PFC, listed in Supplementary Table S3, include probes in the vicinity of several loci previously implicated in psychiatric phenotypes. In the CER, six probes (cg14392966, cg17855963, cg25590492, cg12284382, cg10757978, cg04525580) passed the experiment-wide significance threshold (P < 1E-07) (Fig. 1b). Of interest the top-ranked DMP, cg14392966 (P = 3.06E-11), which is located within the coding region (exon 1) of the PUS3 gene on Chr11, has been previously associated with severe neurodevelopmental disorders46. Supplementary Figure S6 shows that this DMP is hypomethylated in two of the three cohorts in suicide cases compared to healthy controls. A list of the top 20 DMPs in the CER is provided in Supplementary Table S4.
Manhattan plot showing site-specific genome-wide pattern of DNA methylation in a the human prefrontal cortex (PFC) and b the human xerebellum (CER). One CpG site was identified as differentially methylated between suicide cases and healthy controls at experiment-wide significant (P = 1E-07) in the PFC. Six CpG sites were identified as differentially methylated between suicide cases and healthy controls at experiment-wide significant (P = 1E-07) in the CER.
Region-based analysis of altered DNA methylation in suicide completers
We used the python module, Comb-p39, to identify DMRs in suicide cases compared to controls in each brain region. The regional analysis identified three and eight significant (Sidak-corrected P < 0.05) DMRs in the PFC and CER, respectively (see Table 1 for details). In the PFC, the top-ranked DMR was found within the WRB gene (Sidak-corrected P = 5.11E-06) and was consistently hypomethylated across all five CpG sites in suicide cases relative to controls, in all four PFC methylomic studies (Fig. 2). Of interest, the second top-ranked suicide-associated DMR in the PFC (Sidak-corrected P = 3.81E-05) was located downstream of the promoter region of the PSORS1C3 non-coding gene, a DMR previously reported by our group as associated with MDD suicide completers22.
Plot showing the top-ranked DMR in the PFC. This DMR, spanning 5 CpG sites and located in the promoter region of the WRB gene (Sidak-corrected P = 5.11E-06), was found consistently hypomethylated in suicide cases compared to healthy controls, across all 4 suicide brain cohorts. The solid line is for illustration purposes and not indicative that the CpG sites between sites are also methylated.
In the CER, the top-ranked suicide-associated DMR was identified on chromosome 22, distributed along the intronic region of the CERC2 gene and spanning 4 CpG sites. The CERC2-associated DMR (Fig. 3) showed significant hypermethylation (Sidak-corrected P = 5.68E-07) across all 4 CpG sites within the region in suicide cases compared with controls. The direction of this change was found to be consistent across all 3 CER methylomic studies.
Plot showing the top-ranked DMR in the CER. This DMR, spanning 4 CpG sites and located within the coding region of the CECR2 gene (Sidak-corrected P = 5.68E-07), was found consistently hypermethylated in suicide cases compared to healthy controls, across 3 suicide brain cohorts. The solid line is for illustration purposes and not indicative that the CpG sites between sites are also methylated.
Pathway analysis
The biological relevance of our findings was investigated through gene ontology analysis on genes annotated to suicide-associated DMPs (P ≤ 1E-04). Results revealed an enrichment of DNA methylation alterations in genes involved in cognitive processes such as long-term synaptic depression and brain development (See Supplementary Table S5 and S6).
Suicide-associated DMPs identified in the CER are largely independent of comorbid psychiatric disorders
In order to disentangle the relative contribution of psychopathologies from DNA methylation changes specific to suicide, we performed an additional meta-analysis in the CER whereby all the non-psychiatric controls initially included in our meta-analysis were excluded and individuals with documented Axis-I psychiatric disorders (MDD, SZ, BD) and who died by suicide were compared to psychiatric cases without a documented history of SB/suicide fatalities (3 cohorts, N = 130, cases N = 50, controls N = 80). Comparison of the results (effect sizes at the top 500 (P < 0.05) DMPs) from our original CER meta-analysis and the secondary analysis (Supplementary Fig. S7) revealed a strong positive correlation (P = 2.2E-16; R = 0.89). Moreover, 2 of the 6 CER-associated DMPs (P < 1E-07) were nominally significantly differentially methylated in our suicide cases versus psychiatric controls analysis (cg10757978, P = 1.19E-04; cg04525580, P = 0.017) (see Supplementary Table S7 for details) and a similar direction of effect was observed for both analyses for the remaining 4 CER-associated DMPs. We were unable to perform the same analysis in the PFC due to lack of samples from individuals with an axis-I diagnosis who did not die by suicide.
Functional validation and gene expression analysis of the PSORS1C3 DMR
Given our replication of a PSORS1C3 DMR in suicide22 we aimed to functionally evaluate its effect on the expression of nearby genes. First, we examined the effect of DNA methylation at the suicide-associated DMR on nearby gene expression using a CpG-Free Luciferase Reporter (pCpGL vector) gene assay47. Next, we examined gene expression levels of nearby annotated genes (PSORS1C3 and POU5F1) in a subset of samples for which brain tissue was available (two brain regions (BA11 and BA25, N = 70; suicide cases (N = 36), non-psychiatric controls (N = 34)) and examined the correlation between expression levels of our selected target genes and mean DNA methylation at the suicide-associated DMR.
We found a marked increase in the relative expression of firefly luciferase activity normalised to Renilla luciferase in the unmethylated PSORS1C3 cloned pCpGL vector compared to the methylated vector (Fig. 4) (fold change = 206, P = 0.006, N = 3). Next, we quantified gene expression levels of the PSORS1C3 long non-coding gene in a cohort of 70 post-mortem brain samples (BA11, N = 38; BA25, N = 32, both regions obtained from the same individuals). Our analysis showed that in our sample set, PSORS1C3 lncRNA was not expressed in the PFC (Ct Value >31 or undetermined). This result is consistent with findings in Genotype-Tissue Expression (GTEx) portal48 (https://www.gtexportal.org/home/gene/PSORS1C3), which shows little to no expression for this gene in brain samples examined. Next, we examined expression levels of the second closest gene (POU5F1) to the suicide-associated DMR. An LMM was used to compare mean dCt values between suicide cases and non-psychiatric controls and the analysis showed no significant difference in gene expression levels between the two groups (P = 0.598; Supplementary Fig. S8A). Furthermore, we found no significant correlation between mean DNA methylation at the suicide-associated region PSORS1C3 DMR and POU5F1 gene expression levels (Pearson’s R = −0.04, P = 0.67; Supplementary Fig. S8B).
There is a significant increase in firefly luciferase expression when normalised to Renilla luciferase, the normalised expression ratios of firefly activity increases to 3.81 when unmethylated versus 0.0269 when methylated in vitro (Students t-test, P = 0.0057, n = 3) which corresponds to a 206-fold change increase in luciferase activity in the unmethylated PSORS1C3 cloned vector compared to the methylated identical vector. N = 3 experimental repeats with each experiment containing three technical repeats. Error bars represent ± the standard error.
Discussion
In this study, we utilised previously published and unpublished methylomic datasets to perform a meta-analysis of variable DNA methylation in the brain of suicide completers. DNA methylation data were available for two different brain regions (PFC: 4 cohorts, N = 211; CER: 3 cohorts, N = 114) and data were meta-analysed across the suicide cohorts for each brain region separately. To our knowledge, this represents the most extensive methylomic study of suicide completers using post-mortem brain tissue to date.
While several studies suggest the involvement of the PFC in SB22,49,50,51, suicide-associated epigenetic changes in the CER have not been investigated. However, the CER is known to play an important role in motor control, cognition, and emotional processing and is involved in a variety of psychiatric disorders, including depression, bipolar disorder, and schizophrenia52. Suicidal behaviour in those with depression has been associated with a decreased cerebellar volume53 and low regional cerebral blood flow in the cerebellum54. Moreover imaging studies have reported structural abnormalities associated to suicide attempt in MDD in cortical and subcortical regions, including cerebellum55 and alterations in functional cerebellum networks were found in depressed patients with a suicide attempt history56. Altogether these studies support the hypothesis of a potential involvement of CER in the psychopathology of attempted suicide in patients with MDD. We set out to further explore the role of DNA methylation and suicidality in this region as well as to determine if suicidality associated DNA methylation changes are brain region specific.
We first examined site-specific genome-wide patterns of DNA methylation in suicide cases compared with controls in the PFC and CER separately. We identified one DMP (cg00963169, P = 3.30E-08) in the PFC, which reached experiment-wide significance. This probe, located in the intronic region of the ELAVL4 gene, shows consistent hypomethylation in suicide cases compared to controls. The ELAVL4 gene has a known role in translation and stabilisation of mRNA, especially in the brain, and acts as a negative regulator of proliferation, activity, and differentiation in neural stem cells57,58. Through their mRNA stabilising activities, this family of proteins modulate neuronal development and maintenance, and their altered activity has been implicated in neurological conditions59 and disorders including Alzheimer’s disease60, schizophrenia61, and autism62. Of interest, the association with suicide at this site was found to be largely driven by PFC cohorts derived from neuronal nuclei (see Supplementary Fig. S5) and thus future replication of this finding in sorted neuronal cells may yield more significant associations with suicide.
In the CER, we identified six probes (cg14392966, cg17855963, cg25590492, cg12284382, cg10757978, cg04525580) at experiment-wide significance threshold (P < 1E-07). The top-ranked DMP, cg14392966 (P = 3.06E-11), is located in exon 1 of the PUS3 gene. PUS3 encodes a highly conserved enzyme responsible for post-transcriptional modification of tRNA and has previously been associated with intellectual disability63 and severe neurodevelopmental disorders46. The remaining DMPs include probes in the vicinity of several loci previously implicated in pathways relevant to psychiatric phenotypes. For example, ZIC1 is thought to play an important role in neurogenesis and cerebellum differentiation64, whereas RASD2 is known to modulate dopaminergic neurotransmission65. Furthermore, the probe cg04525580 (P = 9.08E-08) is located at the 5′UTR of the interferon regulatory factor 2 (IRF2) gene. This locus plays an important role in transcriptional activation at promoters66 and regulates the expression of a variety of genes involved in immune responses in the brain67, further supporting a role for immune-related pathways in suicide.
To increase the power of our study to identify changes in DNA methylation between cases and controls and given that DNA methylation at adjacent probes is often correlated, we employed the regional-based analysis, Comb-p, to identify DMRs. Our analysis identified three and eight significant (Sidak-corrected P < 0.05) DMRs in the PFC and CER, respectively. In the PFC, the top-ranked DMR, located in intron 1 of the WRB gene, is consistently hypomethylated across all five CpG sites in suicide cases relative to controls, in all cohorts. Recent studies suggest a role for Wrb in photoreceptor synaptic transmission in zebrafish68 and the WRB locus was reported among the differentially expressed genes in a mouse model study looking at cognitive impairment and neuropathology in Down syndrome brain69 further supporting the hypothesis of its involvement in the correct development and functioning of the CNS.
The second top-ranked suicide-associated DMR in the PFC (Sidak-corrected P = 3.81E-05) was located downstream the promoter region of the PSORS1C3 non-coding gene, a DMR previously reported by our group as associated with MDD suicide completers22. Although the function of this gene product remains unclear, it is thought to be a potential regulator of nearby immune-related genes70 and is a known risk gene for psoriasis71,72, supporting a role in immune system regulation. To gain further insight into the role of this suicide-associated PSORS1C3 DMR on nearby gene expression, we first used a dual-luciferase assay to determine if DNA methylation at this region decreases expression of firefly luciferase in a CpG-free vector. The methylated PSORS1C3 DMR construct was significantly associated with decreased expression of the reporter gene product, indicating that the methylation status of this DMR has the potential to modify promoter activity; however, the identity of the modified gene product is unknown. We found no evidence of suicide-associated differential gene expression of nearby genes, PSORS1C3 and POU5F1, in the PFC. We hypothesise that DNA methylation changes at this suicide-associated DMR are associated with a different nearby gene or an unknown splice variant of either the PSORS1C3 or POU5F1 genes. Finally, an additional DMR was identified in the PFC, located on chromosome 22, in the promoter region of the LGALS1 gene. This gene is thought to play a role in immune system functioning65 and DNA methylation changes at this locus have previously been implicated in schizophrenia33.
In the CER, the top-ranked suicide-associated DMR, located in the intronic region of the CERC2 gene on chromosome 22, spans four CpG sites. This locus is known to be involved in the control of the periodic oscillation of cyclin E expression in proliferating cells likely through its histone deacetylase activity73. The CERC2-associated DMR showed significant hypermethylation (Sidak-corrected P = 5.68E-07) across all four CpG sites within the region in suicide cases compared with controls and the direction of this change was found to be consistent across all three independent CER methylomic studies. To the best of our knowledge, this gene has not been previously implicated in the pathology of SB. Seven additional suicide-associated DMRs were identified in the CER. Of interest is the DMR located in exon 10 of the SLC44A4 gene; a gene recently implicated in a study looking at the role of the major histocompatibility complex region in schizophrenia susceptibility74. An additional suicide-associated DMR, worthy of further investigation, is located in exon 3 of the WWTR1 gene, a transcriptional coactivator known for its role in preserving neuronal health75. Furthermore, a missense variant in this gene was recently associated with lower cognitive ability in a GWAS study for infant mental and motor ability76. Finally, we identified a suicide-associated DMR located downstream the promoter region of MED13L gene and genetic variants at this locus have been widely reported as associated with intellectual disability77,78, suggesting that this gene may play an important role in neurological development.
Since SB is often a complication of a psychiatric disorder, distinguishing suicide diathesis-related DNA methylation changes from those associated with mood disorders and other psychiatric disorders has remained a challenge. In order to unravel the relative contribution of psychopathologies from DNA methylation changes specific to suicide, we performed an additional meta-analysis in the CER whereby individuals with documented Axis-I psychiatric disorders (MDD, SZ, BD) and who died by suicide were compared to psychiatric cases without a documented history of SB. Comparison of the results (effect size of the top 500 nominally significant (P < 0.05) DMPs) from our original CER meta-analysis and the secondary analysis revealed a strong positive correlation (P = 2.2E-16; R = 0.89). Moreover, we replicated our findings for two of the six CER-associated DMPs, which reached multiple testing threshold in our suicide cases versus psychiatric controls meta-analysis (cg10757978, P = 1.19E-04; cg04525580, P = 0.017) and we observed similar direction of effect for the remaining loci. Taken together these findings suggest that suicide-associated DMPs identified in the CER are largely independent of comorbid psychiatric disorders. Unfortunately, we were unable to perform the same analysis in the PFC due to the limited number of samples that did not die by suicide but had an Axis-I diagnosis.
Despite the power of the methodological approaches used in this study, there are several caveats. First, the modest number of studies included made this meta-analysis relatively underpowered to detect small changes in DNA methylation. Despite this we were able to identify several statistically significant DMPs and DMRs in both brain regions. Another major limitation is that bulk brain tissue was used in most of the studies included in our meta-analysis and cellular heterogeneity is a well-known confounder in DNA methylation studies. In order to bypass this issue, we used a previously reported in silico method to estimate the neuronal proportion in each sample in bulk PFC cohorts and included these estimates in the statistical models32. This method could not be applied to our analysis of the CER and thus it is plausible that cellular heterogeneity is confounding some of our CER results. Third, recent research has implicated the importance of other DNA modifications (i.e., 5-hydroxymethyl cytosine) in the brain79. Our measure of DNA methylation in this study cannot be distinguished from 5-hydroxymethyl cytosine (5hmC). Of interest, we examined the presence of detectable 5hmC levels at statistical significant DMPs identified in this study in their respective brain regions using the Hydroxymethylation Annotation in Brain Integrative Tool (HABIT) tool (http://epigenetics.iop.kcl.ac.uk/HMC/)41. This tool identified detectable levels of 5hmC at the following suicide-associated DMPs cg00963169 (PFC), and cg17855963 and cg04525580 (CER), suggesting that the majority of DMPs identified in this study are not confounded by 5hmC. However, future studies should attempt to examine the role of 5hmC in SB. Fourth, medication data, smoking information, and method of suicide were not available for all individuals; thus, we cannot rule out the possibility that the observed DNA methylation changes are influenced by these potential confounders.
Fifth, we acknowledge the possibility that many of the associations reported (DMPs/DMRs) could be related to the severity and/or duration of the mental health disorder. From our secondary analysis in the cerebellum we show that for certain top-ranked DMPs the association appears to be suicide associated rather than associated with the underlying mental health disorder. Given the lack of information related to severity and/or duration of mental illness for samples included in this meta-analysis we cannot rule out the contribution of the above-mentioned confounders.
Finally, although our study presents evidence for novel DNA methylation changes associated with suicide, further replication using a larger sample size is required to support these results. In addition, future studies could also examine the transcriptional consequences of the observed DNA methylation changes at the PSORS1C3 DMR on additional nearby genes and/or novel splice variants in the region. There is considerable interest in using DNA methylation-based biomarkers as predictors for suicide risk and previous studies30,80,81,82,83,84 have identified polymorphic CpGs that can act as a unique molecular signature for suicide prediction. The data from this study provide many more candidate regions as potential biomarkers for suicide risk and also identifies genes/networks potentially dysregulated in suicidal brain.
In summary, our data, which utilise several published and unpublished suicide cohorts, have identified DMPs and several DMRs associated with suicide in both the PFC and CER, including the previously identified DMR upstream of the PSORS1C3 non-coding gene. We show that this DMR can influence gene expression using a dual-luciferase assay, but we have yet to identify its target gene.
References
CDC. Suicide Mortality by State Report. https://www.cdc.gov/nchs/pressroom/sosmap/suicide-mortality/suicide.htm (2017).
WHO. Global Health Observatory (GHO) Data. https://www.who.int/news-room/fact-sheets/detail/suicide (2018)
Malone, K. M. et al. Protective factors against suicidal acts in major depression: reasons for living. Am. J. Psychiatry 157, 1084–1088 (2000).
Schosser, A. et al. Genomewide association scan of suicidal thoughts and behaviour in major depression. PLoS ONE 6, e20690 (2011).
Willour, V. L. et al. A genome-wide association study of attempted suicide. Mol. Psychiatry 17, 433–444 (2012).
Galfalvy, H. et al. A pilot genome wide association and gene expression array study of suicide with and without major depression. World J. Biol. Psychiatry 14, 574–582 (2013).
Mullins, N. et al. GWAS of suicide attempt in psychiatric disorders and association with major depression polygenic risk scores. Am. J. Psychiatry. https://doi.org/10.1176/appiajp201918080957 (2019).
Policicchio, S., Dempster, E. L. & Murphy, T. M. Deciphering the epigenetic landscape of suicidal behaviour: a review of current findings, caveats and future directions. OBM Genet. 2, 039 (2018).
Turecki, G. & Brent, D. A. Suicide and suicidal behaviour. Lancet 387, 1227–1239 (2016).
Bell, J. T. et al. Epigenome-wide scans identify differentially methylated regions for age and age-related phenotypes in a healthy ageing population. PLoS Genet. 8, e1002629 (2012).
Feil, R. & Fraga, M. F. Epigenetics and the environment: emerging patterns and implications. Nat. Rev. Genet. 13, 97–109 (2012).
Burns, S. B., Szyszkowicz, J. K., Luheshi, G. N., Lutz, P. E. & Turecki, G. Plasticity of the epigenome during early-life stress. Semin. Cell Dev. Biol. 77, 115–132 (2018).
Mill, J. & Heijmans, B. T. From promises to practical strategies in epigenetic epidemiology. Nat. Rev. Genet. 14, 585–594 (2013).
Wong, C. C. et al. Methylomic analysis of monozygotic twins discordant for autism spectrum disorder and related behavioural traits. Mol. Psychiatry 19, 495–503 (2014).
Pidsley, R. et al. Methylomic profiling of human brain tissue supports a neurodevelopmental origin for schizophrenia. Genome Biol. 15, 483 (2014).
Uddin, M. et al. Epigenetic and immune function profiles associated with posttraumatic stress disorder. Proc. Natl Acad. Sci. USA 107, 9470–9475 (2010).
Fisher, H. L. et al. Methylomic analysis of monozygotic twins discordant for childhood psychotic symptoms. Epigenetics 10, 1014–1023 (2015).
Murphy, T. M. et al. Anxiety is associated with higher levels of global DNA methylation and altered expression of epigenetic and interleukin-6 genes. Psychiatr. Genet. 25, 71–78 (2015).
Dempster, E. L. et al. Genome-wide methylomic analysis of monozygotic twins discordant for adolescent depression. Biol. Psychiatry 76, 977–983 (2014).
Davies, M. N. et al. Hypermethylation in the ZBTB20 gene is associated with major depressive disorder. Genome Biol. 15, R56 (2014).
Uddin, M. et al. Epigenetic and inflammatory marker profiles associated with depression in a community-based epidemiologic sample. Psychol. Med. 41, 997–1007 (2011).
Murphy, T. M. et al. Methylomic profiling of cortex samples from completed suicide cases implicates a role for PSORS1C3 in major depression and suicide. Transl. Psychiatry 7, e989 (2017).
Murphy, T. M. et al. Genetic variation in DNMT3B and increased global DNA methylation is associated with suicide attempts in psychiatric patients. Genes Brain Behav. 12, 125–132 (2013).
Labonte, B. et al. Genome-wide epigenetic regulation by early-life trauma. Arch. Gen. Psychiatry 69, 722–731 (2012).
Ernst, C. et al. Alternative splicing, methylation state, and expression profile of tropomyosin-related kinase B in the frontal cortex of suicide completers. Arch. Gen. Psychiatry 66, 22–32 (2009).
Fiori, L. M. & Turecki, G. Epigenetic regulation of spermidine/spermine N1-acetyltransferase (SAT1) in suicide. J. Psychiatr. Res 45, 1229–1235 (2011).
Keller, S. et al. Increased BDNF promoter methylation in the Wernicke area of suicide subjects. Arch. Gen. Psychiatry 67, 258–267 (2010).
Haghighi, F. et al. Increased DNA methylation in the suicide brain. Dialogues Clin. Neurosci. 16, 430–438 (2014).
Fiori, L. M., Gross, J. A. & Turecki, G. Effects of histone modifications on increased expression of polyamine biosynthetic genes in suicide. Int. J. Neuropsychopharmacol. 15, 1161–1166 (2012).
Maussion, G. et al. Functional DNA methylation in a transcript specific 3’UTR region of TrkB associates with suicide. Epigenetics 9, 1061–1070 (2014).
Kozlenkov, A. et al. DNA methylation profiling of human prefrontal cortex neurons in heroin users shows significant difference between genomic contexts of hyper- and hypomethylation and a younger epigenetic age. Genes (Basel) 8, 152 (2017).
Guintivano, J., Aryee, M. J. & Kaminsky, Z. A. A cell epigenotype specific model for the correction of brain cellular heterogeneity bias and its application to age, brain region and major depression. Epigenetics 8, 290–302 (2013).
Viana, J. et al. Schizophrenia-associated methylomic variation: molecular signatures of disease and polygenic risk burden across multiple brain regions. Hum. Mol. Genet. 26, 210–225 (2017).
Pidsley, R. et al. A data-driven approach to preprocessing Illumina 450K methylation array data. BMC Genomics 14, 293 (2013).
Price, M. E. et al. Additional annotation enhances potential for biologically-relevant analysis of the Illumina Infinium HumanMethylation450 BeadChip array. Epigenet. Chromatin 6, 4 (2013).
Chen, Y. A. et al. Discovery of cross-reactive probes and polymorphic CpGs in the Illumina Infinium HumanMethylation450 microarray. Epigenetics 8, 203–209 (2013).
Sarkar, D. Multivariate Data Visualization with R. http://lmdvr.r-forge.r-project.org. Accessed 2008 (2008).
Hannon, E. et al. An integrated genetic-epigenetic analysis of schizophrenia: evidence for co-localization of genetic associations and differential DNA methylation. Genome Biol. 17, 176 (2016).
Pedersen, B. S., Schwartz, D. A., Yang, I. V. & Kechris, K. J. Comb-p: software for combining, analyzing, grouping and correcting spatially correlated P-values. Bioinformatics 28, 2986–2988 (2012).
Šidák, Z. Rectangular confidence regions for the means of multivariate normal distributions. J. Am. Stat. Assoc. 62, 626–633 (1967).
Lunnon, K. et al. Variation in 5-hydroxymethylcytosine across human cortex and cerebellum. Genome Biol. 17, 27 (2016).
Rydbirk, R. et al. Assessment of brain reference genes for RT-qPCR studies in neurodegenerative diseases. Sci. Rep. 6, 37116 (2016).
Pfaffl, M. W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29, e45 (2001).
Rehli, M. K. M. Functional analysis of promoter CPG-methylation using a CpG-free luciferase reporter vector. Epigenetics 1, 4 (2006).
Jacobs, J. L. & Dinman, J. D. Systematic analysis of bicistronic reporter assay data. Nucleic Acids Res. 32, e160 (2004).
Abdelrahman, H. A., Al-Shamsi, A. M., Ali, B. R. & Al-Gazali, L. A null variant in PUS3 confirms its involvement in intellectual disability and further delineates the associated neurodevelopmental disease. Clin. Genet. 94, 586–587 (2018).
Klug, M. & Rehli, M. Functional analysis of promoter CPGmethylation using a CpG-free luciferase reporter vector. Epigenetics 1, 127–130 (2006).
Consortium, G. T. The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 45, 580–585 (2013).
Schneider, E., El Hajj, N., Muller, F., Navarro, B. & Haaf, T. Epigenetic dysregulation in the prefrontal cortex of suicide completers. Cytogenet. Genome Res 146, 19–27 (2015).
Nagy, C. et al. Astrocytic abnormalities and global DNA methylation patterns in depression and suicide. Mol. Psychiatry 20, 320–328 (2015).
Nagy, C., Torres-Platas, S. G., Mechawar, N. & Turecki, G. Repression of astrocytic connexins in cortical and subcortical brain regions and prefrontal enrichment of H3K9me3 in depression and suicide. Int. J. Neuropsychopharmacol. 20, 50–57 (2017).
Phillips, J. R., Hewedi, D. H., Eissa, A. M. & Moustafa, A. A. The cerebellum and psychiatric disorders. Front. Public Health 3, 66 (2015).
Hwang, J. P. et al. Cortical and subcortical abnormalities in late-onset depression with history of suicide attempts investigated with MRI and voxel-based morphometry. J. Geriatr. Psychiatry Neurol. 23, 171–184 (2010).
Amen, D. G., Prunella, J. R., Fallon, J. H., Amen, B. & Hanks, C. A comparative analysis of completed suicide using high resolution brain SPECT imaging. J. Neuropsychiatry Clin. Neurosci. 21, 430–439 (2009).
Lee, Y. J. et al. Decreased regional gray matter volume in suicide attempters compared to suicide non-attempters with major depressive disorders. Compr. Psychiatry 67, 59–65 (2016).
Jung, J. et al. Alterations in functional brain networks in depressed patients with a suicide attempt history. Neuropsychopharmacology. https://doi.org/10.1038/s41386-019-0560-z (2019).
Stawski, R. et al. Reduced expression of ELAVL4 in male meningioma patients. Brain Tumor Pathol. 30, 160–166 (2013).
Akamatsu, W. et al. The RNA-binding protein HuD regulates neuronal cell identity and maturation. Proc. Natl Acad. Sci. USA 102, 4625–4630 (2005).
Ince-Dunn, G. et al. Neuronal Elav-like (Hu) proteins regulate RNA splicing and abundance to control glutamate levels and neuronal excitability. Neuron 75, 1067–1080 (2012).
Talman, V., Pascale, A., Jantti, M., Amadio, M. & Tuominen, R. K. Protein kinase C activation as a potential therapeutic strategy in Alzheimer’s disease: is there a role for embryonic lethal abnormal vision-like proteins? Basic Clin. Pharm. Toxicol. 119, 149–160 (2016).
Yamada, K. et al. Genome-wide association study of schizophrenia in Japanese population. PLoS ONE 6, e20468 (2011).
Berto, S., Usui, N., Konopka, G. & Fogel, B. L. ELAVL2-regulated transcriptional and splicing networks in human neurons link neurodevelopment and autism. Hum. Mol. Genet. 25, 2451–2464 (2016).
de Paiva, A. R. B. et al. PUS3 mutations are associated with intellectual disability, leukoencephalopathy, and nephropathy. Neurol. Genet. 5, e306 (2019).
Yokota, N. et al. Predominant expression of human zic in cerebellar granule cell lineage and medulloblastoma. Cancer Res. 56, 377–383 (1996).
Vitucci, D. et al. Rasd2 modulates prefronto-striatal phenotypes in humans and ‘schizophrenia-like behaviors’ in mice. Neuropsychopharmacology 41, 916–927 (2016).
Sun, H. et al. Quantitative integration of epigenomic variation and transcription factor binding using MAmotif toolkit identifies an important role of IRF2 as transcription activator at gene promoters. Cell Discov. 4, 38 (2018).
Drew, P. D. et al. Interferon regulatory factor-2 physically interacts with NF-kappa B in vitro and inhibits NF-kappa B induction of major histocompatibility class I and beta 2-microglobulin gene expression in transfected human neuroblastoma cells. J. Neuroimmunol. 63, 157–162 (1995).
Daniele, L. L., Emran, F., Lobo, G. P., Gaivin, R. J. & Perkins, B. D. Mutation of wrb, a component of the guided entry of tail-anchored protein pathway, disrupts photoreceptor synapse structure and function. Invest. Ophthalmol. Vis. Sci. 57, 2942–2954 (2016).
Ling, K. H. et al. Functional transcriptome analysis of the postnatal brain of the Ts1Cje mouse model for Down syndrome reveals global disruption of interferon-related molecular networks. BMC Genomics 15, 624 (2014).
Nair, R. P. et al. Sequence and haplotype analysis supports HLA-C as the psoriasis susceptibility 1 gene. Am. J. Hum. Genet. 78, 827–851 (2006).
Chang, Y. T. et al. Psoriasis vulgaris in Chinese individuals is associated with PSORS1C3 and CDSN genes. Br. J. Dermatol. 155, 663–669 (2006).
Wisniewski, A., Matusiak, L., Szczerkowska-Dobosz, A., Nowak, I. & Kusnierczyk, P. HLA-C*06:02-independent, gender-related association of PSORS1C3 and PSORS1C1/CDSN single-nucleotide polymorphisms with risk and severity of psoriasis. Mol. Genet. Genomics 293, 957–966 (2018).
Polanowska, J. et al. The periodic down regulation of Cyclin E gene expression from exit of mitosis to end of G(1) is controlled by a deacetylase- and E2F-associated bipartite repressor element. Oncogene 20, 4115–4127 (2001).
Yamada, K. et al. Population-dependent contribution of the major histocompatibility complex region to schizophrenia susceptibility. Schizophr. Res. 168, 444–449 (2015).
Pfleger, C. M. The Hippo pathway: a master regulatory network important in development and dysregulated in disease. Curr. Top. Dev. Biol. 123, 181–228 (2017).
Sun, R. et al. Identification of novel loci associated with infant cognitive ability. Mol. Psychiatry. https://doi.org/10.1038/s41380-018-0205-3 (2018).
Hamdan, F. F. et al. De novo mutations in moderate or severe intellectual disability. PLoS Genet. 10, e1004772 (2014).
Nizon, M. et al. Variants in MED12L, encoding a subunit of the mediator kinase module, are responsible for intellectual disability associated with transcriptional defect. Genet. Med. 21, 2713–2722 (2019).
Branco, M. R., Ficz, G. & Reik, W. Uncovering the role of 5-hydroxymethylcytosine in the epigenome. Nat. Rev. Genet. 13, 7–13 (2011).
Clive, M. L. et al. Discovery and replication of a peripheral tissue DNA methylation biosignature to augment a suicide prediction model. Clin. Epigenet. 8, 113 (2016).
Sadeh, N. et al. Epigenetic Variation at Ska2 Predicts Suicide Phenotypes and Internalizing Psychopathology. Depress Anxiety 33, 308–315 (2016).
Kaminsky, Z. et al. Epigenetic and genetic variation at SKA2 predict suicidal behavior and post-traumatic stress disorder. Transl. Psychiatry 5, e627 (2015).
Lockwood, L. E., Su, S. & Youssef, N. A. The role of epigenetics in depression and suicide: a platform for gene-environment interactions. Psychiatry Res 228, 235–242 (2015).
Guintivano, J. et al. Identification and replication of a combined epigenetic and genetic biomarker predicting suicide and suicidal behaviors. Am. J. Psychiatry 171, 1287–1296 (2014).
Acknowledgements
We are grateful to all the patients and control subjects who contributed to this study. The authors would like to acknowledge support of the Brain and Behaviour Research Foundation through a NARSAD Young Investigator Grant to T.M.M. and from the UK Medical Research Council (MRC) (grant number MR/K013807/1) to J.M. Z.K. would like to acknowledge funding from the NIH grant (NIMH 1R21MH094771). The Douglas Bell Canada Brain Bank is supported by the FRQS through the Quebec Network on Suicide, Mood Disorders and Related Disorders, and by Brain Canada through an infrastructure grant. E.L.D. would like to acknowledge the AMS Springboard scheme that supported SW studentship.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Policicchio, S., Washer, S., Viana, J. et al. Genome-wide DNA methylation meta-analysis in the brains of suicide completers. Transl Psychiatry 10, 69 (2020). https://doi.org/10.1038/s41398-020-0752-7
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41398-020-0752-7
This article is cited by
-
Integrative transcriptome- and DNA methylation analysis of brain tissue from the temporal pole in suicide decedents and their controls
Molecular Psychiatry (2023)
-
A sex-specific genome-wide association study of depression phenotypes in UK Biobank
Molecular Psychiatry (2023)