Abstract
In order to determine the role of single nucleotide variants (SNVs) in modulating antidepressant response, we conducted a study, consisting of 929 major depressive disorder (MDD) patients, who were treated with antidepressant drugs (drug-only) or in combination with a repetitive transcranial magnetic stimulation (plus-rTMS), followed by targeted exome sequencing analysis. We found that the “plus-rTMS” patients presented a more effective response to the treatment when compared to the ‘drug-only’ group. Our data firstly demonstrated that the SNV burden had a significant impact on the antidepressant response presented in the “drug-only” group, but was limited in the “plus-rTMS” group. Further, after controlling for overall SNV burden, seven single nucleotide polymorphisms (SNPs) at five loci, IL1A, GNA15, PPP2CB, PLA2G4C, and GBA, were identified as affecting the antidepressant response at genome-wide significance (P < 5 × 10−08). Additional multiple variants achieved a level of correction for multiple testing, including GNA11, also shown as a strong signal for MDD risk. Our study showed some promising evidence on genetic variants that could be used as individualized therapeutic guides for MDD patients.
Similar content being viewed by others
Introduction
Major depressive disorder (MDD) is one of the most common severe mental disorders with a 17.8% lifetime prevalence in global population1. Unfortunately, current antidepressant drugs still have obvious shortcomings, including delayed onset, insufficient effectiveness, and low remission rates2,3,4,5, which combined with a high prevalence have significantly exacerbated the disease to be the second leading cause of disability worldwide according to the 2020 Global Burden of Disease Study6. Therefore, it is imperative in the study of antidepressant treatment to achieve a rapid improvement of depressive symptoms and to increase the recovery rate of the first episode of MDD7,8.
Pharmacogenetic studies have indicated that genomic variations are one of the critical biological factors that prominently affect the individual differences of antidepressant response, in which common single nucleotide polymorphisms (SNPs) are estimated to explain 50% or more of the variation of the antidepressant response9. Nevertheless, previous findings from candidate gene approaches have been poorly replicated, with insufficient predictive power to be useful in clinical practice and analytical problems of uncontrolling population stratification or genetic heterogeneity10,11. In addition, so far a total of eight genome-wide association studies (GWAS) of antidepressant effectiveness have been carried out mainly in Caucasian ancestry populations (eight clinical studies, ten published articles); none of these studies have found a genetic variant that achieved genome-wide significance12,13,14,15,16,17,18,19,20,21. However, several evaluations of the above eight clinical studies, in particular, a recent meta-analysis of Genome Based Therapeutic Drugs for Depression (GENDEP) (n = 738), Sequenced Treatment Alternatives to Relieve Depression (STAR*D) (n = 1409) and European Group for the Study of Resistant Depression (GSRD) (n = 1148), the three most significant pharmacogenomic studies of antidepressant responses, identified two gene sets (GO:0043949 and GO:0000183) and the integrin subunit alpha 9 (ITGA9) and neurexin (NRXN3) genes21,22 while the activator of transcription and developmental regulator (AUTS2) gene was discovered in a Korean sample19. However, there was no pharmacogenetic GWAS from independent Han Chinese populations. Most recently, Cai et al.23 have identified the association of two SNPs with the MDD risk in the most significant samples of Han Chinese, which were sirtuin 1 (SIRT1) gene and phospholysine phosphohistidine inorganic pyrophosphate phosphatase (LHPP) gene.
Exome sequencing, also known as targeted exome capture, is an efficient strategy for the selection of genomic coding sequences as well as UTR and upstream region, particularly for the study of SNPs and small insertions and deletions (indels) of known genes24. Thus far, only two published reports, using whole-exome sequencing, have indicated that the rs41271330 SNP of the bone morphogenetic protein 5 (BMP5) gene strongly associates with worse selective serotonin reuptake inhibitor (SSRI) treatment response25 and exm-rs1321744 located in a methylated DNA immunoprecipitation sequencing site is associated with antidepressant remission26.
Notably, repeated transcranial magnetic stimulation (rTMS) has been widely used in the clinical setting due to its noninvasiveness, convenience, safety, and effectiveness as an antidepressant modality27. It has also demonstrated a competence for rapid treatment responses, in particular, when combined with antidepressant drugs at the beginning of a medical intervention28,29. However, rTMS treatments for MDD still present prominently individual differences, with only about 25% response rate30. A few studies have found that functional serotonin 1A receptor (5-HT1A), serotonin transporter (5-HTT), and brain-derived neurotrophic factor (BDNF) gene polymorphisms affected the rTMS treatment response in MDD groups, although the results were inconsistent31.
In summary, genetic studies of antidepressant effectiveness with outcome follow-up at multiple time points provide a robust study design with the potential to identify factors that could inform individualized medical treatment32. Therefore, we conducted a targeted exome sequencing of patients undergoing antidepressant treatment for MDD, in order to identify the effect of rare and common single nucleotide variants (SNVs) on the antidepressant responses using the largest sample of untreated MDD patients in the Han Chinese population, to focus on the different effects on responses over the period of treatment and remission rates (eight weeks) and identifying genetic variants associated with the difference in response between the “drug-only” group and the “plus-rTMS” group. We identified genetic variants that could be utilized to develop individualized treatments for MDD.
Materials and methods
Overview
This clinical study of antidepressant effectiveness was conducted in a naturalistic clinical setting and primarily enrolled untreated adult in-patients who were experiencing current episodes of nonpsychotic unipolar depression in an ethnically homogeneous population of Han Chinese patients. Meanwhile, the study recruited well-matched healthy controls and sequenced targeted exome to analyze association with the antidepressant response.
Subjects
The subjects enrolled in this study were Han Chinese patients referred to Zhongda Hospital MDD in-patient database. All recruited patients were between 18 and 65 years of age and met the diagnosis criteria for nonpsychotic MDD according to the Diagnostic and Statistical Manual of the American Psychiatric Association (DSM-IV; American Psychiatric Association, 1994). All subjects were newly diagnosed or recently relapsed patients, who had been drug-free for over two weeks and presented a baseline score of 18 or above on the 17-item Hamilton Depression Rating Scale (HAM-D17)33, having presented depressive symptoms for at least two weeks before entry. The patients were diagnosed by two independent senior psychiatrists and confirmed by a third psychiatrist if the diagnosis was inconsistent, and blind to previous evaluations.
The exclusion criteria included other documented diagnostic history on Axis 1 of the DSM-IV, independent diagnosis of personality disorder, mental retardation, pregnancy, lactation, primary organic disease, and other medical illnesses impairing psychiatric evaluation, or a history of electroconvulsive therapy within the previous six months or rTMS treatment or contraindication to rTMS. Newly diagnosed patients were also excluded if they had experienced a manic episode in the 12 months following entry. All subjects gave written informed consent for their participation in the study, which was approved by the hospital ethical committee (2016ZDSYLL100-P01), in accordance with the Declaration of Helsinki. A flowchart of the recruited subjects is shown in Fig. S1. At the end, 929 cases and 438 controls remained for the downstream analysis.
Antidepressant treatment and outcome evaluation
All the MDD patients enrolled in this study were treated with single antidepressant drugs (drug-only) or with concurrent rTMS treatment for the first two weeks (consecutive 14 days) (plus-rTMS) according to local clinical practice. During patient treatment, selected single antidepressant drugs were administered for eight weeks and an increase in the drug dosage prescribed at baseline was allowed if the patient had not achieved “very much improved” or “much improved” symptoms based on the Clinical Global Impression (CGI) scale. Concomitant psychotropic medications were not permitted, except for a low dosage of a benzodiazepine anxiolytic (e.g. alprazolam 0.4–0.8 mg/day, estazolam 1–2 mg/day) for the alleviation of insomnia when necessary. Patients requiring a change in antidepressant drug or demonstrating nonadherence were excluded from the study.
An rTMS with an 8-shaped magnetic coil (TypeMagPro R30, MagVenture Co., Ltd; Farum, Denmark) was used and the coil was centered and fixed over the participant’s right dorsolateral prefrontal cortex (DLPFC). The conventional rTMS was administered at 80% of the motor threshold and at a low frequency of 1 Hz34, which remarkably reduced the attack rate of epilepsy and mania. Stimulation was given 20 min per day with each train lasting 10 min, with an interval between trains of 30 s.
The primary outcome of the study was the severity of depressive symptoms measured by the HAM-D17 symptom scale by the researchers, assessed at the baseline and at scheduled follow-up time points at weeks 2, 4, 6, and 8.
Sample size determination and validation studies
The sample size was determined based on the repeated measures analysis of variance models35,36. Given that our primary goals were to identify how genetic variants affected the antidepressant response over the time of treatment, the between-subject factor in the variance model was a genotype group, which was coded as a binary in the dominant or recessive model. The calculation was performed using the PROC GLMPOWER procedure based on the Hotelling-Lawley F test with a linear exponent autoregressive correlation structure, a base correlation of 0.48, and a decay of 0.70 (see Supplementary III Methods 1 Table SM1 and SM2, Fig. SM1). The calculation of sample size showed that the “drug-only” group would be adequate at a power of 80% and alpha of 0.000006, while the number of subjects was smaller in the “plus-rTMS” group. Since the primary outcome of this study was based on the scheduled clinical evaluation at multiple time points, our findings would have provided stronger evidence than that with a population-based cross-sectional study.
Targeted gene selection and sequencing
The selection of the targeted genes was based on pathways known to be involved in the etiology and pathophysiology of MDD and/or in mediating the antidepressant effect based on the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway. We selected various SNPs or loci that were known to be top signals in previous genetic studies of MDD and other mental disorders at the time of the study design. Altogether, we selected 1309 designable target genes for exome sequencing, with the detailed list of the genes selected for sequencing provided in the Supplementary methods (Tables SM3 and SM4). For each of these genes, both coding and noncoding regulatory regions, comprising of the five prime untranslated region (5′ UTR), 3′ UTR, and intron−exon boundaries (25 bp), were included in the sequencing targets. The sequencing of target gene was performed on an Illumina MiSeq high-throughput sequencing platform (Illumina, San Diego, CA, USA) with the assistance of Shanghai Genesky Bio-Tech Company.
Bioinformatics analysis
Alignment and variant calling
Raw sequencing data were demultiplexed into individual Fastq read files with the Illumina’s Bcl2fastqv2.16.0.10 software program based on unique index pairs. Low-quality (Q < 15) reads/bases were trimmed using the FastX tool developed by the Hannon Lab (http://hannonlab.cshl.edu/fastx_toolkit/index.html). High-quality reads were aligned to the National Center for Biotechnology Information (NCBI) human reference genome (hg19) using the Burrows−Wheeler Aligner (BWA) software37. Subsequently, the aligned reads were processed further using the Picard’s MarkDuplicates, SAM tools38 and the Indel Realignment and Base Quality Score Recalibration platform from the Genome Analysis Toolkit39. The GATK v3.539 and the VarScan v2.3.9 software programs40 were used to generate genotype information of candidate SNVs in the targeted regions for each individual, and the called SNVs were subsequently combined. A call was made to determine whether the SNVs identified had minimal depth coverage higher than (>) 20× and a quality score >30 in more than 80% of the subjects sequenced. After in-house quality control for both clinical and sequencing data, a total of 126,316 SNVs within 1309 genes, comprising 17,870 common SNPs and 108,173 rare SNVs (minor allele frequency [MAF] <0.01), of which 107,875 rare SNVs were successfully annotated.
SNVs annotations
The ANNOVAR software program (released on February 1, 2016) was used for the annotation of SNVs41. SNVs were divided into two groups: SNPs and rare variants. Any SNVs with a minor allele frequency of ≤1% and with a genotyping rate of more than 90% was considered a SNP (SNPs that failed the Hardy-Weinberg equilibrium test at a significance level of 0.00001 were removed). Similarly, SNVs presenting a minor allele frequency of <1% were considered rare variants. In addition, we created a priority score for each rare variant by integrating multiple annotation information, such as conservation, frequency, and protein scores (e.g., PolyPhen). A detailed description of the level-score can be found in Table SM5 (Supplementary Methods).
We then performed an independent validation of the top SNPs that may affect the antidepressant response, using improved multiplex ligation detection reactions (iMLDR)42. The SNPs presenting a 98% accuracy of the genotypes between the raw and validation data were reported here (Supplementary Methods).
Statistical analysis
Common and rare SNVs were separated for further data processing and quality control before subsequent analysis. Rare SNVs (MAF < 1%) were further classified by gene region and function class. For statistical analysis purposes, we also classified the SNVs by comparing them with existing databases, such as the 1000 Genomes, and were consistently called by two computer programs. A sum of the number of minor alleles, called the SNV burden, was obtained for each MDD patient, in order to assess the influence on the antidepressant treatment response. After quality controls were performed, we utilized all common variants for the examination of cryptic relatedness and the control of population stratification. Identity by descent (IBD) analysis of common SNPs was used to remove individuals with cryptic relatedness (estimated kinship coefficient > 0.25) and multidimensional scaling (MDS) components were used for the examination of genetic outliers or population structure. The number of common SNPs in this study was adequate to reveal the population structure43,44. The PLINK v1.9 computer software was used for this analysis.
The analysis of the primary outcome was measured by the HAM-D17 assessment scale, which was first performed on the overall SNV burden and then by gene region and function class for the antidepressant response. After controlling for the global SNV burden (log-transformed), we then examined the common SNPs for the antidepressant response. A general linear model with random effect was used to analyze the primary outcome (HAM-D17)45, which were log-transformed to be close to the normal distribution and evaluated at multiple time points during the clinical study. The main effect was common genetic variants and genotypes by time interaction, while adjusting for covariates of sex, age of onset, number of depressive episodes, duration of illness (months), level of education, type of medication, and first MDS components. All analysis was performed using the PLINK v1.9, SAS release 9.4 and R3.3.2 release software platform (https://www.r-project.org/).
Furthermore, three levels of threshold were defined for significance or as a strong signal. While our study was based on targeted exome sequencing analysis, we still applied a standard of genome-wide significance to assure a robust finding (P < 5 × 10−08). We have also tried to present all possible top SNPs that achieved a Bonferroni correction (α = 9.7 × 10−07) and survived the correction for the number of SNPs (α = 0.05/12,561 = 3.98 × 10−06) for further testing of these top molecular signals.
cis-eQTL analysis, RNA expression, and bioinformatics investigation
Multiple datasets of RNA expression in human tissues were used to validate our top findings at the level of gene or protein expression and cis-regulatory associations. They were shown as follows: (1) the RNA-seq data provided by the HPA (https://www.proteinatlas.org/); (2) the RNA-seq data of the GTEx (https://www.gtexportal.org), which was mapped using the Ensembl gene ID and with the cis-eQTL analysis carried out online; and (3) a microarray RNA expression of postmortem human brains was analyzed for cis-regulatory effects of the rs8092 SNP on the GNA15 locus, which was based on 88 subjects of European ancestry (for consistency with the other samples), older than five years of age (http://braincloud.jhmi.edu/)46. Unless specified, all LD were calculated using the sample of the Han Chinese in Beijing, China (CHB)-Japanese in Tokyo, Japan (JPT) in the 1000 Genomes (N = 404).
Results
Clinical study for the effectiveness of antidepressant intervention
Finally, a total of 929 patients with MDD enrolled in this study (drug-only 530 vs. plus-rTMS 399). The single antidepressant drug includes SSRIs (51%), serotonin-norepinephrine reuptake inhibitors (SNRIs; 24%), and other antidepressant drugs (25%). As shown in Table 1 and Tables S1−S2, there were no significant differences (P > 0.05) in the mean or frequency of sex, family history, type of drugs, duration of illness, and the baseline clinical symptom, which was a log-transformed HAM-D17 (all P > 0.05). We noted a difference (P < 0.05) in the number of depressive episodes, age, education and age of onset between the two treatment groups at baseline. However, in a multiple regression model analysis, only the level of education (P = 0.0472) and age of onset (P = 0.0346) were slightly different between the two treatment groups.
There were no significant differences in the baseline log-HAM-D17 score between the two groups (P = 0.059). However, more patients in the “plus-rTMS” group presented two or fewer depressive episodes (P = 0.0001) (Table 1). Obviously, patients in the “plus-rTMS” group had more effective response (58.65% vs. 40.76%) within the first two weeks (P = 0.00014) and four weeks (P = 0.0054) after treatment as compared with the “drug-only” group. Furthermore, the overall remission rate in the “plus-rTMS” group was 65.20%, higher than the 55.66% of “drug-only” group (P = 0.0035) (Table 2).
The SNV burden modulates the antidepressant response of MDD patients depending on the specific treatment and gene region
A total of 107,875 rare SNVs were found located mainly in intronic, exonic, and UTR3 variant regions (Table S3). Approximately half of the rare SNVs (56,552) found were detected in patients with MDD and not in the other 1000Genomes or the 438 healthy controls. Moreover, out of the total SNVs found, 53.61% of the rare SNVs (57,830) were consistently detected with both the GATK and the Var Scan variant-calling software applications. In addition, exonic variants increased from 24.2 to 36.2% and UTR3 variants increased from 22.7 to 28.99%, while intronic variants decreased (Table S4).
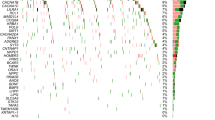
Interestingly, the SNV burden, analyzed by various methods (as shown in Tables S5−S8 and Fig. 1 A1−A2 and B1−B2), was found to modulate the antidepressant response only in the “drug-only” group. Specifically, the antidepressant response was affected by the SNV burden at intronic, upstream, exonic, UTR3, ncRNA intronic, downstream, and splicing regions (P ≤ 5.32 × 10−04) in the “drug-only” group. In contrast, only intergenic SNVs showed a signal in the “plus-rTMS” treatment group (P = 2.37 × 10−03) (Table S6). Notably, MDD patients with a higher level of SNV burden tended to exhibit a slower response to antidepressants only in the “drug-only” group (P = 1.23 × 10−04) (Table S9).
a The burden of all 107,875 SNVs: A1 the “drug-only” group and A2 the “plus- repetitive transcranial magnetic stimulation (rTMS)” treatment group. b The burden of the 56,552 SNVs specifically detected in major depressive disorder (MDD) patients, but not found in either the 1000Genomes or healthy control subjects: B1 the “drug-only” group and B2 the “plus-rTMS” treatment group. c The burden of all 57,830 SNVs consistently called using the GATK and VarScan variant software programs: C1 the “drug-only” group and C2 the “plus-rTMS” treatment group. d The burden of the 27,220 SNVs detected specifically in MDD patients, but not in either the 1000Genomes or healthy control subjects used for this study, which were consistently called by the GATK and VarScan variant software programs: D1 the “drug-only” group and D2 the “plus-rTMS” treatment group (x-axis is the negative logarithm of P value).
Similarly, the SNV burden was also found to modulate the antidepressant response differently by treatment group when our analysis was focused on the 57,830 SNVs that were consistently called by the GATK and VarScan programs, in which the SNV burden markedly affected the antidepressant response only in the “drug-only” group as compared to the “plus-rTMS” group (see Tables S10−S13, Fig. 1 C1−C2 and D1−D2 for detail).
Common variants modulate the antidepressant response
Controlling for the SNV burden, we further performed an analysis of 12,561 SNPs under a dominant and recessive model to search for common variants that may modulate the antidepressant response differently by treatment group (Figs. S2, S3). To avoid too fewer number of homozygotes, this analysis only considered SNPs with MAF > 5%. (1) Importantly, the seven SNPs at five loci were found associated with the antidepressant response at genome-wide significance (5 × 10−08) (Table 3). These included SNPs rs3783553 and rs3783550 at the IL1A loci (minimum P = 1.41x10−08), rs11671393 at GNA15 (P = 4.95 x 10−12), and rs4733201 at PPP2CB (P = 1.07 × 10−08), which were identified in the “drug-only” group, and SNPs rs2303744 at PLA2G4C (minimum P = 2.70 × 10−10), rs9628662 (P = 2.45 × 10−09), and rs12034326 (P = 4.4 × 10−08) at GBA, which were identified in the “plus-rTMS” group. The detailed estimates of treatment responses by the SNP genotype and treatment time indicated that the trend of the responses was different between the genotype groups (Table S14). (2) In addition, we observed ten SNPs, including GNA11 with strong signals for the antidepressant response in the “drug-only” or the “plus-rTMS” group, respectively, achieved a strict Bonferroni correction (α = 9.9 × 10−07) (see Table 3 for detail). (3) Additional SNPs at loci AGXT, ACE, CYP2C8, SGMS1, PLCH2, RPTOR, and GNA11 also achieved a level of threshold for correcting the number of SNPs tested (α = 0.05/12,561 = 3.98 × 10−06) (Table 3).
Common variants associated with both antidepressant response and risk of MDD
Interestingly, only the rs8092 SNP at the UTR3 variant of the GNA11 loci showed the association with both antidepressant response and MDD risk (Table S15). This SNP is also upstream of GNA15 where the rs11671393 SNP showed genome-wide significance (P = 4.85 × 10−12) for the antidepressant response (Fig. 2a). These two SNPs were less likely in linkage disequilibrium (LD; R2 = 0.0144 and D′ = 0.1215) (Fig. 2b, c). Patient carriers of the major risk allele C of rs8092 (Fig. 2d) and allele G of rs11671393 at GNA15 tend to have a slower and poorer response to the antidepressant treatment (Fig. 2e). In addition, the rs8092 SNP had a cis-association with GNA15 in the prefrontal cortex of postmortem human brains (N = 88, P = 7.0 × 10−03 (Fig. 2f) and in other tissues (see Fig. S4). In addition, significant cis-eQTL (expression quantitative trait locus) in human tissue samples of the genotype-tissue expression (GTEx) dataset was found for all five loci involved in the antidepressant response at genome-wide significance (Table S16), which were significant after multiple testing correction (FDR < 0.05).
a Scatter plots depicting the P values obtained for the antidepressant response (X sign, a dot indicates a P value for the MDD risk association). b A linkage disequilibrium (LD) plot of MDD patients (based on common variants from the sequencing analysis). c An LD plot of healthy control subjects. d Differences in the log of the Hamilton Rating Scale for Depression-17-Item (HAM-D17) scale ratings between carriers of the major risk allele C (REC = 0) and homozygous for the minor T allele of the rs8092 single nucleotide polymorphism (SNP) at the GNA11 locus in the “plus-rTMS” treatment group. e Differences in the log-HAM-D17 between carriers of the major allele G (REC = 0) and homozygous for the minor allele of the rs1167393 SNP at the GNA15 locus in the “drug-only” group. f The major risk allele C of the rs8092 SNP was associated with the expression of GNA15 in postmortem human brains of adult subjects of European ancestry (n = 88, subjects five years of age or more, P_ANOVA = 0.0006) adjusted for RIN, postmortem interval (PMI), age, sex, and pH. The plot was made based on post hoc analysis of the least square (LS)-means in the RNA expression of GNA15.
Replication of loci reported in previous genome-wide association studies of MDD and genetic studies of the antidepressant response
Despite the different convergence of individual SNPs between the various genotyping platforms, in this study we made an effort to validate the loci reported in previous GWAS of MDD and genetic studies of the antidepressant response. Out of the seven loci reported in previous GWAS of MDD23,47,48,49 included in our study, seven SNPs at ENOX1, ESR2, PMFBP1, and EP300 showed a significant impact on the antidepressant response in the “drug-only” group (P < 0.05), whereas the rs730210 SNP at ENOX1 and the rs3818121 SNP at EP300 showed a significant impact on the antidepressant response in the “plus rTMS” group (P < 0.05).
Furthermore, out of the suggested loci found to be promising candidates for the antidepressant response in clinical applications after validation34, multiple SNPs at ABCB1, BDNF, CYP2C19, SLC6A2, CYP2D6, and HTR2A showed a trend for influencing antidepressant response at a nominal significance in our study (P < 0.05) (Table S17). However, we did not find a positive validation for SNPs at the GNB3 and FKBP5 loci, although they were included in the present study. Unfortunately, ITGA9 and NRXN3 were not included in this study, and we were not able to replicate any SNPs at loci AUTS2 (P > 0.05).
Discussion
The present study firstly found significant differences in the rate of response to antidepressant treatment after two weeks, with a remission rate after eight weeks treatment between the two subgroups, in which the “plus-rTMS”-treated patients presented a more effective response to treatment as compared to the “drug-only” group, indicating that the concurrent use of rTMS may have an additional rapid improvement in the clinical symptoms.
It is interesting to note that the SNV burden had a significant impact on the antidepressant response primarily in the “drug-only” patients. This could be due to the fact that these patients had more repeated depressive episodes than those receiving TMS, perhaps a consequence of a higher level of SNV burden which may in turn relate to a higher likelihood of having treatment-resistant depression50. On the other hand, the patients in the “plus-rTMS” group presented higher rates of effective responses after being treated for two weeks, which is consistent with previous findings51. Thus, the additional treatment effect may, to some extent, overcome the burden imparted by these SNVs.
Importantly, our study had identified the seven SNPs (MAF > 5%) at the five loci IL1A, GNA15, PPP2CB, PLA2G4C and GBA for treatment response at genome-wide significance in MDD patients. Furthermore, we had identified the additional ten SNPs including rs8092 at GNA11 achieved a threshold for multiple testing correction. Importantly, the SNPs identified here were strongly associated with an increased likelihood of remission at the end of the clinical study for MDD patients.
Some of the above seven SNPs were randomly identified or in LD with other SNPs, noted for their strong signals, but none of these SNPs had been shown to reach genome-wide significance in previous GWAS of related phenotypes. For example, the 4-bp indel at rs3783553 and rs3783550 SNP at IL1A identified in this study have previously been reported to be associated with various cancers and severe inflammatory diseases, such as ankylosing spondylitis mostly in Asian populations, including Chinese Han52,53. The rs3783550 SNP at IL1A was also found to be in perfect LD (R2 = 0.197 and D′ = 1) with the rs11677416 SNP at CKAP2L-IL1A, which has been found to have a strong signal for antipsychotic response in patients with schizophrenia treated with olanzapine54. The rs11671393 SNP at GNA15 was also found to be in perfect LD with the rs2302062 and rs11890198 SNPs, previously noted to have strong signals for glucose homeostasis55 and heart failure56. In addition, one variant (chr8:30765196-T) at the PPP2CB loci had been noted as a signal for daytime sleepiness57. However, rs1549637 SNP in PLA2G4C is reportedly related to negative symptom severity in schizophrenic patients58, and the rs251684 variant of PLA2G4C is associated with autism spectrum disorder59. Notably, multiple variants at the GBA locus have been found associated with Parkinson’s disease in four previous GWAS60,61,62,63.
In addition, all the seven SNPs found here were eQTLs in human tissues. For instance, the IL1A encodes the interleukin 1 alpha (IL-1α) protein, also known as hematopoietin 1, which is a pleiotropic cytokine involved in various immune responses, inflammatory processes, and hematopoiesis64. GNA15, which encodes the G protein subunit alpha 15, is highly expressed in esophageal tissues and bone marrow in the GTEx dataset (https://www.gtexportal.org). PPP2CB, which encodes the phosphatase two catalytic subunit beta enzyme, is highly expressed in the brain and by the immune system, as shown in the HPA dataset. Furthermore, the PLA2G4C locus encodes the protein phospholipase A2 group IVC, which is highly expressed in multiple human tissues, including the brain, the gastrointestinal tract, the bone marrow, and the immune system. The GBA encodes the enzyme glucosylceramidase beta, which is highly expressed in human tissues, including the brain, the immune system, and the gastrointestinal tract. Similarly, the rs12034326 SNP at the GBA locus, demonstrated a strong cis-association in multiple tissues including the esophagus.
Interestingly, one of the SNPs achieving significance following correction for multiple testing—rs8092 SNP at GNA11—is also associated with both the antidepressant response and risk of MDD. First, rs8092 was shown to have a strong signal for the antidepressant response close to the Bonferroni correction. Additionally, it presented the strongest signal for the MDD association among all the top SNPs identified for the antidepressant response. While the association of rs8092 with MDD was not found at genome-wide significance in our study, rs8092 has been previously identified at genome-wide significance analysis for multiple sclerosis, in which depression is a critical complication, as shown in a study of a large homogenous Scandinavian cohort through identity by descent (IBD) mapping65. Further, GNA11 (P < 0.0054) was implicated in association with MDD by a pathway analysis study from a previous GWAS of MDD of European ancestry66. Notably, the rs8092 SNP was previously shown to have a cis-association with GNA11 in human brains from the GTEx dataset and in the prefrontal cortex of postmortem human brains, together with a nearby gene GNA15, in which the rs11671393 SNP was found to be associated with antidepressant response at genome-wide significance in our study. Similarly, mutations in the GNA11 locus have been found associated with hypocalciuric hypercalcemia type II (HHC2), hypocalcemia dominant 2 (HYPOC2), and uveal melanoma conditions67,68. All of this evidence may support the notion that the rs8092 SNP is a candidate for both the antidepressant response and risk of MDD.
There are several limitations to this study. One of its weaknesses is the lack of a replication cohort. The study relied on the self-reporting of several factors including family history, which may be under-reported, although we found this factor to have no influence on the results. In assessing the exome, we may not identify all possible genetic factors, some of which may reside in nonexomic—intergenic or intronic—regulatory sequences.
In summary, our study shows that both rare and common genetic variants can modulate the antidepressant response differently between the two subgroups of antidepressant treatments selected here. These findings generate promising evidence for the use of individualized and precision medicine in patients with MDD and may provide new insights into the underlying molecular mechanism of antidepressant response.
References
Disease, G. B. D., Injury, I. & Prevalence, C. Global, regional, and national incidence, prevalence, and years lived with disability for 310 diseases and injuries, 1990-2015: a systematic analysis for the Global Burden of Disease Study 2015. Lancet 388, 1545–1602 (2016).
Pigott, H. E., Leventhal, A. M., Alter, G. S. & Boren, J. J. Efficacy and effectiveness of antidepressants: current status of research. Psychother. Psychosom. 79, 267–279 (2010).
Rush, A. J. et al. Is prior course of illness relevant to acute or longer-term outcomes in depressed out-patients? A STAR*D report. Psychol. Med. 42, 1131–1149 (2012).
Huynh, N. N. & McIntyre, R. S. What are the implications of the STAR*D trial for primary care? A review and synthesis. Prim. Care Companion J. Clin. Psychiatry 10, 91–96 (2008).
Trivedi, M. H. et al. Evaluation of outcomes with citalopram for depression using measurement-based care in STAR*D: implications for clinical practice. Am. J. Psychiatry 163, 28–40 (2006).
Murray, C. J. & Lopez, A. D. The Global Burden of Diseases: A Comprehensive Assessment of Mortality and Disability from Diseases, Injuries and Risk Factors in 1990 and Projected to 2020 (Harvard School of Public Health, Boston; WHO and World Bank, 1996).
Pazini, F. L., Cunha, M. P. & Rodrigues, A. L. S. The possible beneficial effects of creatine for the management of depression. Prog. Neuro-Psychopharmacol. Biol. Psychiatry 89, 193–206 (2019).
Gerhard, D. M., Wohleb, E. S. & Duman, R. S. Emerging treatment mechanisms for depression: focus on glutamate and synaptic plasticity. Drug Discov. Today 21, 454–464 (2016).
Lin, E. & Lane, H. Y. Genome-wide association studies in pharmacogenomics of antidepressants. Pharmacogenomics 16, 555–566 (2015).
Gonda, X. et al. Genetic variants in major depressive disorder: from pathophysiology to therapy. Pharmacol. Therapeut. 194, 22–43 (2019).
Fabbri, C., Porcelli, S. & Serretti, A. From pharmacogenetics to pharmacogenomics: the way toward the personalization of antidepressant treatment. Can. J. Psychiatry Rev. canadienne de. Psychiatr. 59, 62–75 (2014).
Ising, M. et al. A genome wide association study points to multiple loci that predict antidepressant drug treatment outcome in depression. Arch. Gen. Psychiatry 66, 966–975 (2009).
Uher, R. et al. Genome-wide pharmacogenetics of antidepressant response in the GENDEP project. Am. J. Psychiatry 167, 555–564 (2010).
Garriock, H. A. et al. A genome wide association study of citalopram response in major depressive disorder. Biol. Psychiatry 67, 133–138 (2010).
Tansey, K. E. et al. Genetic predictors of response to serotonergic and noradrenergic antidepressants in major depressive disorder: a genome-wide analysis of individual-level data and a meta-analysis. PLoS Med. 9, e1001326 (2012).
Hunter, A. M. et al. A genome-wide association study of a sustained pattern of antidepressant response. J. Psychiatr. Res. 47, 1157–1165 (2013).
Antypa, N., Drago, A. & Serretti, A. Genomewide interaction and enrichment analysis on antidepressant response. Psychol. Med. 44, 753–765 (2014).
Biernacka, J. M. et al. The International SSRI Pharmacogenomics Consortium (ISPC): a genome-wide association study of antidepressant treatment response. Transl. Psychiatry 5, e553 (2015).
Myung, W. et al. A genome-wide association study of antidepressant response in Koreans. Transl. Psychiatry 5, e633 (2015).
Cocchi, E. et al. Genome-wide association study of antidepressant response: involvement of the inorganic cation transmembrane transporter activity pathway. BMC Psychiatry 16, 106 (2016).
Fabbri, C. et al. Genome-wide association study of treatment-resistance in depression and meta-analysis of three independent samples. Br. J. Psychiatry 214, 36–41 (2019).
Fabbri, C. et al. New insights into the pharmacogenomics of antidepressant response from the GENDEP and STAR*D studies: rare variant analysis and high-density imputation. Pharmacogenomics J. 18, 413–421 (2018).
Consortium C. Sparse whole-genome sequencing identifies two loci for major depressive disorder. Nature 523, 588–591 (2015).
Bamshad, M. J. et al. Exome sequencing as a tool for Mendelian disease gene discovery. Nat. Rev. Genet. 12, 745–755 (2011).
Tammiste, A. et al. Whole-exome sequencing identifies a polymorphism in the BMP5 gene associated with SSRI treatment response in major depression. J. Psychopharmacol. 27, 915–920 (2013).
Wong, M. L. et al. Clinical outcomes and genome-wide association for a brain methylation site in an antidepressant pharmacogenetics study in Mexican Americans. Am. J. Psychiatry 171, 1297–1309 (2014).
Perera, T. et al. The Clinical TMS Society consensus review and treatment recommendations for TMS therapy for major depressive disorder. Brain Stimul. 9, 336–346 (2016).
Berlim, M. T., Van den Eynde, F. & Daskalakis, Z. J. High-frequency repetitive transcranial magnetic stimulation accelerates and enhances the clinical response to antidepressants in major depression: a meta-analysis of randomized, double-blind, and sham-controlled trials. J. Clin. Psychiatry 74, e122–e129 (2013).
Rumi, D. O. et al. Transcranial magnetic stimulation accelerates the antidepressant effect of amitriptyline in severe depression: a double-blind placebo-controlled study. Biol. Psychiatry 57, 162–166 (2005).
Lam, R. W., Chan, P., Wilkins-Ho, M. & Yatham, L. N. Repetitive transcranial magnetic stimulation for treatment-resistant depression: a systematic review and metaanalysis. Can. J. Psychiatry Rev. canadienne de. Psychiatr. 53, 621–631 (2008).
Silverstein, W. K. et al. Neurobiological predictors of response to dorsolateral prefrontal cortex repetitive transcranial magnetic stimulation in depression: a systematic review. Depress Anxiety 32, 871–891 (2015).
Holsboer, F. How can we realize the promise of personalized antidepressant medicines? Nat. Rev. Neurosci. 9, 638–646 (2008).
Hamilton, M. A rating scale for depression. J. Neurol. Neurosurg. Psychiatry 23, 56–62 (1960).
Feffer, K. et al. 1Hz rTMS of the right orbitofrontal cortex for major depression: Safety, tolerability and clinical outcomes. Eur. Neuropsychopharmacol. 28, 109–117 (2018).
Muller, K. E., Edwards, L. J., Simpson, S. L. & Taylor, D. J. Statistical tests with accurate size and power for balanced linear mixed models. Stat. Med. 26, 3639–3660 (2007).
Johnson, J. L. et al. POWERLIB: SAS/IML software for computing power in multivariate linear models. J. Statist. Softw. 30, 1–55 (2009).
Li, H. & Durbin, R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26, 589–595 (2010).
Li, H. et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
McKenna, A. et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303 (2010).
Koboldt, D. C. et al. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 22, 568–576 (2012).
Wang, K., Li, M. & Hakonarson, H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 38, e164 (2010).
Liu, Y. et al. Oligoadenylate synthetase 3 S381R gene polymorphism is associated with severity of EV71 infection in Chinese children. J. Clin. Virol. 101, 29–33 (2018).
Turakulov, R. & Easteal, S. Number of SNPS loci needed to detect population structure. Hum. Heredity 55, 37–45 (2003).
Fischer, M. C. et al. Estimating genomic diversity and population differentiation—an empirical comparison of microsatellite and SNP variation in Arabidopsis halleri. BMC Genomics 18, 69 (2017).
Apud, J. A., Zhang, F., Decot, H., Bigos, K. L. & Weinberger, D. R. Genetic variation in KCNH2 associated with expression in the brain of a unique hERG isoform modulates treatment response in patients with schizophrenia. Am. J. Psychiatry 169, 725–734 (2012).
Colantuoni, C. et al. Temporal dynamics and genetic control of transcription in the human prefrontal cortex. Nature 478, 519–523 (2011).
Hyde, C. L. et al. Identification of 15 genetic loci associated with risk of major depression in individuals of European descent. Nat. Genet. 48, 1031–1036 (2016).
Wray, N. R. et al. Genome-wide association analyses identify 44 risk variants and refine the genetic architecture of major depression. Nat. Genet. 50, 668–681 (2018).
Howard, D. M. et al. Genome-wide association study of depression phenotypes in UK Biobank identifies variants in excitatory synaptic pathways. Nat. Commun. 9, 1470 (2018).
Fabbri, C. et al. The genetics of treatment-resistant depression: a critical review and future perspectives. Int. J. Neuropsychopharmacol. 22, 93–104 (2019).
Valiulis, V. et al. Electrophysiological differences between high and low frequency rTMS protocols in depression treatment. Acta Neurobiologiae Experimentalis 72, 283–295 (2012).
Hashemi, M. et al. 4-bp insertion/deletion (rs3783553) polymorphism within the 3’UTR of IL1A contributes to the risk of prostate cancer in a sample of Iranian population. J. Cell. Biochem. 119, 2627–2635 (2018).
Li, L. et al. Association of IL-1A and IL-1B polymorphisms with ankylosing spondylitis among the Chinese Han population: a case-control study. Oncotarget 8, 28278–28284 (2017).
McClay, J. L. et al. Genome-wide pharmacogenomic study of neurocognition as an indicator of antipsychotic treatment response in schizophrenia. Neuropsychopharmacology 36, 616–626 (2011).
Palmer, N. D. et al. Genetic variants associated with quantitative glucose homeostasis traits translate to type 2 diabetes in Mexican Americans: the GUARDIAN (Genetics Underlying Diabetes in Hispanics) Consortium. Diabetes 64, 1853–1866 (2015).
Smith, N. L. et al. Association of genome-wide variation with the risk of incident heart failure in adults of European and African ancestry: a prospective meta-analysis from the cohorts for heart and aging research in genomic epidemiology (CHARGE) consortium. Circulation Cardiovascular Genet. 3, 256–266 (2010).
Spada, J. et al. Genome-wide association analysis of actigraphic sleep phenotypes in the LIFE Adult Study. J. Sleep Res. 25, 690–701 (2016).
Nadalin, S. & Buretić-Tomljanović, A. An association between PLA2G6 and PLA2G4C gene polymorphisms and schizophrenia risk and illness severity in a Croatian population. Prostaglandins Leukot. Ess. Fat. Acids 121, 57–59 (2017).
Liu, S. et al. The rs251684 variant of PLA2G4C is associated with autism spectrum disorder in the Northeast Han Chinese population. Genet Test. Mol. Biomark. 20, 747–752 (2016).
Chang, D. et al. A meta-analysis of genome-wide association studies identifies 17 new Parkinson’s disease risk loci. Nat. Genet. 49, 1511–1516 (2017).
Nalls, M. A. et al. Large-scale meta-analysis of genome-wide association data identifies six new risk loci for Parkinson’s disease. Nat. Genet. 46, 989–993 (2014).
Do, C. B. et al. Web-based genome-wide association study identifies two novel loci and a substantial genetic component for Parkinson’s disease. PLoS Genet. 7, e1002141 (2011).
Pickrell, J. K. et al. Detection and interpretation of shared genetic influences on 42 human traits. Nat. Genet. 48, 709–717 (2016).
de Mooij, C. E. M., Netea, M. G., van der Velden, W. & Blijlevens, N. M. A. Targeting the interleukin-1 pathway in patients with hematological disorders. Blood 129, 3155–3164 (2017).
Westerlind, H. et al. GNA11和MS -Identity-by-descent mapping in a Scandinavian multiple sclerosis cohort. Eur. J. Hum. Genet. 23, 688–692 (2015).
Kao, C. F., Jia, P., Zhao, Z. & Kuo, P. H. Enriched pathways for major depressive disorder identified from a genome-wide association study. Int. J. Neuropsychopharmacol. 15, 1401–1411 (2012).
Van Raamsdonk, C. D. et al. Mutations in GNA11 in uveal melanoma. N. Engl. J. Med. 363, 2191–2199 (2010).
Nesbit, M. A. et al. Mutations affecting G-protein subunit alpha11 in hypercalcemia and hypocalcemia. N. Engl. J. Med. 368, 2476–2486 (2013).
Acknowledgements
We would like to thank all study participants, without whom this research would not have been possible. We thank the colleagues who participated in the collection of the clinical data, Yingying Yin, Ying Chen, Liting Dong, and Wenxiang Liao in the Department of Neurology and Psychiatry of the Affiliated ZhongDa Hospital and Medical School of the Southeast University, and Beijing GenoMed Data Technology Co. Ltd. This study has been supported by grants from the National Key Research and Development Program of China (2016YFC1306700 to Z.Z., and 2016YFC1306702 to Y.Y.), the National Key Science and Technology Program in the 12th Five Year Plan of China-The Project of New Drug Formulation (2012ZX09506-001-009 to Z.Z.), the National Natural Science Foundation of China (81420108012, 31371074 and 81830040 to Z.Z. and 81301167 to Z.X.), Program of Excellent Talents in Medical Science of Jiangsu Province (JCRCA2016006 to Z.Z.), Science and Technology Program of Guangdong (2018B030334001 to Z.Z.), the Jiangsu Provincial Medical Youth Talent (QNRC2016825 to Z.X.), Nature Science Foundation of Jiangsu Province (BK20181272 to Z.X.) as well as the support from the Global Clinical and Translational Research Institute, Bethesda, MD and the Beijing “Haiju” Scholarship (to F.Z.).
Author information
Authors and Affiliations
Contributions
Z.Z. conceived and designed the study and F.Z. designed the study analysis plan. Z.Z. was in charge of the clinical studies and Z.X., Y.Y., H.Z., X.H. and C.X. carried out the clinical study protocols and participated in the recruitment of the study subjects, biospecimen collection, and clinical evaluation. X.Q., C.L. and Y.T. conducted the DNA extraction, designed the experiments, and performed the sequencing analysis, as well as the bioinformatics annotations. F.Z. and L.X. conducted all statistical analysis, bioinformatics investigation of genes identified, RNA-seq data analysis, and network and pathway analysis. F.Z. and Z.Z. carried out the interpretation of results, while Z.X., L.X. and C.L. prepared the tables and figures. Z.Z., F.Z., Z.X. and Y.T. wrote the manuscript and were responsible for the data interpretation.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Xu, Z., Xie, C., Xia, L. et al. Targeted exome sequencing identifies five novel loci at genome-wide significance for modulating antidepressant response in patients with major depressive disorder. Transl Psychiatry 10, 30 (2020). https://doi.org/10.1038/s41398-020-0689-x
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41398-020-0689-x