Abstract
Anhedonia is a core symptom of several psychiatric disorders but its biological underpinnings are poorly understood. We performed a genome-wide association study of state anhedonia in 375,275 UK Biobank participants and assessed for genetic correlation between anhedonia and neuropsychiatric conditions (major depressive disorder, schizophrenia, bipolar disorder, obsessive compulsive disorder and Parkinson’s Disease). We then used a polygenic risk score approach to test for association between genetic loading for anhedonia and both brain structure and brain function. This included: magnetic resonance imaging (MRI) assessments of total grey matter volume, white matter volume, cerebrospinal fluid volume, and 15 cortical/subcortical regions of interest; diffusion tensor imaging (DTI) measures of white matter tract integrity; and functional MRI activity during an emotion processing task. We identified 11 novel loci associated at genome-wide significance with anhedonia, with a SNP heritability estimate (h2SNP) of 5.6%. Strong positive genetic correlations were found between anhedonia and major depressive disorder, schizophrenia and bipolar disorder; but not with obsessive compulsive disorder or Parkinson’s Disease. Polygenic risk for anhedonia was associated with poorer brain white matter integrity, smaller total grey matter volume, and smaller volumes of brain regions linked to reward and pleasure processing, including orbito-frontal cortex. In summary, the identification of novel anhedonia-associated loci substantially expands our current understanding of the biological basis of state anhedonia and genetic correlations with several psychiatric disorders confirm the utility of this phenotype as a transdiagnostic marker of vulnerability to mental illness. We also provide the first evidence that genetic risk for state anhedonia influences brain structure, including in regions associated with reward and pleasure processing.
Similar content being viewed by others
Introduction
Anhedonia refers to reduced capacity to experience pleasure in situations that individuals would normally enjoy, and has been a focus of psychiatry research for decades1,2. It constitutes a core symptom of several neuropsychiatric disorders, including major depressive disorder (MDD), schizophrenia, bipolar disorder and obsessive compulsive disorder (OCD), as well as Parkinson’s Disease (PD)3,4,5,6. Along with a direct negative association with quality of life and subjective wellbeing, anhedonia is associated with multiple negative health-related behaviours, such as smoking, illicit drug use, and low physical activity, even in the absence of psychiatric disorder7,8. In line with a Research Domain Criteria (RDoC) approach9, state anhedonia can be measured and studied as a dimensional psychopathological trait.
Anhedonia has been closely linked to the function and structure of reward circuitry in the brain (primarily frontal, striatal and limbic regions). These neurobiological associations are consistent with the view that anhedonia reflects dysfunction in reward processing10,11. Measures of anhedonia have been associated with altered functional activity during reward-based tasks within frontal cortical regions (medial frontal cortex) and subcortical striatal regions (caudate and putamen)12, and with reduced volumes in a similar set of frontal and striatal regions13,14. Anhedonia is also associated with reduced white matter integrity15,16.
The genetic underpinnings of anhedonia are largely unknown. Several GWAS of disorders where anhedonia is a feature have been reported, such as MDD and schizophrenia17,18. However, to date only small underpowered GWAS studies of anhedonia have been published. A study of 759 patients with MDD identified 18 SNPs associated with an ‘interest-activity’ measure of anhedonia19. The largest study to date is a mega-analysis of three studies of young people from the UK and Sweden, with a total sample size of 6579; a single locus was associated with anhedonia in the discovery sample, but not in the replication sample20. A Finnish study examined genetic associations with physical and social anhedonia, as assessed with the Chapman scales21, in 3820 individuals but no genome-wide significant loci were identified22. Genetic loci associated with anhedonia have therefore not yet been reliably identified in large clinical or general population samples23, and association tests of genetic risk for anhedonia with brain structure and function have not yet been performed.
Here we report a large GWAS of state anhedonia within the UK Biobank cohort. We also use a polygenic risk score (PRS) approach to assess whether genetic loading for anhedonia is associated with brain structure and brain function.
Methods
UK Biobank sample
UK Biobank is a large cohort of over half a million UK residents, aged between 39 and 73 years at baseline assessment24. The cohort was designed to assess how genetic, environmental and lifestyle factors influence a range of morbidities in middle and older age. Baseline assessments occurred over a 4-year recruitment period (from 2006 to 2010) across 22 UK centres. These assessments covered a wide range of social, cognitive, lifestyle and physical health measures. Informed consent was obtained from all participants, and this study was conducted under generic approval from the NHS National Research Ethics Service (approval letter dated 13 May 2016, Ref 16/NW/0274) and under UK Biobank approvals for application #6553 ‘Genome-wide association studies of mental health’ (PI Smith).
Genotyping, imputation and quality control
In March 2018 UK Biobank released genetic data for 487,409 individuals, genotyped using the Affymetrix UK BiLEVE Axiom or the Affymetrix UK Biobank Axiom arrays (Santa Clara, CA, USA), which have over 95% of content in common22. Pre-imputation quality control, imputation (both 1000Genomes and HRC Reference Panels) and post-imputation cleaning were conducted centrally by UK Biobank (described in the UK Biobank release documentation, please see URLs in appendix for details).
Phenotyping
As part of the comprehensive baseline assessment participants were asked: “Over the past two weeks, how often have you had little interest or pleasure in doing things?” (Data field 2060). Respondents could choose from the following answers: “not at all”; “several days”; “more than half the days”; and “nearly every day”. These responses were coded as 0, 1, 2 and 3 respectively. This question on anhedonia is derived from the Patient Health Questionnaire-9 (PHQ-9), a well-validated screening instrument for MDD25. To maximise numbers available for downstream magnetic resonance imaging (MRI) analyses, we excluded from the primary GWAS those participants with any available MRI data (n = 20,174). Additional exclusion criteria for the GWAS included individuals in whom: over 10% of genetic data were missing; self-reported sex did not match genetic sex; sex chromosome aneuploidy was reported; where heterozygosity value was a clear outlier; and participants not of European ancestry (n = 73,385).
Genetic association and heritability
Genetic association with the measure of anhedonia was performed using BOLT-LMM26,27, which accounts for population structure and sample relatedness by including a genetic relatedness matrix within the models. Models were further adjusted for age, sex, and genotyping array. SNPs included in the analysis were filtered by MAF > 0.01, Hardy–Weinberg Equilibrium p > 1 × 10–6, and imputation score > 0.3. BOLT-LMM was also used to provide a SNP-heritability estimate and an estimate of λGC.
Genetic correlations
Linkage Disequilibrium Score Regression (LDSR)28 was carried out using LDSC on GWAS summary statistics from several published studies, to obtain genetic correlations with psychiatric disorders where anhedonia is known to be a feature (MDD29, schizophrenia30, bipolar disorder30, and OCD31), as well as Parkinson’s Disease32. We left the intercept unconstrained to allow for sample overlap between the sample used here and the outputs of the other GWAS.
Polygenic risk score generation
Polygenic risk scores (PRS) were created using LDpred33. LDpred differs from the more common pruning and threshold (P&T) PRS because it generates a single risk score for the trait of interest derived from as many loci as possible. A training set was created to obtain the LD structure by using 1000 unrelated Biobank participants who had passed the same genetic QC as those used in the GWAS (but who were excluded from the GWAS because they did not respond to the anhedonia question and had not provided brain imaging data). These training data were then used for the construction of anhedonia PRS in those participants for who brain imaging data were available.
Brain imaging variables
A number of structural and functional brain MRI measures have been made available by UK Biobank as Imaging Derived Phenotypes (IDPs)34. These measures are: total volume (mm3) of brain grey matter, white matter and cerebrospinal fluid (CSF), each normalised for head size; volumes (grey matter or total) of 15 cortical and subcortical regions of interest (ROIs); diffusion tensor imaging (DTI) measures of white matter integrity (fractional anisotropy (FA) and mean diffusivity (MD)); and functional MRI activity during an emotion processing task (the Hariri face shape task)35 in an amygdala mask and group-defined mask consisting of occipito-temporal and amygdala regions. For a more detailed description of these variables, and for details of MRI acquisition and pre-processing and IDP selection, please see supplementary methods.
Polygenic risk score and brain imaging analyses
MRI data were available for 20,174 UK Biobank participants. PRS/MRI analyses were conducted in a subset of 17,120 participants who had available MRI data and who were not included in the GWAS, after exclusion of participants who did not meet genetic quality control criteria (n = 2479) or who self-reported a developmental or neurological disorder at either the baseline assessment or the imaging visit (n = 575) (please see Table S1 for exclusions). For each MRI outcome, data points with values more than 3 standard deviations from the sample mean were excluded.
Models were adjusted for age at MRI visit, (age at MRI)2, sex, genotype array, the first eight genetic principal components, and lateral, transverse and longitudinal scanner position covariates. Total tissue volume measures (total grey matter, white matter and ventricular CSF volumes) were normalised for head size prior to adjustment for the above covariates. ROI analyses were additionally adjusted for total brain volume (calculated by summing total grey matter, white matter and ventricular cerebrospinal fluid (CSF) volume); and fMRI analyses were also adjusted for head motion during the emotion processing task. False Discovery Rate (FDR) correction was applied36,37.
Results
Demographics
The GWAS was performed on 375,724 UK Biobank participants, of whom 203,322 (54.1%) were female. The age of the sample ranged from 39 to 73 and the mean age was 57 years (S.D. = 8.01). In response to the anhedonia question, 299,232 (79.64%) answered “not at all”; 60,212 (16.0%) reported “several days”; 9405 (2.5%) reported “more than half the days” and 6876 (1.8%) reported “nearly every day”.
MRI analyses were conducted in 17,120 participants, 52.4% (8978) of whom were female. The mean age (at the time of MRI) of these participants was 62.7 years (S.D. = 7.46; range = 45–80 years). Of the 16,783 participants with available MRI data who answered the anhedonia question, 13,810 (82.3%) responded “not at all”, 2469 (14.7%) reported feelings of anhedonia for “several days”, 297 (1.8%) for “more than half the days”, and 207 (1.2%) “nearly every day”.
Genome-wide association study findings
GWAS results are presented as a Manhattan plot in Fig. 1, and details of genome-wide significant loci are provided in Table S2. In all, there were 1100 SNPs that were genome-wide significant (p < 5 × 10–8), and, of these, represented 11 independent loci on 9 different chromosomes (Table S2; Figs. S1–S11). An independent signal was defined as the region of r2 > 0.1 within a 500 MB window from the most significant SNP below genome-wide significance.
Results are presented as a Manhattan Plot and as a QQ plot (inset). Association analysis p-values for each SNP are plotted (as –log10(p)) vs. chromosomal position. The red and blue lines indicate the genome-wide significant and suggestive p-value thresholds, respectively. The QQ plot shows observed vs expected p-values for every SNP.
Some inflation of the GWAS results was observed (λGC = 1.15), however considering the sample size this is expected to have had a negligible impact on findings. There was evidence for a polygenic component (LDSR intercept = 1.03, S.E. = 0.005) and no evidence for undue inflation of the test statistics due to unaccounted population stratification.
Genome-wide association study replication
To help validate the phenotype, we attempted a replication in the sample that was excluded from the GWAS. Only one of the eleven loci identified achieved nominal significance (CHR6:27731402, β = −0.02, p = 0.015). This would not survive correction for multiple testing. Of the 11 top hit SNPs, 8 of 11 coefficients were in the same direction as in the discovery set (Table S3).
Additionally, we determined if the PRS of anhedonia in those excluded from the GWAS was associated with their state anhedonia scores. The model was adjusted for the first 4 genetic principle components, genotyping chip, age, sex and Townsend score. We detected a small but highly significant association between the anhedonia PRS and the anhedonia phenotype scores (β = 0.0007, p = 1.17 × 10–14).
Genetic correlations
Using LDSR we assessed whether genetic predisposition to anhedonia overlapped with that for several psychiatric disorders or traits. Anhedonia had significant genetic correlation with MDD (rg 0.77, q = 1.84 × 10–139), schizophrenia (rg 0.28, q = 5.28 × 10–15) and bipolar disorder (rg 0.12, q = 0.002) but not with OCD or Parkinson’s disease (q = 0.6 and q = 0.97 respectively) (Table 1).
PRS and brain MRI structural and functional outcomes
Associations between PRS for anhedonia and total brain tissue volumes are presented in Table S4. Greater polygenic risk for anhedonia was associated with lower total grey matter volume, but not with total white matter volume or with total ventricular CSF volume.
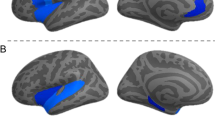
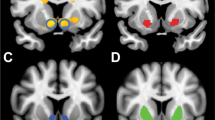
Associations were then assessed between PRS for anhedonia and volumes of 15 cortical and subcortical regions of interest (ROIs were derived a priori from meta-analysis and literature review; please see Table S5 and Supplementary Methods). Greater genetic risk score for anhedonia was associated with smaller volumes for insular cortex, orbitofrontal cortex, middle frontal gyrus and anterior temporal fusiform cortex. Uncorrected associations were also observed with nucleus accumbens, medial frontal cortex and caudate volumes, but these did not survive FDR correction. The locations of the ROIs showing significant association with the PRS are displayed in Supplementary Fig. S12, and a point-range plot showing the association evidence for ROI volumes is displayed in Fig. 2a.
In subsequent analyses of association with white matter integrity, greater PRS for anhedonia was not associated with general factors of either higher mean diffusivity (MD) or fractional anisotropy (FA) (Table S6). PRS associations were assessed with 15 individual white matter tracts (bilateral tracts were combined in the same models and models were adjusted for hemisphere; see Supplementary Methods, Supplementary Tables S7 and S8 for tract-specific FA and MD results, respectively). Higher PRS for anhedonia was associated with lower FA in four of the fifteen tracts (anterior thalamic radiation, forceps minor, medial lemniscus, posterior thalamic radiation) (Fig. 2b). Higher PRS for anhedonia was also associated with higher MD in 9 of the 15 tracts (all apart from acoustic radiation, corticospinal tract, forceps major, inferior longitudinal fasciculus, medial lemniscus and parahippocampal part of cingulum) (Fig. 2c). There was no association between PRS for anhedonia and functional MRI activity during the emotion-processing task (median Blood Oxygen Level Dependent signal for the face vs. shape contrast) (Table S9).
Discussion
These analyses represent the largest genetic association study of state anhedonia performed to date. We identified eleven genetic loci associated with anhedonia in the UK general population. Within each associated region there were a number of genes that could have a functional impact on anhedonia and the pleasure cycle (considered in detail below). Consistent with an RDoC approach focusing on transdiagnostic symptoms and traits, we found strong genetic correlations between anhedonia and MDD, schizophrenia and bipolar disorder, but not between anhedonia and OCD or Parkinson’s Disease. Despite the lack of correlation with Parkinson’s Disease, several of the loci identified in our GWAS include genes with known association with Parkinson’s Disease (see below). We also report the first investigation of associations between genetic loading for anhedonia and both brain structure and brain function.
Genes within anhedonia-associated loci
There are multiple genes in each QTL identified in the GWAS. Here we discuss potentially relevant genes based on their known function in the scientific literature. Within the chromosome 1 locus there are multiple RGS genes, most notably RGS1 and RGS2, encoding regulators of G-protein signalling that show prominent expression in the brain38. RGS2 has previously been identified as a modulator of LRRK239 expression, a gene known to be a genetic cause of Parkinson’s Disease. RGS2 has also been associated with symptom severity in schizophrenia40 and lower expression of RGS2 may be related to depression-like behaviours in animal models41.
EPHB1 on chromosome 3 encodes an ephrin receptor tyrosine kinase identified in a GWAS of antidepressant response42, and is associated with symptoms of schizophrenia in Chinese Han populations43, and with susceptibility to Parkinson’s Disease44.
At the more centromeric locus on chromosome 11, GRM5 encodes a metabotropic glutamate receptor that has been extensively studied in relation to MDD45,46 and schizophrenia47,48. Assessments in mice have also found that agonists of GRM5 attenuate Parkinsonian motor deficits via striatal dopamine depletion49. The Genotype Tissue Expression (GTEx) database38 shows prominent expression of GRM5 in the brain, especially within the nucleus accumbens, a region with a major role in the prediction of reward50,51.
Another likely candidate gene at this chromosome 11 locus is DISC1FP1. DISC1 (Disrupted in Schizophrenia 1), a gene located on chromosome 1, is part of a chromosome 1:11 translocation that increases risk of schizophrenia, schizoaffective disorder and bipolar disorder52. DISC1FP1 is disrupted by this translocation, impacting on both intracellular NADH oxidoreductase activities and protein translation53.
At the more telomeric locus on chromosome 11, NCAM1 (neural cell adhesion molecule 1) has been implicated as a potential link between depressive symptoms and brain structure54, specifically decreased FA. NCAM1 has been reported to be present at increased levels in the amygdala38, another brain region associated with the pleasure cycle, in depressed subjects55. However, we did not find any association between PRS for anhedonia and either amygdala volume or functional activity in the amygdala during an emotion processing task. It is possible that NCAM1 may exert its effects via expression in other brain regions.
Another gene of interest at this second chromosome 11 locus is DRD2, encoding the dopamine D2 receptor. There is an extensive literature on the importance of DRD2 for the psychopharmacology of both schizophrenia and MDD56,57.
The chromosome 12 locus contains only LOC and LINC genes. These encode different classes of functional RNA but little is known about their function beyond possible post-transcriptional regulation of other gene products.
The more centromeric locus on chromosome 14 contains PRKD1, which encodes a serine/threonine-protein kinase identified in a GWAS of schizophrenia18. The more telomeric locus on chromosome 14 contains the gene SLC8A3, a gene involved in maintaining Ca2+ homoeostasis within a variety of tissues, including neurons. SLC8A3 may also play a role in Parkinson’s Disease58.
ISLR2 on chromosome 15 is involved in neurodevelopment59 but its role in psychiatric and neurological traits is not well characterised. Another candidate gene within this locus, NRG4, encodes a neuregulin protein that activates type-1 growth factor receptors. Recent work has shown that NRG4 acts a regulator of the growth and elaboration of pyramidal neuron dendrites in the developing neocortex60. Pyramidal neurons have been directly implicated in the pathophysiology of schizophrenia61.
Finally, on chromosome 18, DCC (the most significant hit) encodes a netrin 1 receptor with a role in axon guidance, and has been previously reported to be associated with anhedonic phenotypes in mice and humans62 and potentially with schizophrenia pathogenesis63. We have previously identified DCC in GWAS of mood instability64, suicidality65 and multisite chronic pain66.
Brain structure and function
Our findings on the relationship between genetic loading for anhedonia and brain structure and function are of considerable interest. Greater levels of anhedonia, in healthy and clinical populations, have been linked to altered functional activity (and less consistently to reduced volume) in frontal/striatal regions involved in reward or pleasure processing12,13,14,67,68,69,70,71. The regions most consistently implicated include nucleus accumbens, caudate, putamen, medial frontal cortex and orbitofrontal cortex.
We found that increased genetic risk for anhedonia was associated with smaller volumes of the orbitofrontal cortex the (Fig. 2a; Supplementary Fig. 1) a region involved in representation of reward value and social reward dependence70,72. We also noted uncorrected associations of PRS for anhedonia and smaller volumes of nucleus accumbens (involved in reward value and pleasure processing69,73) and medial frontal cortex (linked to pleasure processing69), but these associations did not survive correction for multiple comparisons. These cortical/subcortical volume findings are therefore consistent with an association between genetic risk for anhedonia and reward/pleasure processing73.
Increased genetic risk for anhedonia was also associated with smaller volumes of insular cortex (associated with emotion processing74) and fusiform cortex. Most ROIs in our analysis were selected on the basis that they were associated with smaller volumes in MDD vs. healthy controls in the most recent MDD brain structure meta-analysis75. Our findings are consistent with two scenarios: either, genes for anhedonia partly mediate the association between the ROI volumes and MDD, or these genes exhibit horizontal pleiotropy and affect both phenotypes via separate mechanisms. Consistent with our finding of widespread associations of genetic risk for anhedonia with cortical/subcortical volumes, we also found an association with reduced total grey matter volume, adjusted for head size.
Evidence of reduced white matter integrity (FA) in several tracts in individuals scoring higher on measures of anhedonia has been reported16,76. We found that higher values for the anhedonia PRS were associated with higher MD (reflecting poorer white matter integrity), in most individual tracts, but not in a general factor of MD. Several tracts also showed reduced FA.
We did not, however, find any association between PRS for state anhedonia and functional brain activity. UK Biobank ROIs were selected based on average responses during an emotion processing task; it is therefore plausible that effects would emerge with the use of a reward processing task, or by applying whole-brain voxel-wise analyses, to include further reward/pleasure processing regions.
Strengths and limitations
This study is the largest GWAS of state anhedonia to date, and substantially contributes new knowledge on the biology of this important transdiagnostic symptom. We conducted analyses within a large population-based cohort. Our primary analysis included individuals with mental health histories, but these represent only a small proportion of the total sample, which is an order of magnitude larger than any previous study of this kind. Subclinical anhedonia is common, and associated with increased risk of later mental illness77. This idea is reinforced by the significant genetic correlations with other psychiatric disorders. Additionally, we used an ordered ordinal phenotype, resulting in more power to detect associations than with the more common dichotomised (‘non-anhedonic’ vs. ‘anhedonic’) analyses78. Identification of loci associated with population-level anhedonia may be important from a personalised medicine perspective, for example, in terms of developing stratified medicine approaches to identify individuals at high-risk of developing psychiatric disorders.
Notably, however, the UK Biobank cohort has a degree of selection bias. In general, volunteers are typically healthier and of higher socioeconomic status and higher education level than the general population79, so reported levels of anhedonia in this sample may be lower than the population rate. Therefore, it is plausible that the strength of associations we identified may be an underestimate of the true population value; there may also be an inflated type 2 error rate.
The measure of anhedonia employed here was a single question from a depression screening instrument, the PHQ-9, assessing frequency of symptoms anhedonia within the preceding two weeks. This item therefore measures state anhedonia at a single time point, and clearly could be influenced by environmental factors, such as season or current health status80,81. However, we have assumed here that despite transient environmental and physical factors, individuals who are prone to trait anhedonia will be more likely to report higher frequency of anhedonia at any given time point, and it seems likely that a recent anhedonia phenotype will be enriched for individuals with stronger genetic predisposition towards anhedonia. In line with this assumption, the anhedonia item of the PHQ-9 at a single time point has demonstrated utility in predicting longitudinal brain structural change82.
Additionally, our attempts at replication of the GWAS results met with limited success, with no real replication of the top hits. However, the PRS analysis was able to detect a significant correlation with state anhedonia in the replication sample. We attribute the lack of replication success in the GWAS to lack of power (small replication sample size) and small effect sizes for individual SNPs.
The GWAS treated the ordered ordinal responses to the frequency of anhedonia question as linear. This approach is likely to have had a minimal impact on the BOLT-LMM GWAS results. Although the distance between points on the anhedonia scale is not evenly spaced, each point can reasonably be considered to be at least several days of anhedonia in the preceding two weeks than the point before it.
The anhedonia measure did not enable determination of specific anhedonia subtypes. Existing validated instruments typically divide anhedonia into physical and social subscales21, or into anticipatory vs. consummatory components of pleasure83. Future studies using more detailed anhedonia scales may be of use in examining the extent of genetic overlap between anhedonia subtypes.
Conclusion
We report the largest GWAS to date of state anhedonia, a common symptom associated with several psychiatric disorders. We identified 11 novel genetic loci and our findings indicate substantial genetic overlap between anhedonia and several psychiatric disorders including MDD, schizophrenia and bipolar disorder. PRS analyses revealed association between genetic loading for anhedonia and smaller volumes of several brain regions, and poorer white matter integrity. Taken together, these findings provide important insights into the neurobiology of an important but under-studied psychiatric symptom and strongly support the proposition that genetic predisposition to anhedonia influences brain structure and function.
URLs
UK Biobank genetic data release information - https://data.bris.ac.uk/datasets/3074krb6t2frj29yh2b03x3wxj/UK%20Biobank%20Genetic%20Data_MRC%20IEU%20Quality%20Control%20version%201.pdf. Genotyping and quality control information – http://www.ukbiobank.ac.uk/wp-content/uploads/2014/04/UKBiobank_genotyping_QC_documentation-web.pdf.
Code availability
BOLT-LMM software is available from https://data.broadinstitute.org/alkesgroup/BOLT-LMM/downloads/. LDpred software is available from https://bitbucket.org/bjarni_vilhjalmsson/ldpred/src/master/
References
Andreasen, N. C. & Olsen, S. Negative v positive schizophrenia. Definition and validation. Arch. Gen. Psychiatry 39, 789–794 (1982).
Heinz, A., Schmidt, L. G. & Reischies, F. M. Anhedonia in schizophrenic, depressed, or alcohol-dependent patients-neurobiological correlates. Pharmacopsychiatry 27(Suppl 1), 7–10 (1994).
Dichter, G. Anhedonia in Unipolar Major Depressive Disorder: A Review. Open Psychiatry J. 4, 1–9 (2010).
Pelizza, L. & Ferrari, A. Anhedonia in schizophrenia and major depression: state or trait? Ann. Gen. Psychiatry 8, 22 (2009).
Abramovitch, A., Pizzagalli, D. A., Reuman, L. & Wilhelm, S. Anhedonia in obsessive-compulsive disorder: beyond comorbid depression. Psychiatry Res 216, 223–229 (2014).
Loas, G., Krystkowiak, P. & Godefroy, O. Anhedonia in Parkinson's disease: an overview. J. Neuropsychiatry Clin. Neurosci. 24, 444–451 (2012).
Leventhal, A. M. et al. Anhedonia associated with stimulant use and dependence in a population-based sample of American adults. Exp. Clin. Psychopharmacol. 18, 562–569 (2010).
Leventhal, A. M. Relations between anhedonia and physical activity. Am. J. health Behav. 36, 860–872 (2012).
Cuthbert, B. N. & Insel, T. R. Toward the future of psychiatric diagnosis: the seven pillars of RDoC. BMC Med. 11, 126 (2013).
Der-Avakian, A. & Markou, A. The neurobiology of anhedonia and other reward-related deficits. Trends Neurosci. 35, 68–77 (2012).
Heshmati, M. & Russo, S. J. Anhedonia and the brain reward circuitry in depression. Curr. Behav. Neurosci. Rep. 2, 146–153 (2015).
Zhang, B. et al. Mapping anhedonia-specific dysfunction in a transdiagnostic approach: an ALE meta-analysis. Brain Imaging Behav. 10, 920–939 (2016).
Pizzagalli, D. A. et al. Reduced caudate and nucleus accumbens response to rewards in unmedicated individuals with major depressive disorder. Am. J. Psychiatry 166, 702–710 (2009).
Wacker, J., Dillon, D. G. & Pizzagalli, D. A. The role of the nucleus accumbens and rostral anterior cingulate cortex in anhedonia: integration of resting EEG, fMRI, and volumetric techniques. Neuroimage 46, 327–337 (2009).
Henderson, S. E. et al. A preliminary study of white matter in adolescent depression: relationships with illness severity, anhedonia, and irritability. Front Psychiatry 4, 152 (2013).
Keedwell, P. A. et al. Cingulum white matter in young women at risk of depression: the effect of family history and anhedonia. Biol. Psychiatry 72, 296–302 (2012).
Howard, D. M. et al. Genome-wide meta-analysis of depression identifies 102 independent variants and highlights the importance of the prefrontal brain regions. Nat. Neurosci. 22, 343–352 (2019).
Schizophrenia Working Group of the Psychiatric Genomics C. Biological insights from 108 schizophrenia-associated genetic loci. Nature 511, 421–427 (2014).
Ren, H. et al. Genes associated with anhedonia: a new analysis in a large clinical trial (GENDEP). Translational. Psychiatry 8, 150 (2018).
Pain, O. et al. Genome-wide analysis of adolescent psychotic-like experiences shows genetic overlap with psychiatric disorders. Am. J. Med. Genet. Part B, Neuropsychiatr. Genet. : Off. Publ. Int. Soc. Psychiatr. Genet. 177, 416–425 (2018).
Chapman, L. J., Chapman, J. P. & Raulin, M. L. Scales for physical and social anhedonia. J. Abnorm. Psychol. 85, 374–382 (1976).
Ortega-Alonso, A. et al. Genome-Wide Association Study of Psychosis Proneness in the Finnish Population. Schizophrenia Bull. 43, 1304–1314 (2017).
Ronald, A. & Pain, O. A systematic review of genome-wide research on psychotic experiences and negative symptom traits: new revelations and implications for psychiatry. Hum. Mol. Genet. 27(R2), R136–R152 (2018).
Sudlow, C. et al. UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 12, e1001779 (2015).
Kroenke, K., Spitzer, R. L. & Williams, J. B. The PHQ-9: validity of a brief depression severity measure. J. Gen. Intern Med. 16, 606–613 (2001).
Loh, P. R. et al. Efficient Bayesian mixed-model analysis increases association power in large cohorts. Nat. Genet 47, 284–290 (2015).
Loh, P.-R., Kichaev, G., Gazal, S., Schoech, A. P. & Price, A. L. Mixed-model association for biobank-scale datasets. Nat. Genet. 50, 906–908 (2018).
Bulik-Sullivan, B. K. et al. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat. Genet. 47, 291–295 (2015).
Wray, N. R. et al. Genome-wide association analyses identify 44 risk variants and refine the genetic architecture of major depression. Nat. Genet. 50, 668–681 (2018).
Bipolar Disorder and Schizophrenia Working Group of the Psychiatric Genomics Consortium. Genomic Dissection of Bipolar Disorder and Schizophrenia, Including 28 Subphenotypes. Cell 173, 1705–1715.e1716 (2018).
International Obsessive Compulsive Disorder Foundation Genetics Collaborative (IOCDF-GC) and OCD Collaborative Genetics Association Studies (OCGAS). Revealing the complex genetic architecture of obsessive-compulsive disorder using meta-analysis. Mol. Psychiatry 23, 1181–1188 (2018).
Nalls M. A. et al. Expanding Parkinson’s disease genetics: novel risk loci, genomic context, causal insights and heritable risk. bioRxiv 2019: 388165.
Vilhjálmsson Bjarni, J. et al. Modeling Linkage Disequilibrium Increases Accuracy of Polygenic Risk Scores. Am. J. Hum. Genet. 97, 576–592 (2015).
Elliott L. T. et al. The genetic basis of human brain structure and function: 1,262 genome-wide associations found from 3,144 GWAS of multimodal brain imaging phenotypes from 9,707 UK Biobank participants. bioRxiv 2017: 178806.
Hariri, A. R., Tessitore, A., Mattay, V. S., Fera, F. & Weinberger, D. R. The amygdala response to emotional stimuli: a comparison of faces and scenes. Neuroimage 17, 317–323 (2002).
Benjamini, Y. & Hochberg, Y. Controlling False Discov. Rate - A Practical Powerful Approach Mult. Test. 57, 289–300 (1995).
Kim, K. I. & van de Wiel, M. A. Effects of dependence in high-dimensional multiple testing problems. BMC Bioinforma. 9, 114 (2008).
Consortium, G. T. The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 45, 580–585 (2013).
Dusonchet, J. et al. A Parkinson’s disease gene regulatory network identifies the signaling protein RGS2 as a modulator of LRRK2 activity and neuronal toxicity. Hum. Mol. Genet 23, 4887–4905 (2014).
Campbell, D. B. et al. Association of RGS2 and RGS5 variants with schizophrenia symptom severity. Schizophr. Res. 101, 67–75 (2008).
Lifschytz, T. et al. Relationship between Rgs2 gene expression level and anxiety and depression-like behaviour in a mutant mouse model: serotonergic involvement. Int J. Neuropsychopharmacol. 15, 1307–1318 (2012).
Ising, M. et al. A genomewide association study points to multiple loci that predict antidepressant drug treatment outcome in depression. Arch. Gen. psychiatry 66, 966–975 (2009).
Su, L. et al. Association of EPHB1 rs11918092 and EFNB2 rs9520087 with psychopathological symptoms of schizophrenia in Chinese Zhuang and Han populations. Asia Pac. Psychiatry 8, 306–308 (2016).
Kim, J. M. et al. SNPs in axon guidance pathway genes and susceptibility for Parkinson’s disease in the Korean population. J. Hum. Genet 56, 125–129 (2011).
Chandley, M. J. et al. Elevated gene expression of glutamate receptors in noradrenergic neurons from the locus coeruleus in major depression. Int J. Neuropsychopharmacol. 17, 1569–1578 (2014).
Paul, I. A. & Skolnick, P. Glutamate and depression: clinical and preclinical studies. Ann. N. Y Acad. Sci. 1003, 250–272 (2003).
Devon, R. S. et al. The genomic organisation of the metabotropic glutamate receptor subtype 5 gene, and its association with schizophrenia. Mol. Psychiatry 6, 311–314 (2001).
Matosin, N. et al. Alterations of mGluR5 and its endogenous regulators Norbin, Tamalin and Preso1 in schizophrenia: towards a model of mGluR5 dysregulation. Acta Neuropathol. 130, 119–129 (2015).
Black, Y. D. et al. Protective effect of metabotropic glutamate mGluR5 receptor elimination in a 6-hydroxydopamine model of Parkinson's disease. Neurosci. Lett. 486, 161–165 (2010).
Knutson, B., Fong, G. W., Adams, C. M., Varner, J. L. & Hommer, D. Dissociation of reward anticipation and outcome with event-related fMRI. Neuroreport 12, 3683–3687 (2001).
McClure, S. M., York, M. K. & Montague, P. R. The neural substrates of reward processing in humans: the modern role of FMRI. Neuroscientist 10, 260–268 (2004).
Hodgkinson, C. A. et al. Disrupted in schizophrenia 1 (DISC1): association with schizophrenia, schizoaffective disorder, and bipolar disorder. Am. J. Hum. Genet. 75, 862–872 (2004).
Ji, B. et al. Inhibition of protein translation by the DISC1-Boymaw fusion gene from a Scottish family with major psychiatric disorders. Hum. Mol. Genet. 23, 5683–5705 (2014).
Petrovska, J. et al. The NCAM1 gene set is linked to depressive symptoms and their brain structural correlates in healthy individuals. J. Psychiatr. Res. 91, 116–123 (2017).
Irwin, W. et al. Human amygdala activation detected with echo-planar functional magnetic resonance imaging. Neuroreport 7, 1765–1769 (1996).
Gluskin, B. S. & Mickey, B. J. Genetic variation and dopamine D2 receptor availability: a systematic review and meta-analysis of human in vivo molecular imaging studies. Transl. psychiatry 6, e747–e747 (2016).
Glantz, L. A. et al. Pro-apoptotic Par-4 and dopamine D2 receptor in temporal cortex in schizophrenia, bipolar disorder and major depression. Schizophr. Res. 118, 292–299 (2010).
Wood-Kaczmar, A., Deas, E., Wood, N. W. & Abramov, A. Y. The role of the mitochondrial NCX in the mechanism of neurodegeneration in Parkinson's disease. Adv. Exp. Med. Biol. 961, 241–249 (2013).
Abudureyimu, S. et al. Essential Role of Linx/Islr2 in the Development of the Forebrain Anterior Commissure. Sci. Rep. 8, 7292 (2018).
Paramo, B., Wyatt, S. & Davies, A. M. An essential role for neuregulin-4 in the growth and elaboration of developing neocortical pyramidal dendrites. Exp. Neurol. 302, 85–92 (2018).
Garey, L. When cortical development goes wrong: schizophrenia as a neurodevelopmental disease of microcircuits. J. Anat. 217, 324–333 (2010).
Torres-Berrío, A. et al. DCC Confers Susceptibility to Depression-like Behaviors in Humans and Mice and Is Regulated by miR-218. Biol. Psychiatry 81, 306–315 (2017).
Wang, Z. et al. Axon guidance pathway genes are associated with schizophrenia risk. Exp. Ther. Med. 16, 4519–4526 (2018).
Ward, J. et al. Genome-wide analysis in UK Biobank identifies four loci associated with mood instability and genetic correlation with major depressive disorder, anxiety disorder and schizophrenia. Transl. Psychiatry 7, 1264 (2017).
Strawbridge, R. J. et al. Identification of novel genome-wide associations for suicidality in UK Biobank, genetic correlation with psychiatric disorders and polygenic association with completed suicide. EBioMedicine 41, 517–525 (2019).
Johnston K. J. A. et al. Genome-wide Association Study of Multisite Chronic Pain in UK Biobank. bioRxiv 2018: 502807.
Auerbach, R. P. et al. Neuroanatomical Prediction of Anhedonia in Adolescents. Neuropsychopharmacol. : Off. Publ. Am. Coll. Neuropsychopharmacol. 42, 2087–2095 (2017).
Harvey, P. O., Pruessner, J., Czechowska, Y. & Lepage, M. Individual differences in trait anhedonia: a structural and functional magnetic resonance imaging study in non-clinical subjects. Mol. Psychiatry 12, 767–775 (2007). 703.
Sabatinelli, D., Bradley, M. M., Lang, P. J., Costa, V. D. & Versace, F. Pleasure rather than salience activates human nucleus accumbens and medial prefrontal cortex. J. Neurophysiol. 98, 1374–1379 (2007).
Lebreton, M. et al. The brain structural disposition to social interaction. Eur. J. Neurosci. 29, 2247–2252 (2009).
Pornpattananangkul, N., Leibenluft, E., Pine, D. S. & Stringaris, A. Association Between Childhood Anhedonia and Alterations in Large-Scale Resting-State Networks and Task-Evoked Activation. JAMA Psychiatry 76, 624–633 (2019).
Kringelbach, M. L. The human orbitofrontal cortex: linking reward to hedonic experience. Nat. Rev. Neurosci. 6, 691–702 (2005).
Gorwood, P. Neurobiological mechanisms of anhedonia. Dialogues Clin. Neurosci. 10, 291–299 (2008).
Murphy, F. C., Nimmo-Smith, I. & Lawrence, A. D. Functional neuroanatomy of emotions: A meta-analysis. Cogn., Affect., Behav. Neurosci. 3, 207–233 (2003).
Wise, T. et al. Common and distinct patterns of grey-matter volume alteration in major depression and bipolar disorder: evidence from voxel-based meta-analysis. Mol. Psychiatry 22, 1455–1463 (2017).
Yang, X. H. et al. White matter microstructural abnormalities and their association with anticipatory anhedonia in depression. Psychiatry Res. Neuroimaging 264, 29–34 (2017).
Horwath, E., Johnson, J., Klerman, G. L. & Weissman, M. M. What are the public health implications of subclinical depressive symptoms? Psychiatr. Q 65, 323–337 (1994).
Auerbach, R. P., Pagliaccio, D. & Pizzagalli, D. A. Toward an Improved Understanding of Anhedonia. JAMA Psychiatry 76, 571–573 (2019).
Fry, A. et al. Comparison of Sociodemographic and Health-Related Characteristics of UK Biobank Participants With Those of the General Population. Am. J. Epidemiol. 186, 1026–1034 (2017).
Husain, M. & Roiser, J. P. Neuroscience of apathy and anhedonia: a transdiagnostic approach. Nat. Rev. Neurosci. 19, 470–484 (2018).
Lyall, L. M. et al. Seasonality of depressive symptoms in women but not in men: A cross-sectional study in the UK Biobank cohort. J. Affect Disord. 229, 296–305 (2018).
Grool, A. M. et al. Location and progression of cerebral small-vessel disease and atrophy, and depressive symptom profiles: the Second Manifestations of ARTerial disease (SMART)-Medea study. Psychol. Med. 42, 359–370 (2012).
Gooding, D. C. & Pflum, M. J. The assessment of interpersonal pleasure: introduction of the Anticipatory and Consummatory Interpersonal Pleasure Scale (ACIPS) and preliminary findings. Psychiatry Res. 215, 237–243 (2014).
Acknowledgements
J.W. is supported by the JMAS Sim Fellowship for depression research from the Royal College of Physicians of Edinburgh (173558). A.F. is supported by an MRC Doctoral Training Programme Studentship at the University of Glasgow (MR/K501335/1). R.J.S. is supported by a UKRI Innovation- HDR-UK Fellowship (MR/S003061/1). K.J.A.J. is supported by an MRC Doctoral Training Programme Studentship at the Universities of Glasgow and Edinburgh. D.J.S. acknowledges the support of a Lister Prize Fellowship (173096) and the MRC Mental Health Data Pathfinder Award (MC_PC_17217). R.A.I.B. is supported by a British Academy Post-Doctoral Fellowship.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Ward, J., Lyall, L.M., Bethlehem, R.A.I. et al. Novel genome-wide associations for anhedonia, genetic correlation with psychiatric disorders, and polygenic association with brain structure. Transl Psychiatry 9, 327 (2019). https://doi.org/10.1038/s41398-019-0635-y
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41398-019-0635-y
This article is cited by
-
Roles of the medial and lateral orbitofrontal cortex in major depression and its treatment
Molecular Psychiatry (2024)
-
Dimensional and transdiagnostic phenotypes in psychiatric genome-wide association studies
Molecular Psychiatry (2023)
-
Neurophysiological stratification of major depressive disorder by distinct trajectories
Nature Mental Health (2023)
-
Social Anhedonia: (f)MRI Studies
Neuroscience and Behavioral Physiology (2023)
-
On the etiology of aesthetic chills: a behavioral genetic study
Scientific Reports (2022)