Abstract
Attention deficit hyperactive disorder (ADHD) is a highly heritable neurodevelopmental disorder, and excessive daytime sleepiness is frequently observed in ADHD patients. Excessive daytime sleepiness is also a core symptom of narcolepsy and essential hypersomnia (EHS), which are also heritable conditions. Psychostimulants are effective for the symptomatic control of ADHD (primary recommended intervention) and the two sleep disorders (frequent off-label use). However, the common biological mechanism for these disorders has not been well understood. Using a previously collected genome-wide association study of narcolepsy and EHS, we calculated polygenic risk scores (PRS) for each individual. We investigated a possible genetic association between ADHD and narcolepsy traits in the Hamamatsu Birth Cohort for mothers and children (HBC study) (n = 876). Gene-set enrichment analyses were used to identify common pathways underlying these disorders. Narcolepsy PRS were significantly associated with ADHD traits both in the hyperactivity domain (e.g., P-value threshold < 0.05, β [SE], 5.815 [1.774]; P = 0.002) and inattention domain (e.g., P-value threshold < 0.05, β [SE], 5.734 [1.761]; P = 0.004). However, EHS PRS was not significantly associated with either domain of ADHD traits. Gene-set enrichment analyses revealed that pathways related to dopaminergic signaling, immune systems, iron metabolism, and glial cell function involved in both ADHD and narcolepsy. Findings indicate that ADHD and narcolepsy are genetically related, and there are possible common underlying biological mechanisms for this relationship. Future studies replicating these findings would be warranted to elucidate the genetic vulnerability for daytime sleepiness in individuals with ADHD.
Similar content being viewed by others
Introduction
Attention deficit hyperactive disorder (ADHD) is a common neurodevelopmental disorder with onset occurring in childhood. Genetic factors play an important role in the development of ADHD, with a heritability of up to 75%1. Both rare and common genetic variants have been associated with ADHD. Recent genome-wide association studies (GWAS) have identified several genomic regions associated with ADHD, with a modest contribution of common variants to its development2.
Recently, based on a polygenic model for complex disorders, polygenic risk scores (PRS) have been calculated and widely used to explain the genetic liability of individuals for certain diseases or phenotypes, using common genetic variants tested in GWAS3. PRS can also be used to examine the genetic association between two different diseases or phenotypes. To date, the association of ADHD PRS has been reported with depressive symptoms4, body mass index5, substance use disorder6, and educational attainment7, but no examination has thus far been conducted with sleep disorders. Nevertheless, patients with ADHD frequently show excessive daytime sleepiness (EDS)8. Narcolepsy is characterized by EDS with or without cataplexy (sudden and uncontrollable muscle weakness or paralysis triggered by a strong emotion), hypnagogic hallucinations, and sleep paralysis (vivid and dream-like hallucinations and muscle paralysis while falling asleep), and disturbed nocturnal sleep. The prevalence of ADHD in individuals with narcolepsy is (~35%)9 higher than in the general population (~5%)10. Furthermore, psychostimulants, such as methylphenidate, are effective for controlling symptoms of ADHD (first-line approved treatment) and narcolepsy11 (frequently used off-line with good effects). However, the nature of the relationship between the two conditions and the possibility that underlying biological mechanisms connect the two have not been elucidated yet.
One recent study conducted in Japan demonstrated that the relative risk of narcolepsy in affected first-degree family members is 10- to 40-fold higher than in the general population, suggesting that genetic factors play an important role in this disease12. Of note, the HLA DQB1*15:01 and DQB1*06:02 genes have been shown to be associated with narcolepsy, and additional genes may be involved13. Another recent GWAS study, using Japanese individuals, identified some genomic loci related to narcolepsy outside of the major histocompatibility complex (MHC) region, such as CPT1B, TCRA, and P2RY1112. Capitalizing on the much higher prevalence of narcolepsy in the Japanese population (0.16–0.18%)14 compared to the Caucasian population (0.02–0.06%)15, and using the previous Japanese GWAS study of narcolepsy as a discovery cohort, we investigated a possible genetic association between ADHD traits and narcolepsy in our birth cohort sample from the HBC study. We also conducted a polygenic risk analysis of ADHD traits using a summary from the previous GWAS study of essential hypersomnia (EHS), another highly heritable sleep disorder16, conducted in a Japanese population17. They identified genome-wide significance in single nucleotide polymorphism (SNP) on the CRAT gene located in 9q34.1117. Although there has been no direct evidence showing the high-concordance rate in twins and familial accumulation of EHS and narcolepsy, it has been reported that these disorders share several susceptibility genes16, and ~40% of the heritability of EHS was explained by the genetic background of narcolepsy12.
Furthermore, in order to learn more about the common biological mechanisms underlying ADHD and narcolepsy, a gene-set enrichment analysis was performed.
Methods
Participants
The discovery cohort for narcolepsy consisted of 409 patients with narcolepsy-cataplexy (narcolepsy type 1: NT1) and 1562 healthy controls. The discovery cohort for EHS was 125 EHS patients without HLA-DQB1*06:02 and 562 controls without HLA-DQB1*06:02. Details are available on the NBDC Human Database (https://biosciencedbc.jp/en/). The target population from the HBC study included infants (n = 876) born between December 2007 and June 2011. Recruitment procedures are fully described elsewhere18. The study procedures were approved by the Hamamatsu University School of Medicine and the University Hospital Ethics Committee (research ID:17-037 and 19-145). Written informed consent was obtained from each caregiver for his/her infant’s participation. Individuals with intellectual disability (FSIQ < 80, n = 66), measured by Wechsler Preschool and Primary Scale of Intelligence (WPPSI), were excluded from the analysis. Also, individuals with parents of non-Japanese descent (n = 8) were removed from the study to minimize the effect of population stratification. This study followed the STrengthening the Reporting of OBservational studies in Epidemiology (STROBE) reporting guideline.
Measurement
ADHD traits were ascertained by interview at age 8 using the Japanese version of the ADHD-RS, previously confirmed for its validity and reliability19. ADHD-RS consists of two subscales: inattention (nine items) and hyperactivity-impulsivity (nine items). Each item was rated from 0 (never or rarely) to 3 (very often). The sum of raw scores for inattention, hyperactivity-impulsivity domains, and total scores were converted to percentile scores using the scoring procedure provided in the manufacture’s manual20. Full-scale IQ was measured using the Japanese version of the WPPSI, a reliable and child-friendly measure of cognitive development, administered at age 4. All psychological evaluations were conducted by trained psychologists.
Genotyping, quality control, and imputation
Genotyping was conducted using Japonica array, designed specifically for 650,000 SNP for a Japanese population21. In brief, the quality controls for retaining SNPs and individuals were as follows: missing data for SNP < 0.02, Pi-hat calculated by pairwise identify-by-descent (IBD) analysis <0.2, SNP Hardy Weinberg equilibrium of p > 10−6, and minor allele frequency > 0.01. Seventy-six individuals were removed from the analysis as “related individuals” by IBD analysis. Genotyping imputation was performed using BEAGLE 5.0 to the 1000 Genome Project reference panel phase 3 of the Japanese population22. SNPs with an imputation INFO score <0.8 were excluded. We also excluded SNPs located within the MHC region because of high linkage equilibrium in this region. The number of SNPs analyzed for PRS was 5,606,655.
PRS analysis
We used PRSice-2 to generate PRS, according to the developers’ protocol23. The summary GWAS data used to determine the PRS for narcolepsy and EHS was obtained from the NBDC Human Database (https://humandbs.biosciencedbc.jp/en/). To account for population stratification, we included four principle components (PCs). The PCs were calculated based on the pruned data with PLINK 1.924. The criteria for SNP clumping was pairwise LD r2 < 0.1 within the 1 Mb window. PRSs were calculated with different P-value thresholds at 0.05, 0.05, 0.1, 0.5, and 1. Standardized PRS scores (mean = 0 and standard deviation = 1) were used for the analyses.
Gene-set enrichment analysis
Gene-set enrichment analyses were conducted using PRSet implemented in PRSice-2 software23. Gene-sets collections were obtained from the MSigDB database, and GO gene sets (c5) were used for the analyses (http://software.broadinstitute.org/gsea/msigdb/index.jsp). A total GO gene sets used for analysis was 7350. The P-value threshold for PRSet was set at 1 since gene-set PRSs containing a small portion of SNPs may be unrepresentative of the whole gene-sets25. The obtained P-value was corrected for 10,000 permutation tests, and the significance was set at 0.05.
Genetic correlation analysis
Genetic correlations (rg) between narcolepsy and EHS were calculated using LD (linkage disequilibrium) Score regression26. Pre-computed LD scores for the East Asian population from the 1000 Genomes Project were used in the analysis (LD scores available on https://github.com/bulik/ldsc) and the summary statistics from the NBDC Human Database was used. Genetic correlation between ADHD and narcolepsy was analyzed using the recent large ADHD GWAS study2. Since the difference in the ethnic population between ADHD and narcolepsy GWAS study, we utilized the “Popcorn” program, which can calculate trans-ethnic genetic correlation27.
Results
PRS analysis
Participant characteristics are summarized in Table 1. A total of 726 participants (373 males, 353 females) were analyzed. The narcolepsy PRS was significantly associated with ADHD traits at various P-value thresholds either in the hyperactivity (e.g., P-value threshold < 0.05, β [SE], 5.815 [1.774]; P = 0.002), inattentive domains (e.g., P-value threshold < 0.05, β [SE], 5.734 [1.761]; P = 0.004) and total score (e.g., P-value threshold < 0.05, β [SE], 6.496 [1.676]; P = 0.001) (Table 2, Fig. 1a, b, Supplemental figure). EHS PRS was not associated with ADHD traits at any P-value threshold analyzed (e.g., P-value threshold < 0.05), in the hyperactivity domain (β [SE], 2.206 [1.726]; P = 0.2088) and inattentive domain (β [SE], 2.025 [1.715]; P = 0.3753); total score (β [SE], 1.903 [1.649]; P = 0.4174) (Table 3).
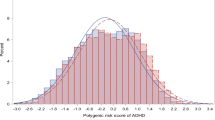
a Quantile plot of Narcolepsy PRS and ADHD traits (hyperactivity). This figure provides the percentile of ADHD-RS hyperactivity score against the strata of polygenic risk score for narcolepsy after the cohort was divided into 100 groups. The error bars mean standard deviation. b Quantile plot of Narcolepsy PRS and ADHD traits (inattention). This figure provides the percentile of ADHD-RS inattention score against the strata of polygenic risk score for narcolepsy after the cohort was divided into 100 groups. The error bar means standard deviation. ADHD attention deficit hyperactivity disorder, PRS polygenic risk score.
Gene-set enrichment analysis
Gene-set enrichment analyses found that several gene-sets, such as “dopaminergic neuron differentiation” (GO: 0071542; β [SE], −5.680 [1.872]; P = 0.0034) and “interleukin 6 production” (GO: 0032635; β [SE], 6.007 [1.834]; P = 0.0006), were associated with hyperactivity traits and narcolepsy (Table 4), while gene-sets, such as “iron coordination entity transport” (GO: 190167; β [SE], −6.686 [1.795]; P = 0.0002) and “positive regulation of glial cell differentiation” (GO: 0045687; β [SE], 5.457 [1.808]; P = 0.00350) were significantly associated with both inattention traits and narcolepsy (Table 5).
Exploratory analysis using the Comparative Toxicogenomics Database (http://ctdbase.org) indicated that several compounds, including N-acetylcysteine, baicalein, and fatty acids, were modulators for hyperactivity-related gene ontologies, and dextroamphetamine was a modulator for inattention-related gene-sets.
Genetic correlation analysis
LD score regression analysis using summary data of previous GWAS studies of narcolepsy and EHS showed that these disorders were positively correlated (rg [SE] = 0.500 [0.062], P = 4.42 × 10−16). Genetic correlation analysis using summary data of Japanese narcolepsy GWAS and ADHD GWAS conducted by psychiatric genomics consortium showed that these disorders were also positively correlated (rg [SE] = 0.165 [0.114], P = 1.11 × 10−5).
Discussion
To our knowledge, this is the first study that has examined the genetic association of ADHD traits and narcolepsy. We found that the genetic risk for narcolepsy has a significant association with ADHD traits using data from the general population. Specifically, both hyperactivity and inattention traits were significantly associated with narcolepsy PRS, suggesting that these disorders have a close genetic association. Genetic correlation analysis using previous GWAS studies of narcolepsy and ADHD also support the present findings.
Managing EDS among individuals with ADHD is important since EDS is directly related to academic performance and teachers’ ratings of classroom behavior and performance28. However, EDS in ADHD has usually been considered the result of delayed sleep phase or poor quality of sleep29, meaning that the nature of the relationship and the possibility of common underlying biological determinants have not been investigated. Based on the present findings, clinicians, caregivers, and teachers should be informed that there could be a genetic vulnerability for daytime sleepiness in children and adolescents with ADHD. Demontis et al.2 found a significant association between ADHD and insomnia. Their finding is not directly linked to our present findings; however, it is also known that subjects with narcolepsy frequently experience disrupted nighttime sleep, which could exacerbate EDS30. Future studies examining the relationship between genetic risks for narcolepsy and the quality of nighttime sleep in the present population is needed.
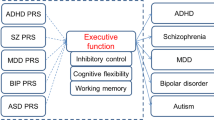
Interestingly, we identified pathways related to common molecular mechanisms underlying ADHD traits and narcolepsy. Several gene-sets were identified as having an association with ADHD symptoms and narcolepsy in the gene-set enrichment analysis. The results are intriguing and bear further study. Notably, gene-sets relevant to dopaminergic signaling were associated with both disorders, consistent with the recognition that most medications currently used for ADHD, narcolepsy and EHS are thought to increase synaptic dopamine9. Immune systems have long been implicated in the mechanism of narcolepsy31, but recent evidence suggests that neuroinflammation is involved in the development of ADHD as well32. Furthermore, neuronal inflammation has been shown to not only alter levels of monoamine neurotransmitters, including dopamine and noradrenaline32 but also to affect the expression of histamine and orexin33,34, which are promising targets for novel drug treatments for narcolepsy. In addition, gene-sets relevant to glial cell function and iron transport were found to be associated with both ADHD traits and narcolepsy. In line with these findings, recent genetics and brain imaging studies have suggested that altered myelin and oligodendrocyte related signaling were implicated in both ADHD2,35 and narcolepsy36,37. Changes in iron homeostasis have been shown to affect dopamine synthesis38 and also been proposed as a biological mechanism for ADHD39,40. Although there has been no direct evidence of iron deficiency in narcolepsy subjects, restless leg syndrome (RLS) was shown to be more prevalent in subjects with narcolepsy compared to general population41. The exact pathophysiology of RLS is still unclear. However, it has been repeatedly reported that RLS is also common in ADHD subjects42, and iron homeostasis and dopamine signaling have been implicated in the development of this condition43. These gene-set enrichment analyses suggest that ADHD traits and narcolepsy share common biological mechanisms such as dopamine signaling, immune system regulation, glial cell function, and iron homeostasis.
We did not find a significant association between EHS PRS and ADHD traits, although genetic correlation analysis showed that narcolepsy and EHS were genetically correlated. One possible explanation for this is that the sample size of the discovery cohort of EHS (Case = 125, control = 1562) was smaller than that of narcolepsy (Case = 409, control = 1562). Thus the calculated EHS PRS did not have adequate power for detecting association with ADHD traits in this study.
Our exploratory analysis using gene-sets enriched in ADHD traits and narcolepsy suggests that medications currently used for ADHD and narcolepsy, such as the psychostimulants, target common biological pathways underlying both disorders. We also found that N-acetylcysteine, baicalein, and fatty acid could be investigated as novel therapeutic agents for both ADHD and narcolepsy symptoms. Although very preliminary, reports suggest the efficacy of N-acetylcysteine for both hyperactivity and inattention symptoms of ADHD44,45. N-acetylcysteine has also been shown to be protective for dopamine neurons46. Baicalein is a flavonoid purified from the plant Scutellaria baicalensis. A recent study using an animal model of ADHD demonstrated the efficacy of core ADHD symptoms by regulating dopaminergic signaling in the brain47. More recently, the reduction of fatty acids in the blood48 among children and adolescents with ADHD, and similarly, the efficacy of a high dose of eicosapentaenoic acid (EPA) for ADHD symptoms have been reported49. Future clinical trials using both existing and novel compounds for ADHD and narcolepsy are warranted.
Limitations
There are some limitations to this study. First, we targeted a general population, leveraging our longitudinal birth cohort HBC. The cohort is representative of the general population, and thus caution is needed when applying the findings to clinically diagnosed ADHD patients. Second, the original GWAS study of narcolepsy included individuals only with NT1, but not narcolepsy type 2 (NT2), which lacks cataplexy. It is unknown whether the genetic architectures of NT1 and NT2 are the same. Thus, a future GWAS study using NT2 is needed. Third, the prevalence of narcolepsy is known to be higher in the Japanese than the Caucasian population, and future studies using different ethnic populations are needed to replicate these findings.
Conclusion
In this study, using PRS analysis, we found that genetic risk factors for narcolepsy contribute to ADHD traits in the general Japanese population, and identified common molecular pathways in these conditions. We hope that this discovery will be replicated and lead to an understanding that EDS in ADHD might have a biological foundation.
References
Sullivan, P. F. & Geschwind, D. H. Defining the genetic, genomic, cellular, and diagnostic architectures of psychiatric disorders. Cell 177, 162–183 (2019).
Demontis, D. et al. Discovery of the first genome-wide significant risk loci for attention deficit/hyperactivity disorder. Nat. Genet. 51, 63–75 (2019).
Martin, A. R., Daly, M. J., Robinson, E. B., Hyman, S. E. & Neale, B. M. Predicting polygenic risk of psychiatric disorders. Biol. Psychiatry 86, 97–109 (2019).
Brikell, I. et al. The contribution of common genetic risk variants for ADHD to a general factor of childhood psychopathology. Mol. Psychiatry 25, 1809–1821 (2020).
Barker, E. D. et al. Do ADHD-impulsivity and BMI have shared polygenic and neural correlates? Mol, Psychiatry (2019).
Wimberley, T. et al. Genetic liability to ADHD and substance use disorders in individuals with ADHD. Addiction 115, 1368–1377 (2020).
Verhoef, E. et al. Disentangling polygenic associations between attention-deficit/hyperactivity disorder, educational attainment, literacy and language. Transl. Psychiatry 9, 35 (2019).
Carpena, M. X. et al. The Role of Sleep Duration and Sleep Problems During Childhood in the Development of ADHD in Adolescence: Findings from a Population-Based Birth Cohort. J. Atten. Disord. 1087054719879500 (2019).
Lecendreux, M. et al. Attention-Deficit/Hyperactivity Disorder (ADHD) Symptoms in Pediatric Narcolepsy: A Cross-Sectional Study. Sleep 38, 1285–1295 (2015).
Association A. P. Diagnostic and Statistical Manual of Mental Disorders. 5th edn (Washington, DC, 2013).
Sharbaf Shoar, N., Marwaha, R. & Molla, M. Dextroamphetamine-Amphetamine. StatPearls: Treasure Island (FL), (2019).
Yamasaki, M. et al. Evaluation of polygenic risks for narcolepsy and essential hypersomnia. J. Hum. Genet. 61, 873–878 (2016).
Miyagawa, T. & Tokunaga, K. Genetics of narcolepsy. Hum. Genome Var. 6, 4 (2019).
Overeem, S., Mignot, E., van Dijk, J. G. & Lammers, G. J. Narcolepsy: clinical features, new pathophysiologic insights, and future perspectives. J. Clin. Neurophysiol. 18, 78–105 (2001).
Mignot, E. Genetic and familial aspects of narcolepsy. Neurology 50(2 Suppl 1), S16–S22 (1998).
Miyagawa, T. et al. Polymorphism located in TCRA locus confers susceptibility to essential hypersomnia with HLA-DRB1*1501-DQB1*0602 haplotype. J. Hum. Genet. 55, 63–65 (2010).
Miyagawa, T. et al. A variant at 9q34.11 is associated with HLA-DQB1*06:02 negative essential hypersomnia. J. Hum. Genet. 63, 1259–1267 (2018).
Takagai, S. et al. Cohort Profile: Hamamatsu Birth Cohort for Mothers and Children (HBC Study). Int J. Epidemiol. 45, 333–342 (2016).
Tani, I., Okada, R., Ohnishi, M., Nakajima, S. & Tsujii, M. Japanese version of home form of the ADHD-RS: an evaluation of its reliability and validity. Res. Dev. Disabil. 31, 1426–1433 (2010).
Pappas, D. ADHD Rating Scale-IV: Checklists, Norms, and Clinical Interpretation. J. Psychoeducational Assess. 24, 172–178 (2006).
Kawai, Y. et al. Japonica array: improved genotype imputation by designing a population-specific SNP array with 1070 Japanese individuals. J. Hum. Genet. 60, 581–587 (2015).
Browning, B. L., Zhou, Y. & Browning, S. R. A one-penny imputed genome from next-generation reference panels. Am. J. Hum. Genet. 103, 338–348 (2018).
Choi, S. W. & O’Reilly, P. F. PRSice-2: Polygenic Risk Score software for biobank-scale data. Gigascience 8, giz082 (2019).
Chang, C. C. et al. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience 4, 7 (2015).
Fanelli, G. et al. Higher polygenic risk scores for schizophrenia may be suggestive of treatment non-response in major depressive disorder. medRxiv (2020).
Bulik-Sullivan, B. et al. An atlas of genetic correlations across human diseases and traits. Nat. Genet. 47, 1236–1241 (2015).
Brown, B. C. Asian Genetic Epidemiology Network Type 2 Diabetes C, Ye, C. J., Price, A. L. & Zaitlen, N. Transethnic genetic-correlation estimates from summary statistics. Am. J. Hum. Genet. 99, 76–88 (2016).
Langberg, J. M., Dvorsky, M. R., Marshall, S. & Evans, S. W. Clinical implications of daytime sleepiness for the academic performance of middle school-aged adolescents with attention deficit hyperactivity disorder. J. Sleep. Res. 22, 542–548 (2013).
Hvolby, A. Associations of sleep disturbance with ADHD: implications for treatment. Atten. Defic. Hyperact. Disord. 7, 1–18 (2015).
Roth, T. et al. Disrupted nighttime sleep in narcolepsy. J. Clin. Sleep. Med. 9, 955–965 (2013).
Barateau, L. & Dauvilliers, Y. Recent advances in treatment for narcolepsy. Ther. Adv. Neurol. Disord. 12, 1756286419875622 (2019).
Dunn, G. A., Nigg, J. T. & Sullivan, E. L. Neuroinflammation as a risk factor for attention deficit hyperactivity disorder. Pharm. Biochem. Behav. 182, 22–34 (2019).
Cacabelos, R., Torrellas, C., Fernandez-Novoa, L. & Lopez-Munoz, F. Histamine and immune biomarkers in CNS disorders. Mediators Inflamm. 2016, 1924603 (2016).
Zhan, S. et al. Molecular mechanism of tumour necrosis factor alpha regulates hypocretin (orexin) expression, sleep and behaviour. J. Cell Mol. Med. 23, 6822–6834 (2019).
Lesch, K. P. Editorial: Can dysregulated myelination be linked to ADHD pathogenesis and persistence? J. Child Psychol. Psychiatry 60, 229–231 (2019).
Hor, H. et al. A missense mutation in myelin oligodendrocyte glycoprotein as a cause of familial narcolepsy with cataplexy. Am. J. Hum. Genet. 89, 474–479 (2011).
Gool, J. K. et al. Widespread white matter connectivity abnormalities in narcolepsy type 1: a diffusion tensor imaging study. Neuroimage Clin. 24, 101963 (2019).
Daubner, S. C., Le, T. & Wang, S. Tyrosine hydroxylase and regulation of dopamine synthesis. Arch. Biochem. Biophys. 508, 1–12 (2011).
Adisetiyo, V., Gray, K. M., Jensen, J. H. & Helpern, J. A. Brain iron levels in attention-deficit/hyperactivity disorder normalize as a function of psychostimulant treatment duration. Neuroimage Clin. 24, 101993 (2019).
Adisetiyo, V. et al. Multimodal MR imaging of brain iron in attention deficit hyperactivity disorder: a noninvasive biomarker that responds to psychostimulant treatment? Radiology 272, 524–532 (2014).
Plazzi, G. et al. Restless legs syndrome is frequent in narcolepsy with cataplexy patients. Sleep 33, 689–694 (2010).
Walters, A. S., Silvestri, R., Zucconi, M., Chandrashekariah, R. & Konofal, E. Review of the possible relationship and hypothetical links between attention deficit hyperactivity disorder (ADHD) and the simple sleep related movement disorders, parasomnias, hypersomnias, and circadian rhythm disorders. J. Clin. Sleep. Med. 4, 591–600 (2008).
Lopez, R. et al. Restless legs syndrome and iron deficiency in adults with attention-deficit/hyperactivity disorder. Sleep 42, zsz027 (2019).
Garcia, R. J. et al. Attention deficit and hyperactivity disorder scores are elevated and respond to N-acetylcysteine treatment in patients with systemic lupus erythematosus. Arthritis Rheum. 65, 1313–1318 (2013).
Deepmala et al. Clinical trials of N-acetylcysteine in psychiatry and neurology: a systematic review. Neurosci. Biobehav. Rev. 55, 294–321 (2015).
Feio-Azevedo, R. et al. Toxicity of the amphetamine metabolites 4-hydroxyamphetamine and 4-hydroxynorephedrine in human dopaminergic differentiated SH-SY5Y cells. Toxicol. Lett. 269, 65–76 (2017).
Zhou, R. et al. Baicalin regulates the dopamine system to control the core symptoms of ADHD. Mol. Brain 12, 11 (2019).
Chang, J. P., Su, K. P., Mondelli, V. & Pariante, C. M. Omega-3 polyunsaturated fatty acids in youths with attention deficit hyperactivity disorder: a systematic review and meta-analysis of clinical trials and biological studies. Neuropsychopharmacology 43, 534–545 (2018).
Chang, J. P. et al. High-dose eicosapentaenoic acid (EPA) improves attention and vigilance in children and adolescents with attention deficit hyperactivity disorder (ADHD) and low endogenous EPA levels. Transl. Psychiatry 9, 303 (2019).
Acknowledgements
We are grateful to the individuals who participated in the study. We would like to thank Ms. Chikako Nakayasu, Ms. Yuko Anma, Ms. Haruka Suzuki for data collection. We would like to thank Ms. Noriko Kodera and Ms. Emi Higashimoto for administration. This work was supported by grants from the Ministry of Education, Culture, Sports, Science & Technology in Japan (grant number 19H03582 to K.J.T.) and National Institute of Mental Health (grant number NIMH R01 MH102729 to Y.N.).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Takahashi, N., Nishimura, T., Harada, T. et al. Polygenic risk score analysis revealed shared genetic background in attention deficit hyperactivity disorder and narcolepsy. Transl Psychiatry 10, 284 (2020). https://doi.org/10.1038/s41398-020-00971-7
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41398-020-00971-7
This article is cited by
-
Understanding zebrafish sleep and wakefulness physiology as an experimental model for biomedical research
Fish Physiology and Biochemistry (2024)
-
Sleep disturbances in ADHD: investigating the contribution of polygenic liability for ADHD and sleep-related phenotypes
European Child & Adolescent Psychiatry (2023)
-
Genome-wide association study reveals ethnicity-specific SNPs associated with ankylosing spondylitis in the Taiwanese population
Journal of Translational Medicine (2022)
-
Genetic links between narcolepsy and ADHD
Translational Psychiatry (2021)