Abstract
Developmental dyslexia (DD) is one of the most prevalent learning disorders, with high impact on school and psychosocial development and high comorbidity with conditions like attention-deficit hyperactivity disorder (ADHD), depression, and anxiety. DD is characterized by deficits in different cognitive skills, including word reading, spelling, rapid naming, and phonology. To investigate the genetic basis of DD, we conducted a genome-wide association study (GWAS) of these skills within one of the largest studies available, including nine cohorts of reading-impaired and typically developing children of European ancestry (N = 2562–3468). We observed a genome-wide significant effect (p < 1 × 10−8) on rapid automatized naming of letters (RANlet) for variants on 18q12.2, within MIR924HG (micro-RNA 924 host gene; rs17663182 p = 4.73 × 10−9), and a suggestive association on 8q12.3 within NKAIN3 (encoding a cation transporter; rs16928927, p = 2.25 × 10−8). rs17663182 (18q12.2) also showed genome-wide significant multivariate associations with RAN measures (p = 1.15 × 10−8) and with all the cognitive traits tested (p = 3.07 × 10−8), suggesting (relational) pleiotropic effects of this variant. A polygenic risk score (PRS) analysis revealed significant genetic overlaps of some of the DD-related traits with educational attainment (EDUyears) and ADHD. Reading and spelling abilities were positively associated with EDUyears (p ~ [10−5–10−7]) and negatively associated with ADHD PRS (p ~ [10−8−10−17]). This corroborates a long-standing hypothesis on the partly shared genetic etiology of DD and ADHD, at the genome-wide level. Our findings suggest new candidate DD susceptibility genes and provide new insights into the genetics of dyslexia and its comorbities.
Similar content being viewed by others
Introduction
Developmental dyslexia (DD) is a neurodevelopmental disorder affecting the ability of learning to read and to spell, in spite of adequate intelligence, educational opportunities, and in the absence of overt neurological and sensorial deficits1. It shows a prevalence of 5–12% among school-aged children, implying life-long learning difficulties for most of the affected individuals1. DD is characterized by a high rate of comorbidity with other neuropsychiatric conditions like attention-deficit hyperactivity disorder (ADHD), depression, and anxiety disorders2. Dyslexic individuals usually have severe and persistent problems in accurate and fluent reading and spelling, and in reading comprehension3. These problems are often associated with early deficits in neurocognitive skills, such as the ability to recognize and manipulate the phonemic constituents of speech (also known as phoneme awareness, PA), the ability to store such phonemes while reading (also known as phonological short-term memory), or the ability to fast map known visual symbols onto spoken word representations (known as naming speed)4. All these abilities show moderate-to-high heritability (40–80%)5,6,7 and significant genetic correlations with DD5. Hence, they represent cognitive indicators of dyslexia risk that are optimally suited for investigating the genetic mechanisms at its basis.
In the last two decades, several studies investigating both DD and the underlying cognitive skills have been carried out to better understand the genetic and neurobiological basis of dyslexia. On the one hand, linkage and targeted association analyses have suggested different candidate DD susceptibility genes (reviewed in refs. 1,8,9). Only a few of these genes have been implicated in DD, reading ability, and underlying cognitive skills in at least two independent studies or datasets1. These include DYX1C1 (15q21)10, KIAA0319 (6p22)11,12,13,14, DCDC2 (6p22)15,16,17,18, MRPL19/GCFC2 (2p12)19, ROBO1 (3p12)20,21,22, GRIN2B23,24, FOXP225,26,27 and CNTNAP227,28,29.
On the other hand, most of the genome-wide association studies (GWAS) published so far have identified mainly suggestive associations with DD and related cognitive traits (p < 10−5)30,31,32,33,34, with only one recent study reporting a genome-wide significant association (p < 5 × 10−8; see below)35. The first GWAS for reading ability used DNA pooling of low vs. high reading ability groups in ~1500 7-year-old children, which were genotyped with a low-density single-nucleotide polymorphism (SNP) microarray (∼107,000 SNPs)34. The SNPs showing the largest allele frequency differences between low- and high-ability groups were tested in an additional follow-up cohort of 4258 children, finally identifying 10 SNPs showing nominally significant associations with continuous variation in reading ability34. However, Luciano et al.36 later found no evidence of replication of these findings in an adolescent population sample of Australian twins and siblings (N = 1177). A later genome-wide linkage and association scan on ∼133,000 SNPs, in a sample of 718 subjects from 101 dyslexia-affected families, identified an association with dyslexia status at rs9313548, near FGF18 (5q35.1)33. More recently, three GWAS studies with different designs were carried out with the aim of identifying shared genetic contributions to reading and language abilities. Luciano et al.32 performed a GWAS on quantitative reading- and language-related traits in two population-based cohorts (N ∼ 6500), analyzing word reading, nonword repetition, and a composite score of reading and spelling abilities. They reported a suggestive association of rs2192161 (ABCC13; 21q11.2) with nonword repetition and of rs4807927 (DAZAP1, 19p13.3) with both the word reading and the reading–spelling score. A case-control GWAS comparing dyslexic (N = 353), language impaired (LI) (N = 163), and comorbid cases (N = 174) to a population-based control dataset (N = 4117) identified nominally significant associations with comorbid DD-LI cases drawn from the same population cohort used by Luciano et al.32. The most significant associations were detected at rs12636438 and rs1679255, mapping to ZNF385D (3p24.3)30. Another GWAS analyzed the first principal component from various reading- and language-related traits (both with and without IQ adjustment) in three datasets comprising children with reading or language problems and their siblings (N = 1862), and reported suggestive associations at rs59197085, upstream of CCDC136/FLNC (7q32.1), and at rs5995177, within RBFOX2 (22q12.3)31. More recently, Truong et al.35 reported a genome-wide significant multivariate association of rs1555839 (10q23.31) with two skills predicting DD risk, namely rapid automatized naming (RAN) and rapid alternating stimulus (RAS), in a multisite case-control study of DD made up of individuals of non-European ancestry (N = 1263). This SNP, located upstream of the pseudogene RPL7P34, was also associated with measures of word reading and was replicated with RAN traits in an independent cohort from Colorado35, partially overlapping with the Colorado dataset analyzed by Gialluisi et al.31.
Although many of the genes suggested by these GWAS studies showed interesting potential biological links to DD and underlying skills, most of these associations did not reach genome-wide significance and were not replicated in independent datasets36,37. Prominently, an analysis of 17 candidate SNPs mostly identified by these GWAS, did not manage to replicate the associations previously reported, with word/nonword reading and fluency, PA and RAN traits, in an independent family-based Dutch population dataset comprising 483 children and 505 parents from 307 nuclear families37. This might have different reasons, including the low statistical power of the original GWAS studies implied by the relatively small sample sizes, and the heterogeneity of recruitment criteria and phenotypic assessment of the cohorts involved. In addition, the candidate susceptibility genes identified and replicated so far explain only a minor part of the genetic variance underlying dyslexia and related cognitive traits, and a big proportion of this heritability remains unexplained.
To help unravel the genetic basis of DD and related neurocognitive skills, we conducted a large international collaborative GWAS. We analyzed the cognitive traits word reading, spelling, decoding skills, phoneme awareness, verbal short-term memory, and naming speed, in nine cohorts of reading impaired and typically developing participants of European ancestry (maximum N = 3468). We observed a genome-wide significant association at 18q12.2 and an association approaching genome-wide significance at 8q12.3, both with rapid automatized naming (RAN, N = 2563). These genetic effects extended beyond the RAN domain, to other DD-related skills. Also, we detected significant genetic overlaps of the traits analyzed with educational attainment and ADHD risk. Overall, our findings provide new insights in the genetic etiology of dyslexia and related cognitive traits.
Subjects and methods
Datasets
Table 1 reports the main details on the datasets involved in this study and on the recruitment criteria.
Unrelated DD cases and controls were recruited across seven different European countries, namely Austria (N = 374), Germany (N = 1061), Finland (N = 336), France (N = 165), Hungary (N = 243), The Netherlands (N = 311), and Switzerland (N = 67). Cases were defined as subjects showing more than 1.25 standard deviations (SD) below grade level on a standardized word reading test, while controls were defined as subjects with less than 0.85 SD below grade level on the same test38. In addition, we included two family-based datasets in the study. One of these, from Colorado, United States (USA), contained children showing a school history of reading difficulties as well as their siblings (N = 585; 266 independent nuclear families)31,39. The other one, from the United Kingdom (UK), consisted of subjects with a formal diagnosis of dyslexia and their unaffected siblings (N = 983; 608 independent nuclear families)31,40. Ethical approval was obtained for each cohort at the local level, and written informed consent was obtained for all the participants or their parents, as described elsewhere31,41,42.
Although the family-based datasets have been previously investigated in GWAS studies31,40,43, the European datasets have been analyzed in a candidate (SNP) association study42, and part of the German dataset has been investigated in relation to mathematical abilities23 and to neurophysiological DD endophenotypes44, such datasets were never analyzed jointly in a GWAS of neurocognitive traits related to dyslexia. In the present study, samples from Austria, Germany, and Switzerland were merged into a single dataset (hereafter called AGS), since they shared language, genetic ancestry, phenotypic measures, and selection criteria38,42,45. No other cohorts were approached for the present study, and all the cohorts contacted presented no refusal or lack of requirements for inclusion criteria.
Phenotypic measures
We focused on the core phenotypes of dyslexia, namely word reading (WRead), nonword reading (NWRead), and word spelling (WSpell), and on five neurocognitive measures underlying reading ability and dyslexia (as well as other comorbid learning disabilities, e.g., dyscalculia). These skills included phoneme awareness (PA), digit span (DigSpan, a measure of verbal short-term memory), and rapid automatized naming of letters (RANlet), digits (RANdig), and pictures (RANpic). These traits showed moderate-to-high cross-trait correlations (see Table S1a, b in Supplementary Methods). A brief explanation of these measures is reported in Table 2, while details on statistical elaboration are reported in Supplementary Methods and elsewhere31,38,45. Briefly, raw scores from psychometric tests were grade-normed (age-adjusted in Colorado) and then z-standardized to reduce skewness, with the exception of the DigSpan score, which was only z-normalized38,45. No phenotypic outliers were detected in any of the datasets analyzed (see Supplementary Methods for details).
Genotype quality control (QC) and imputation
Individuals were genotyped using Illumina HumanHap 300 k, 550 k, 660 k, HumanOmniExpress, and HumanCoreExome BeadChips (see Table S2 for details). Genotype QC was carried out in PLINK v1.90b3s46 and QCTOOL v1.4 (see URLs), as described in Supplementary Methods and elsewhere47. Within each dataset, SNPs were filtered out if they showed a variant call rate < 98%; a minor allele frequency (MAF) <5%, or a Hardy–Weinberg Equilibrium (HWE) test p-value <10−6. Moreover, samples showing a genotyping rate <98%, cryptic relatedness (in datasets of unrelated subjects), identity-by-descent (IBD) not corresponding to the available pedigree information (in sibling-based datasets), and mismatches between genetic and pedigree-based sex were discarded. Furthermore, genetic ancestry outliers—detected in a multidimensional scaling (MDS) analysis of pairwise genetic distance—and samples showing significant deviations in genome-wide heterozygosity were also filtered out (see Table S3).
For imputation, autosomal variants were aligned to the 1000 Genomes phase I v3 reference panel (ALL populations, June 2014 release)48 and pre-phased using SHAPEIT v2 (r837)49. Imputation was performed using IMPUTE2 v2.3.250 in 5 Mb chunks with 500 kb buffers, filtering out variants that were monomorphic in the 1000 Genomes EUR (European) samples. Chunks with < 51 genotyped variants or concordance rates < 92% were fused with neighboring chunks and re-imputed. Finally, imputed variants (genotype probabilities) were filtered out for IMPUTE2 INFO metric < 0.8, MAF < 5% and HWE test p-values <10, using QCTOOL v1.4. We checked again for the absence of genetic ancestry and genome-wide heterozygosity outliers after imputation, which revealed substantial concordance with pre-imputation QC. Further details on the filters used in genotype QC are reported in Table S3, while summary statistics are reported for each dataset in Table S2.
Genetic association testing and meta-analysis
After genotype QC and imputation, autosomal genotype probabilities were tested for association with the continuous traits available within each dataset. In the datasets containing only unrelated subjects—namely AGS, Finland, France, Hungary, and The Netherlands—association with genotype dosage was tested through linear regression in PLINK v1.9, using the first 10 genetic ancestry (MDS) components as covariates. In the sibling-based datasets (Colorado and UK), a generalized linear mixed-effects model association test was carried out through FastLMM v2.0751, using a genetic relationship matrix (GRM) of samples as a random effect while disabling normalization to unit variance for tested SNPs.
Following separate GWAS analyses for each dataset, variant associations with each of the eight univariate traits available were combined using a fixed-effects model based on inverse-variance-weighted effect size in METASOFT v2.0.152. Following the software guidelines, pooled analysis was conducted in two steps: a first run was carried out to compute genomic inflation factors, which were then used to correct meta-analysis statistics in a second run. The numbers of subjects involved in our pooled analysis were 3468 for WRead, 3399 for WSpell, 3409 for NWRead, 3093 for PA, 2591 for DigSpan, 2563 for RANlet and RANdig, and 2562 for RANpic (see Table S4 for detailed sample size by dataset). RAN measures and DigSpan were not available in the UK dataset, which was therefore not included in the pooled analyses of those traits. The numbers of variants analyzed in two or more datasets were 6,952,813 for RANlet, RANdig, RANpic, and DigSpan and 6,969,139 for WRead, WSpell, NWRead, and PA. The common genome-wide significance threshold α = 5 × 10−8 was corrected for multiple testing of five independent latent variables, as computed through MatSpD53 on the correlation matrix of the eight univariate traits analyzed (Table S1a, b). This adjustment resulted in a final Bonferroni-corrected significance level α = 1 × 10−8.
We also carried out a genome-wide multivariate genetic association analysis through TATES54, combining the univariate associations of single traits while taking into account their cross-trait correlation matrix (Table S1a). This analysis was aimed at the detection of vertical (or relational) pleiotropic genetic effects, i.e., those effects which are shared across traits due to their reciprocal relations55. For this analysis, the classical genome-wide significance threshold was used (α = 5 × 10−8).
The most significant associations detected were further investigated to assess their robustness through a permutation-based test. Moreover, we computed their effect size (regression R2) and tested potential epistatic effects of the variants identified. Similarly, we looked for effects of these variants on the other cognitive traits tested in our study through a horizontal pleiotropy test, aimed at detecting effects that were independent on the one observed on RANlet. Also, we looked for independent genetic effects in the genomic regions where these variants lay (18q12.2 and 8q12.3). Finally, we tested them for association with structural neuroimaging measures, which may be potentially correlated with reading and language abilities, namely subcortical volumes (see below). These analyses are reported in details in the Supplementary Methods section.
Assessment of genes and SNPs previously associated with DD and related cognitive traits
We investigated single-variant associations for candidate SNPs and genes previously implicated in DD and related cognitive traits.
First, we assessed all the variants mapping to nine candidate genes (up to 10 kb from the 5′- or 3′-UTR): DYX1C1, DCDC2, KIAA0319, C2ORF3, MRPL19, ROBO1, GRIN2B, FOXP2, and CNTNAP2. For these genes, association with DD and related cognitive traits was previously reported in at least two independent studies (as reviewed in ref. 1). Of note, most of the candidate variants identified in these genes have been already tested in studies showing a variable degree of overlap with our cohorts (reviewed in refs. 1,8,9), hence they cannot be formally replicated within the scope of the current study. For this reason, we focused our replication effort on six candidate SNPs among these variants, for which a statistically significant association (p < 0.05 after correction for multiple testing) has been reported in the past in datasets other than ours, but was never formally replicated. These SNPs included rs6803202, rs4535189, rs331142 and rs12495133 in ROBO121,22, rs7782412 in FOXP227, and rs5796555 in GRIN2B24.
We next tested all the variants showing the strongest associations with DD and related cognitive traits in previous GWAS30,31,32,33,34,35. These included all those variants reported to be associated in previous GWAS papers, including genome-wide significant associations (p < 5 × 10−8), suggestive associations (p < 1 × 10−5), or variants reported as the most significant associations (top 10 or top 100 list, depending on the associations reported in each paper; see Results section for a complete list). Again, some of these variants were identified by studies partially overlapping with our datasets31, while for other SNPs tested the statistics from the original papers were not fully available or not always directly comparable, due to either different design of the study or to different traits analyzed30,31,32,33,34,35. Therefore, a direct comparison was possible only for few variants (see relevant Results section).
Gene- and pathway-based enrichment tests
Gene-based association analyses for the phenotypic traits tested were performed using MAGMA v1.0656. First, genetic variants were assigned to protein-coding genes based on their position according to the NCBI 37.3 (hg19) build, extending gene boundaries by 10 kb from the 3′- and 5′-UTR. A total of 18,033 genes (out of 19,427 genes available) included at least one variant that passed internal QC, and were thus tested in gene-based enrichment analysis. Gene-based statistics were computed using the single-variant association statistics calculated in the GWAS of each phenotype, using default settings. To account for linkage disequilibrium (LD) among the variants tested, we used genetic data from all the datasets pooled together. Given the number of genes (18,033) and of independent latent traits (5) tested, the Bonferroni-corrected genome-wide significance threshold for this analysis was set to α = 0.05 / (18,033 × 5) = 5.5 × 10−7.
Using the results of the gene-based association analysis, we carried out a pathway-based enrichment test for each trait analyzed in the study, through a competitive gene-set analysis in MAGMA v1.06. We tested for enrichment 1329 canonical pathways (i.e., classical representations of biological processes compiled by domain experts) from the Molecular Signatures Database website (MSigDB v5.2, collection C2, subcollection CP; see URLs). To correct enrichment statistics for testing of multiple pathways, we used an adaptive permutation procedure with default settings (up to a maximum of 10,000 permutations). Hence, for gene-set analysis we corrected the significance threshold only for the number of independent latent traits tested (α = 0.05/5 = 0.01).
Polygenic risk score analysis
To assess the genetic overlap of common variants between the dyslexia-related skills tested here and other correlated phenotypes, we carried out a polygenic risk score (PRS) analysis using PRSice v1.2557. This analysis tests genetic overlap between two traits by making use of GWAS summary statistics: a training GWAS is used to build the PRS, which is then tested as a linear predictor of another trait in an independent study (target GWAS). We used the eight-univariate GWAS carried out here as a target, namely WRead, WSpell, NWRead, PA, RANlet, RANdig, RANpic, and DigSpan. As training GWAS, we selected 12 different studies, involving seven subcortical volumes previously tested in a large GWAS (N~13,000)58; an educational attainment trait (expressed in years of education completed, EDUyears; N~293,000)59; and four neuropsychiatric disorders. These included ADHD (N~55,000)60; autism spectrum disorder (ASD; N~16,000)61; major depressive disorder (MDD; N~19,000)62; and schizophrenia (SCZ; N~150,000)63, and were selected in light of their comorbidity with dyslexia reported by previous literature4,64,65,66. Similarly, the choice to test subcortical volumes was driven by the increasing evidence implicating subcortical structures in reading and language abilities (as reviewed in refs. 1,67,68).
We performed a Summary–Summary Statistic Based Analysis using only SNPs with association p-values ≤ 0.05 in each training GWAS, and in linkage equilibrium (r2 < 0.05) with the local top hit within a 300 kb window. Only SNPs which had been tested both in the training and in the target GWAS were tested. The number of SNPs meeting these criteria ranged from 11,017 for MDD vs. DigSpan and RAN traits, to 25,409 for SCZ vs. WRead, WSpell, NWRead, and PA. To verify the robustness of our results, we repeated the analysis at increasing association significance (PT) thresholds in the training GWAS (with PT = 0.001, 0.05, 0.1, 0.2, 0.3, 0.4, 0.5, 0.6, 0.7, 0.8, 0.9, 1.0).
To have an indication on the concordance of shared genetic effects for each pair of traits, we selected variants with association p-values ≤ 0.05 in each training GWAS and computed Pearson’s correlation of effect sizes (hereafter called rβ) with each of the target GWAS analyzed. The significance threshold for these analyses was corrected for multiple testing of five independent target GWAS (i.e., the number of independent latent traits computed through MatSpD, see Table S1b), 12 different training GWAS and 12 different PT thresholds tested (α = 0.05/(5 × 12 × 12) = 6.94 × 10−5.
Results
For each analysis presented below, we report the empirical p-values, along with significance thresholds adequately corrected for multiple testing (see Subjects and methods section).
Single-variant genome-wide associations
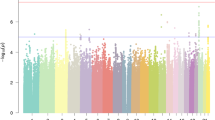
Among the eight traits analyzed in the present GWAS, only RANlet showed genome-wide significant associations withstanding correction for multiple testing (p < 1 × 10−8), mapped to chromosome 18q12.2. The most significant association was observed for rs17663182 (G/T; MAF = 7.7%; p-value = 4.73 × 10−9, major allele (G) β (SE) = 0.35 (0.06)). All the SNPs significantly associated on 18q12 were located within the non-coding gene MIR924HG (micro-RNA 924 host gene, also known as LINC00669; see Fig. 1a) and were in high LD with each other (r2 > 0.9). An additional, independent association approaching genome-wide significance was observed with RANlet at rs16928927 (C/T; MAF = 6.5%; p-value = 2.25 × 10−8, major allele (C) β (SE) = −0.4 (0.07)) on 8q12.3. This SNP was located within the first intron of NKAIN3 (Na+/K+ transporting ATPase interacting 3; see Fig. 1b). Further details on these associations are reported in Fig. 2 and Table 3.
Regional association plots of a 18q12.2 and b 8q12.3 with the RANlet trait. The most significantly associated variants are highlighted in violet. Plots were made using LocusZoom v0.4.8112
Boxplots of the RANlet trait as a function of genotype of the lead variants rs17663182 (left side, major allele G) and rs16928927 (right side, major allele C). Genotype counts are G/G = 2,092; T/G = 307; T/T = 16; missing = 148 for rs17663182 and C/C = 1,965; T/C = 259; T/T = 7; missing =332 for rs16928927 (Note: missing counts include Finland, where rs16928927 was not available). To generate these plots, all datasets were pooled together. RANlet Z-scores plotted here are residualized against the first 10 MDS covariates in all datasets except for Colorado, where we adjusted the phenotypic measure for pairwise genetic relatedness in GenABEL113 (see Supplementary Methods section)
Although neither of the two top SNPs was genotyped, imputation quality was high in all datasets (IMPUTE2 INFO metric 0.89–0.94 for rs17663182 and ~0.99 for rs16928927, respectively). These variants showed consistent allelic trends (Fig. 3a, b), but explained a variable proportion of RANlet variance in the different datasets ([0.03–1.8]% for rs17663182 and [0.067–2.96]% for rs16928927, respectively; Table S5a, b). Both our lead SNPs showed evidence of an association with many of the traits analyzed, especially with RAN traits (see Fig. 4a, b). Indeed, a genome-wide multivariate association analysis with the eight cognitive skills detected a significant association at rs17663182 (p = 3.07 × 10−8), and a suggestive association at rs16928927 (p = 1.46 × 10−7). Similarly, a multivariate association test focused on the three RAN traits revealed a genome-wide significant association of rs17663182 (p = 1.15 × 10−8), while rs16928927 association only approached significance (p = 5.45 × 10−8). However, neither of these two SNPs showed significant effects independent from RANlet on any other trait (Table S5c, d). Similarly, we observed no significant independent genetic influence on RANlet at 18q12.2 and 8q12.3, in a 100 kb window surrounding rs17663182 and rs16928927 (Table S5e, f), as well as no significant epistatic effect of these two variants on RANlet (Table S5g). These SNPs did not show any statistically significant association with volumes of seven different subcortical structures (Table S5h, i).
Detailed results of the GWAS analyses for each univariate trait are reported in Supplementary Figure S1a–p and Table S6a–h, while the most significant multivariate associations detected genome-wide are reported in Table S6i.
Genes and SNPs previously associated with DD and related cognitive traits
In total, 12,785 variants were annotated to nine candidate genes previously implicated in dyslexia by at least two independent studies, namely DYX1C1, DCDC2, KIAA0319, C2ORF3, MRPL19, ROBO1, GRIN2B, FOXP2, and CNTNAP2. We reported associations for all these variants in Table S7a–h. Among these variants, a detailed assessment of six candidate SNPs previously associated with DD or related cognitive measures in independent studies did not reveal any strong evidence of replication in our cohorts (see Table S7i), although we found marginal evidence of association of the ROBO1 variant rs12495133 with WSpell (C/A; MAF = 40%; p-value = 0.045, major allele (C) β (SE) = −0.06 (0.03)), with an allelic trend concordant with the original report22.
Similarly, among variants associated with DD and related cognitive measures in previous GWAS efforts (see Table S8a–i), we identified a few nominally significant associations (p < 0.05) that were comparable with those reported by previous independent studies (Table S8j). The most significant associations were observed at rs10485609, an intronic SNP located within the CSE1L gene (20q13.13), with both word (A/G; MAF = 12%; p-value = 2.6 × 10−3, major allele (A) β (SE) = −0.12 (0.04)) and nonword reading (p-value = 6.5 × 10−3, major allele (A) β (SE) = −0.1 (0.04)). These associations showed the same direction of effect as in the original report34.
Gene- and pathway-based associations
Gene-level analyses of single-variant association signals in MAGMA revealed no significant enrichment of genes after correcting for testing of 18,033 protein-coding genes and of five independent latent traits (α = 5.5 × 10−7; see Table S9a–h). The most significant association was observed for the gene ADCYAP1R1 (adenylate cyclase activating polypeptide 1 receptor type I; 7p14.3) with NWRead (Z-score = 4.6; p = 2 × 10−6). Similarly, also in the gene-set analysis of 1329 canonical pathways from the MSigDB website, no pathway was significantly enriched (α = 0.01 for permutation-based enrichments, already corrected for testing of multiple pathways; see Table S10a–h). However, we found a nominally significant enrichment of associations with WSpell for genes in the BioCarta RAS pathway (Bonferroni-corrected p = 0.045; β (SE) = 0.64 (0.16); see Table S10i for a complete list of genes leading the pathway-based association).
Genetic overlap with neuroimaging, neurodevelopmental, and neuropsychiatric phenotypes
PRS analysis revealed the presence of a significant proportion of shared genetic variance between the different DD-related traits analyzed in our GWAS and some of the neuroimaging, educational, and neuropsychiatric phenotypes investigated in previous large GWAS studies (see Fig. 5; Table S11a–c). In particular, we observed significant genetic overlaps withstanding Bonferroni correction (p < 6.94 × 10−5) with ADHD risk, and with educational attainment (EDUyears). The ADHD PRS was negatively associated with WRead, WSpell, and NWRead (at PT = 0.05: Nagelkerke’s R2 ranging from 0.004 for NWRead to 0.007 for WRead; p ~ [10−5–10−7]), while EDUyears polygenic score was positively associated with WRead, WSpell, NWRead, DigSpan, and PA (at PT = 0.05: R2 ranging from 0.011 for DigSpan to 0.019 for WRead and PA; p ~ [10−8–10−17]). These results were substantially confirmed at different PT thresholds (see Figure S11a–h).
Results of the polygenic risk score (PRS) analysis on the eight traits analyzed in this work (target traits), which were compared with different neuropsychiatric, educational, and neuroimaging phenotypes (training traits). In the heatmap, –log(p) of the R2 computed by PRSice57 at an association p-value threshold (PT) of 0.05 is reported. Complete summary statistics are reported in Tables S11a, b, c
Discussion
In the present study, we investigated genetic effects on eight different neurocognitive skills, including behavioral features and predictors of dyslexia. We conducted a GWAS of up to 3468 subjects from nine different countries, speaking six different languages. Hence, our study represents the most detailed GWAS in the field in terms of phenotypes investigated, countries and languages involved, and one of the largest reported so far.
We identified a genome-wide significant effect on rapid automatized naming of letters, which showed a (relational) pleiotropic influence on the whole RAN domain and, to a lesser extent, on reading abilities. Rapid naming reflects the automaticity of visual–verbal access necessary for efficient word decoding, and accounts for a significant proportion of variance in word reading ability, especially reading fluency, which is independent of the well-established language and phonological processes implicated in reading, like phoneme awareness69. This association between RAN and reading (fluency) has been reported across different orthographies45 and over the life span70. Furthermore, RAN turned out to be a significant predictor of poor reading skills across orthographies38 and is also used in kindergarten to identify children at risk of dyslexia71. Good predictivity of reading performance has been reported for both alphanumeric and non-alphanumeric RAN measures, although correlations are usually stronger between alphanumeric RAN and reading fluency, compared with non-alphanumeric RAN (as reviewed by Kirby et al.72). The correlations between alphanumeric (letters and numbers) RAN and reading skills are also significant through adulthood73. Importantly, RAN predicts later reading performance independently from reading experience or early differences in reading ability74,75,76, and from phoneme awareness. While RAN has been shown to be an important predictor for reading problems, it should be noted that, in line with multiple-deficit models of dyslexia77, RAN represents one of several predictors of DD risk and reading abilities (reviewed in ref. 78). More recently, RAN has been also associated with other learning disorders, like dyscalculia, and it has been hypothesized that RAN deficits in dyslexic children may be independent from those detected in dyscalculic children79,80.
The most significant association signal with RANlet was observed for rs17663182, a variant located within MIR924HG (18q12.2; micro-RNA 924 host gene, or LINC00669). Additional significant associations were detected in the same region for other variants, all in high LD with the lead SNP, which suggests that they identified the same genetic effect on RANlet. This observation was supported by the absence of strong independent genetic effects on RANlet within a 100 kb window surrounding the strongest signal at rs17663182. An extensive lookup of these 18q12.2 variants in common online gene expression databases—including the Genotype-Tissue Expression portal (GTEx)81, the Brain eQTL Almanac (Braineac)82, the Blood eQTL browser83, and the seeQTL database84 —revealed weak evidence of expression quantitative trait loci (eQTL) involving rs17663182 and neighboring associated SNPs. Braineac reports nominally significant eQTL effects (p-value < 0.05) for these SNPs on MIR924HG expression in the occipital cortex, thalamus, and substantia nigra. In addition, HaploReg v4.1 indicated the presence of histone marks usually associated with transcriptional activity in the same region, such as H3K4me1, H3K27ac, and H3K9ac85. To the best of our knowledge, no regulatory role is known for MIR924HG, and MIR924 has not been functionally characterized so far. Nonetheless, the significant associations on 18q12.2 represent an interesting genetic effect for three main reasons:
First and foremost, evidence of genetic linkage to dyslexia-related cognitive traits has been reported for this region in previous studies, although not always reaching statistical significance86,87,88,89. In a genome-wide linkage analysis of a German cohort partly overlapping with our AGS dataset, a linkage peak to a principal component of RAN scores was observed in a region encompassing the microsatellite marker D18S1102, located ~2.1 Mb downstream of rs1766318289. Similarly, a linkage signal was later reported for the same marker with a composite RAN score, in a Dutch sib-pair sample. However, this association was weaker after including parents of the sib-pairs in the analysis86. Early evidence for linkage in 18q12 has been reported with word reading and orthographic coding, in samples partially overlapping with our Colorado and UK datasets87,88. In line with these findings, rs17663182 showed associations with traits other than RANlet in our analysis, including RANdig, RANpic, WRead, and NWRead (further discussed below). It would be tempting to connect the linkage signals mentioned above with the SNP associations at rs17663182, but it is important to point out that this association likely represents only a small fraction of these linkage signals or even a distinct genetic effect, because linkage and association analyses tend to detect different effects90.
Second, a search for binding sites through the online database TargetScanHuman v7.191 allowed us to identify a series of interesting candidate target genes which MIR924 could regulate. These include candidate dyslexia susceptibility genes like MRPL19 and KIAA0319L, although these did not show the highest predicted binding scores to MIR924 (cumulative weighted context++ scores −0.08 and −0.07; ranked 1615 and 1626 over 3472 potential targets).
Third, MIR924HG is expressed in a number of cancer cell lines, but consistently in samples representing iPS differentiation into neurons, according to the FANTOM5 miRNA promoter analysis92. This is interesting in the context that at least three dyslexia candidate genes (namely DCDC2, DYX1C1, and KIAA0319) have been implicated in regulating neuronal migration and cilia functions in model systems9.
In the analysis of RANlet, we observed an additional association approaching genome-wide significance at rs16928927 (8q12.3). This intronic variant is located within NKAIN3 (Na+/K+ transporting ATPase interacting 3), a gene which is widely and specifically expressed in the brain, especially in the fetal temporal lobe, in newborn and in adult hippocampal regions93.
Of note, both our lead SNPs showed associations with different cognitive measures analyzed in this study, especially with RAN traits. This multitrait association trend is particularly noticeable for rs17663182, which showed convincing evidence of influence within and even beyond the RAN domain, extending to reading abilities, as suggested by a genome-wide significant multivariate association with all the cognitive traits analyzed. However, a horizontal pleiotropy test on both variants did not reveal any significant effect specific to cognitive traits other than RANlet. This suggests that these variants likely exert their genetic influence on the common phenotypic variance underlying these traits, with different magnitude of effect on each measure, rather than on trait-specific phenotypic variance.
Despite the biological appeal of the top association signals mentioned above, an imaging genetic assessment of these SNPs did not reveal any significant effect on variation in seven different subcortical volumes58. Considering the sample size of this neuroimaging genetic analysis (N~13,000), we deem it unlikely that this lack of support is caused by a lack of power. However, this negative result does not rule out genetic effects of the variants detected here on other brain structures involved in reading networks, such as the inferior frontal gyrus and the temporal and parietal gyri. These potential associations should be tested in the future, as was previously done for other variants associated with reading-related traits94,95.
An assessment of candidate genes and SNPs implicated in dyslexia and related traits by other studies provided weak or (in most cases) no evidence of replication. Several possible factors may account for these apparently contrasting results. First, the heterogeneity of recruitment of the samples analyzed may lead to discrepant results across different studies: some genetic variants may have stronger effects in the lower tail of the reading and language skills distributions (i.e., in selected DD samples) and negligible effects in a broader range of variation (i.e., in general population samples). Second, the heterogeneity of assessment of the phenotypes may result in traits that ostensibly tap into the same cognitive domain but actually represent slightly different abilities. This applies not only to continuous DD-related measures, but also to the classification of dyslexia cases and controls, for which a consensus is far from being reached in the scientific community3,96,97. Third, different genetic backgrounds of the populations analyzed may be a factor when comparing or meta-analyzing different association studies. The haplotype structure in a specific region may differ between populations, and so may change the LD between the tag SNP (where the association is detected) and the genuine causal SNP (which determines the association). In the presence of substantial population stratification, this could even result in contrasting directions of effect for the same SNP in different studies37,98,99. Fourth, the inconsistent results from association studies may be due to different age ranges of the samples analyzed, e.g., when comparing an adult population with a datasets made up of children37. An alternative explanation may be that the original findings were type I errors, since false-positive results may easily occur in analyses of relatively small samples100. While this is a less likely explanation for those associated SNPs which have been functionally investigated, it may reasonably account for spurious associations, which are more likely to be affected by publication biases (i.e., significant results tend to be favored for publication) and reporting biases (i.e., investigators tend to report only positive findings).
Another interesting finding of our study is the significant genetic overlap that some of the traits analyzed showed with educational attainment (EDUyears) and ADHD. Educational attainment was already reported to share a significant proportion of genetic variance with word reading ability101,102. In a PRS analysis comparing educational attainment with reading efficiency and comprehension, the same EDUYears score used here59 accounted for 2.1% (at the age of 7) to 5.1% (at the age of 14) of the variance in such reading measures in a UK sample (N = 5825), and this association remained significant even after correcting for general cognitive ability and socioeconomic status101. More recently, Luciano et al.102 used the results of a previous GWAS on reading and language-related traits32 to test genetic correlations with several health, socioeconomic, and brain structure measures collected in adults from the UK (maximal N = 111,749; age range 40–69 years). Polygenic scores increasing these traits—namely word reading, nonword repetition, and a reading–spelling score— were all positively associated with a binary index of educational attainment (college or university degree)102. In our paper, we replicate these findings by reporting that variants nominally associated with EDUyears explain almost 2% of the total variance in WRead (used here as a target trait), and extend the evidence of genetic overlap to other behavioral features—WSpell and NWRead—and to cognitive predictors of dyslexia risk like PA and DigSpan.
Our PRS analysis also revealed a shared genetic basis for ADHD risk and the core dyslexia features WRead, WSpell, and NWRead. This long-standing hypothesis was originally supported by behavioral genetics studies of twins103,104,105, and has been later corroborated by molecular genetic studies. The existence of overlapping risk loci between DD and ADHD suggests that these regions could be the potential sites of liability underlying ADHD–DD comorbidity1. Candidate DD susceptibility genes like DYX1C1, DCDC2, and KIAA0319 have been associated with inattention and hyperactivity/impulsivity106,107,108, and candidate ADHD genes like DRD4 have been investigated for linkage and association with DD, with inconsistent results109,110. More recently, Mascheretti et al.111 found significant main and interactive associations upon hyperactivity/impulsivity involving DCDC2 and KIAA0319, while Sánchez-Móran et al.25 reported stronger associations of candidate KIAA0319 and FOXP2 variants with ADHD–DD comorbid cases, compared with simple dyslexic subjects. In line with this evidence, our findings provide further support to a partly shared genetic etiology of DD and ADHD at the genome-wide level.
Among the limitations of our study are a certain variability in the inclusion criteria and phenotypic assessment of some cohorts18, the absence of a follow-up cohort to replicate the genome-wide significant associations detected, and the modest power to detect small effect sizes (see Supplementary Methods). These are counterbalanced by strengths of our study, which include the variety of continuous neurocognitive traits analyzed, covering all the most relevant dyslexia-related behavioral phenotypes, and the homogeneity of QC procedures among datasets, which are fundamental to improve statistical power. Indeed, most of our samples were collected in the context of a large international consortium for studying the neurobiological/genetic basis of dyslexia (Neurodys), whose main purpose is to homogenize traits and datasets to allow for comparable analyses across different countries38,42. Overall, this study represents an early step of one of the largest international collaborations aimed at clarifying the genetic basis of reading abilities and disabilities, which will hopefully contribute to shed a light on the neurobiology of dyslexia.
URLs
Human Integrated Protein Expression Database: http://www.genecards.org/. FANTOM5 Zenbu database: http://fantom.gsc.riken.jp/zenbu/. PLINK: https://www.cog-genomics.org/plink2. QCTOOL: http://www.well.ox.ac.uk/~gav/qctool/. MatSpD: http://gump.qimr.edu.au/general/daleN/matSpD/. MAGMA: http://ctg.cncr.nl/software/magma. MSigDB: http://software.broadinstitute.org/gsea/msigdb; PRSice: http://prsice.info/. Genotype-Tissue Expression portal (GTEx): http://www.gtexportal.org/home/. Brain eQTL Almanac (Braineac): http://www.braineac.org/. Blood eQTL: http://genenetwork.nl/bloodeqtlbrowser/. seeQTL: http://www.bios.unc.edu/research/genomic_software/seeQTL. HaploReg: http://archive.broadinstitute.org/mammals/haploreg/haploreg.php. TargetScan: http://www.targetscan.org/. LocusZoom: http://www.locuszoom.org/
References
Mascheretti, S. et al. Neurogenetics of developmental dyslexia: from genes to behavior through brain neuroimaging and cognitive and sensorial mechanisms. Transl. Psychiatry 7, e987 (2017).
Margari, L. et al. Neuropsychopathological comorbidities in learning disorders. Bmc. Neurol. 13, 198 (2013).
Raskind, W. H., Peter, B., Richards, T., Eckert, M. M. & Berninger, V. W. The genetics of reading disabilities: From phenotypes to candidate genes. Front. Psychol. 3, 1–20 (2013).
Willcutt, E. G. et al. Etiology and neuropsychology of comorbidity between RD and ADHD: The case for multiple-deficit models. Cortex 46, 1345–1361 (2010).
Davis, C. J. et al. Etiology of reading difficulties and rapid naming: the Colorado twin study of reading disability. Behav. Genet. 31, 625–635 (2001).
Gayan, J. & Olson, R. K. Genetic and environmental influences on orthographic and phonological skills in children with reading disabilities. Dev. Neuropsychol. 20, 483–507 (2001).
Gayan, J. & Olson, R. K. Genetic and environmental influences on individual differences in printed word recognition. J. Exp. Child Psychol. 84, 97–123 (2003).
Carrion-Castillo, A., Franke, B. & Fisher, S. E. Molecular genetics of dyslexia: an overview. Dyslexia 19, 214–240 (2013).
Kere, J. The molecular genetics and neurobiology of developmental dyslexia as model of a complex phenotype. Biochem. Biophys. Res. Commun. 452, 236–243 (2014).
Taipale, M. et al. A candidate gene for developmental dyslexia encodes a nuclear tetratricopeptide repeat domain protein dynamically regulated in brain. Proc. Natl. Acad. Sci. USA 100, 11553–11558 (2003).
Francks, C. et al. A 77-kilobase region of chromosome 6p22.2 is associated with dyslexia in families from the United Kingdom and from the United States. Am. J. Hum. Genet. 75, 1046–1058 (2004).
Cope, N. et al. Strong evidence that KIAA0319 on chromosome 6p is a susceptibility gene for developmental dyslexia. Am. J. Hum. Genet. 76, 581–591 (2005).
Harold, D. et al. Further evidence that the KIAA0319 gene confers susceptibility to developmental dyslexia. Mol. Psychiatry 11, 1085–1091 (2006).
Dennis, M. Y. et al. A common variant associated with dyslexia reduces expression of the KIAA0319 gene. PLoS Genet 5, e1000436, https://doi.org/10.1371/journal.pgen.1000436 (2009).
Meng, H. et al. DCDC2 is associated with reading disability and modulates neuronal development in the brain. Proc. Natl. Acad. Sci. USA 102, 17053–17058 (2005).
Schumacher, J. et al. Strong genetic evidence of DCDC2 as a susceptibility gene for dyslexia. Am. J. Hum. Genet. 78, 52–62 (2006).
Marino, C. et al. DCDC2 genetic variants and susceptibility to developmental dyslexia. Psychiatr. Genet. 22, 25–30 (2013).
Riva V. et al. The influence of DCDC2 risk genetic variants on reading: testing main and haplotypic effects. Neuropsychologia.https://doi.org/10.1016/j.neuropsychologia.2018.05.021(2018) in press.
Anthoni, H. et al. A locus on 2p12 containing the co-regulated MRPL19 and C2ORF3 genes is associated to dyslexia. Hum. Mol. Genet. 16, 667–677 (2007).
Hannula-Jouppi, K. et al. The axon guidance receptor gene ROBO1 is a candidate gene for developmental dyslexia. PLoS. Genet. 1, 0467–0474 (2005).
Bates, T. C. et al. Genetic variance in a component of the language acquisition device: ROBO1 polymorphisms associated with phonological buffer deficits. Behav. Genet. 41, 50–57 (2011).
Tran, C. et al. Association of the ROBO1 gene with reading disabilities in a family-based analysis. Genes, Brain Behav. 13, 430–438 (2014).
Ludwig, K. U. et al. Variation in GRIN2B contributes to weak performance in verbal short-term memory in children with dyslexia. Am. J. Med Genet Part B Neuropsychiatr. Genet 153, 503–511 (2010).
Mascheretti, S. et al. GRIN2B mediates susceptibility to intelligence quotient and cognitive impairments in developmental dyslexia. Psychiatr. Genet. 25, 9–20 (2015).
Sánchez-Morán, M. et al. Genetic association study of dyslexia and ADHD candidate genes in a Spanish cohort: Implications of comorbid samples. PLoS ONE 13, e0206431 (2018).
Wilcke, A. et al. Imaging genetics of FOXP2 in dyslexia. Eur. J. Hum. Genet. 20, 224–229 (2012).
Peter, B. et al. Replication of CNTNAP2 association with nonword repetition and support for FOXP2 association with timed reading and motor activities in a dyslexia family sample. J. Neurodev. Disord. 3, 39–49 (2011).
Vernes, S. C. et al. A functional genetic link between distinct developmental language disorders. N. Engl. J. Med. 359, 2337–2345 (2008).
Newbury, D. F. et al. Investigation of dyslexia and SLI risk variants in reading- and language-impaired subjects. Behav. Genet. 41, 90–104 (2011).
Eicher, J. D. et al. Genome-wide association study of shared components of reading disability and language impairment. Genes. Brain. Behav. 12, 792–801 (2013).
Gialluisi, A. et al. Genome-wide screening for DNA variants associated with reading and language traits. Genes. Brain. Behav. 13, 686–701 (2014).
Luciano, M. et al. A genome-wide association study for reading and language abilities in two population cohorts. Genes, Brain Behav. 12, 645–652 (2013).
Field, L. L. et al. Dense-map genome scan for dyslexia supports loci at 4q13, 16p12, 17q22; suggests novel locus at 7q36. Genes. Brain. Behav. 12, 56–69 (2013).
Meaburn, E. L., Harlaar, N., Craig, I. W., Schalkwyk, L. C. & Plomin, R. Quantitative trait locus association scan of early reading disability and ability using pooled DNA and 100K SNP microarrays in a sample of 5760 children. Mol. Psychiatry 13, 729–740 (2008).
Truong D. et al. Multivariate genome-wide association study of rapid automatized naming and rapid alternating stimulus in Hispanic and African American youth. bioRxiv 2017; 202929. https://doi.org/10.1101/202929.
Luciano, M., Montgomery, G. W., Martin, N. G., Wright, M. J. & Bates, T. C. SNP sets and reading ability: testing confirmation of a 10-SNP set in a population sample. Twin. Res. Hum. Genet. 14, 228–232 (2011).
Carrion-Castillo, A. et al. Evaluation of results from genome-wide studies of language and reading in a novel independent dataset. Genes, Brain Behav. 15, 531–541 (2016).
Landerl, K. et al. Predictors of developmental dyslexia in European orthographies with varying complexity. J. Child Psychol. Psychiatry 54, 686–694 (2013).
Willcutt, E. G., Pennington, B. F., Olson, R. K., Chhabildas, N. & Hulslander, J. Neuropsychological analyses of comorbidity between reading disability and attention deficit hyperactivity disorder: in search of the common deficit. Dev. Neuropsychol. 27, 35–78 (2005).
Brandler, W. M. et al. Common variants in left/right asymmetry genes and pathways are associated with relative hand skill. PLoS. Genet. 9, e1003751 (2013).
Moll, K., Kunze, S., Neuhoff, N., Bruder, J. & Schulte-Körne, G. Specific learning disorder: prevalence and gender differences. PLoS ONE 9, e103537 (2014).
Becker, J. et al. Genetic analysis of dyslexia candidate genes in the European cross-linguistic NeuroDys cohort. Eur. J. Hum. Genet. 22, 675–680 (2014).
Gialluisi, A. Investigating the effects of copy number variants on reading and language performance. J. Neurodev. Disord. 8, 17 (2016).
Roeske, D. et al. First genome-wide association scan on neurophysiological endophenotypes points to trans-regulation effects on SLC2A3 in dyslexic children. Mol. Psychiatry 16, 97 (2009).
Moll, K. et al. Cognitive mechanisms underlying reading and spelling development in five European orthographies. Learn Instr. 29, 65–77 (2014).
Chang, C. C. et al. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience 4, 7 (2015).
Andlauer, T. F. et al. Novel multiple sclerosis susceptibility loci implicated in epigenetic regulation. Sci. Adv. 2, e1501678 (2016).
The Genomes Project C. A global reference for human genetic variation. Nature 526, 68 (2015).
Delaneau, O., Zagury, J.-F. & Marchini, J. Improved whole-chromosome phasing for disease and population genetic studies. Nat. Meth 10, 5–6 (2013).
Howie, B. N., Donnelly, P. & Marchini, J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS. Genet. 5, e1000529 (2009).
Lippert, C. et al. FaST linear mixed models for genome-wide association studies. Nat. Meth 8, 833–835 (2011).
Han, B. & Eskin, E. Random-effects model aimed at discovering associations in meta-analysis of genome-wide association studies. Am. J. Hum. Genet. 88: 586–598.
Li, J. & Ji, L. Adjusting multiple testing in multilocus analyses using the eigenvalues of a correlation matrix. Hered. (Edinb.) 95, 221–227 (2005).
van der Sluis, S., Posthuma, D. & Dolan, C. V. TATES: efficient multivariate genotype-phenotype analysis for genome-wide association studies. PLoS. Genet. 9, e1003235 (2013).
Paaby, A. B. & Rockman, M. V. The many faces of pleiotropy. Trends Genet. 29, 66–73 (2013).
de Leeuw, C. A., Mooij, J. M., Heskes, T. & Posthuma, D. MAGMA: generalized gene-set analysis of GWAS data. PLoS. Comput. Biol. 11, e1004219 (2015).
Euesden, J., Lewis, C. M. & O’Reilly, P. F. PRSice: polygenic risk Score software. Bioinformatics 31, 1466–1468 (2015).
Hibar, D. P. et al. Common genetic variants influence human subcortical brain structures. Nature 520, 224–229 (2015).
Okbay, A. et al. Genome-wide association study identifies 74 loci associated with educational attainment. Nature 533, 539–542 (2016).
Demontis, D. et al. Discovery of the first genome-wide significant risk loci for attention deficit/hyperactivity disorder. Nat Genet 51, 63–75 (2019).
The Autism Spectrum Disorders Working Group of The Psychiatric Genomics Consortium. Meta-analysis of GWAS of over 16,000 individuals with autism spectrum disorder highlights a novel locus at 10q24.32 and a significant overlap with schizophrenia. Mol. Autism 8, 21 (2017).
Sullivan, P. F. et al. A mega-analysis of genome-wide association studies for major depressive disorder. Mol. Psychiatry 18, 497–511 (2013).
Schizophrenia Working Group of the Psychiatric Genomics Consortium. Biological insights from 108 schizophrenia-associated genetic loci. Nature 511, 421–427 (2014).
Russell G. & Pavelka Z. Co-occurrence of developmental disorders: children who share symptoms of autism, dyslexia and attention deficit hyperactivity disorder. In Recent Advances in Autism Spectrum Disorders - Volume I. (ed. Fitzgerald M.). 17 (InTech: Rijeka, 2013).
Mugnaini, D., Lassi, S., La Malfa, G. & Albertini, G. Internalizing correlates of dyslexia. World J. Pediatr. 5, 255–264 (2009).
Whitford, V., O'Driscoll, G. A. & Titone, D. Reading deficits in schizophrenia and their relationship to developmental dyslexia: A review.Schizophr Res 193, 11–22 (2017).
Eicher, J. D. & Gruen, J. R. Imaging-genetics in dyslexia: connecting risk genetic variants to brain neuroimaging and ultimately to reading impairments. Mol. Genet. Metab. 110, 201–212 (2013).
Krishnan, S., Watkins, K. E. & Bishop, D. V. M. Neurobiological basis of language learning difficulties. Trends Cogn. Sci. 20, 701–714 (2016).
Wolf, M. & Bowers, P. G. The double-deficit hypothesis for the developmental dyslexias. J. Educ. Psychol. 91, 415–438 (1999).
Scarborough, H. S. Predicting the future achievement of second graders with reading disabilities: Contributions of phonemic awareness, verbal memory, rapid naming, and IQ. Ann. Dyslexia 48, 115–136 (1998).
Schatschneider, C., Fletcher, J. M., Francis, D. J., Carlson, C. D. & Foorman, B. R. Kindergarten prediction of reading skills: a longitudinal comparative analysis. J. Educ. Psychol. 96, 265–282 (2004).
Kirby, J. R. et al. Naming Speed and Reading: From Prediction to Instruction. Reading Research Quarterly 45, 341–362 (2010).
van den bos, K. P., BJH, Zijlstra & lutje Spelberg, H. C. Life-span data on continuous-naming speeds of numbers, letters, colors, and pictured objects, and word-reading speed. Sci. Stud. Read. 6, 25–49 (2002).
Lervåg, Arne & Hulme, Charles Rapid automatized naming (RAN) Taps a mechanism that places constraints on the development of early reading fluency. Psychol. Sci. 20, 1040–1048 (2009).
Parrila, R., Kirby, J. R. & McQuarrie, L. Articulation rate, naming speed, verbal short-term memory, and phonological awareness: longitudinal predictors of early reading development? Sci. Stud. Read. 8, 3–26 (2004).
Bowey, J. A., McGuigan, M. & Ruschena, A. On the association between serial naming speed for letters and digits and word-reading skill: Towards a developmental account. J. Res. Read. 28, 400–422 (2005).
Pennington, B. F. From single to multiple deficit models of developmental disorders. Cognition 101, 385–413 (2006).
Vellutino, F. R., Fletcher, J. M., Snowling, M. J. & Scanlon, D. M. Specific reading disability (dyslexia): what have we learned in the past four decades? J. Child Psychol. Psychiatry 45, 2–40 (2004).
Willburger, E., Fussenegger, B., Moll, K., Wood, G. & Landerl, K. Naming speed in dyslexia and dyscalculia. Learn Individ Differ. 18, 224–236 (2008).
Mazzocco, M. M. M. & Grimm, K. J. Growth in rapid automatized naming from grades K to 8 in children with math or reading disabilities. J. Learn. Disabil. 46, 517–533 (2013).
Consortium TGte. The genotype-tissue expression (GTEx) project. Nat. Genet. 45, 580–585 (2013).
Ramasamy, A. et al. Genetic variability in the regulation of gene expression in ten regions of the human brain. Nat. Neurosci. 17, 1418–1428 (2014).
Westra, H. J. et al. Systematic identification of trans eQTLs as putative drivers of known disease associations. Nat. Genet. 45, 1238–1243 (2013).
Xia, K. et al. seeQTL: a searchable database for human eQTLs. Bioinformatics 28, 451–452 (2012).
Ward, L. D. & Kellis, M. HaploReg v4: systematic mining of putative causal variants, cell types, regulators and target genes for human complex traits and disease. Nucleic Acids Res. 44, D877–D881 (2016).
De Kovel, C. G. F. et al. Confirmation of dyslexia susceptibility loci on chromosomes 1p and 2p, but not 6p in a Dutch sib-pair collection. Am. J. Med Genet Part B Neuropsychiatr. Genet. 147, 294–300 (2008).
Fisher, S. E. & DeFries, J. C. Developmental dyslexia: genetic dissection of a complex cognitive trait. Nat. Rev. Neurosci. 3, 767–780 (2002).
Scerri, T. S. et al. Identification of candidate genes for dyslexia susceptibility on chromosome 18. PLoS One 5, e13712 (2010).
Schulte-Korne, G. et al. Interrelationship and familiality of dyslexia related quantitative measures. Ann. Hum. Genet. 71, 160–175 (2007).
Ott, J., Kamatani, Y. & Lathrop, M. Family-based designs for genome-wide association studies. Nat. Rev. Genet. 12, 465 (2011).
Agarwal, V., Bell, G. W., Nam, J.-W. & Bartel, D. P. Predicting effective microRNA target sites in mammalian mRNAs. eLife 4, e05005 (2015).
de Rie, D. et al. An integrated expression atlas of miRNAs and their promoters in human and mouse. Nat. Biotechnol. 35, 872 (2017).
The FC, the RP, Clst. A promoter-level mammalian expression atlas. Nature 507, 462 (2014).
Eicher, J. D. et al. Dyslexia and language impairment associated genetic markers influence cortical thickness and white matter in typically developing children. Brain. Imaging Behav. 10, 272–282 (2016).
Gialluisi, A., Guadalupe, T., Francks, C. & Fisher, S. E. Neuroimaging genetic analyses of novel candidate genes associated with reading and language. Brain Lang. 172, 9–15 (2017).
Peterson, R. L. & Pennington, B. F. Developmental dyslexia. Annu. Rev. Clin. Psychol. 11, 283–307 (2015).
Pennington, B. F. & Bishop, D. V. M. Relations among speech, language, and reading disorders. Annu. Rev. Psychol. 60, 283–306 (2009).
Luciano, M. et al. A haplotype spanning KIAA0319 and TTRAP is associated with normal variation in reading and spelling ability. Biol. Psychiatry 62, 811–817 (2007).
Lin, P.-I., Vance, J. M., Pericak-Vance, M. A. & Martin, E. R. No gene is an island: the flip-flop phenomenon. Am. J. Hum. Genet. 80, 531–538 (2007).
Colhoun, H. M., McKeigue, P. M. & Davey Smith, G. Problems of reporting genetic associations with complex outcomes. Lancet (Lond., Engl.) 361, 865–872 (2003).
Selzam, S. et al. Genome-wide polygenic scores predict reading performance throughout the school years. Sci. Stud. Read. 21, 334–349 (2017).
Luciano, M. et al. Single nucleotide polymorphisms associated with reading ability show connection to socio-economic outcomes. Behav Genet 47, 469–479 (2017).
Willcutt, E. G., Pennington, B. F., Olson, R. K. & DeFries, J. C. Understanding comorbidity: a twin study of reading disability and attention-deficit/hyperactivity disorder. Am. J. Med Genet Part B Neuropsychiatr. Genet. 144B, 709–714 (2007).
Willcutt, E. G., Pennington, B. F. & DeFries, J. C. Twin study of the etiology of comorbidity between reading disability and attention-deficit/hyperactivity disorder. Am. J. Med. Genet. 96, 293–301 (2000).
Norton, E. S. & Wolf, M. Rapid automatized naming (Ran) and reading fluency: implications for understanding and treatment of reading disabilities. Annu Rev Psychol. 63, 427–452 (2012).
Couto, J. M. et al. Association of attention-deficit/hyperactivity disorder with a candidate region for reading disabilities on chromosome 6p. Biol. Psychiatry 66, 368–375 (2009).
Wigg, K. G. et al. Support for EKN1 as the susceptibility locus for dyslexia on 15q21. Mol. Psychiatry 9, 1111–1121 (2004).
Wigg, K. G. et al. Association of ADHD and the Protogenin gene in the chromosome 15q21.3 reading disabilities linkage region. Genes. Brain. Behav. 7, 877–886 (2008).
Hsiung, G.-Y. R., Kaplan, B. J., Petryshen, T. L., Lu, S. & Field, L. L. A dyslexia susceptibility locus (DYX7) linked to dopamine D4 receptor (DRD4) region on chromosome 11p15.5. Am. J. Med. Genet. B. Neuropsychiatr. Genet. 125B, 112–119 (2004).
Marino, C. et al. No evidence for association and linkage disequilibrium between dyslexia and markers of four dopamine-related genes. Eur. Child Adolesc. Psychiatry 12, 198–202 (2003).
Mascheretti, S. et al. Complex effects of dyslexia risk factors account for ADHD traits: evidence from two independent samples. J. Child Psychol. Psychiatry Allied Discip. 58, 75–82 (2017).
Pruim, R. J. et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics 26, 2336–2337 (2010).
Aulchenko, Y. S., Ripke, S., Isaacs, A. & van Duijn, C. M. GenABEL: an R library for genome-wide association analysis. Bioinformatics 23, 1294–1296 (2007).
Wechsler D. The Wechsler intelligence scale for children. 3rd edn. London: The Psychological Corporation., 1992.
Wechsler D. Wechsler intelligence scale for children. 4th edn San Antonio, TX: Psychological Corporation., 2003.
Wechsler D. Manual for the Wechsler Intelligence Scale for Children – Revised. The Psychological Corporation, New York, NY, 1974.
Wechsler D. Manual for the Wechsler Adult Intelligence Scale – Revised. Psychological Corporation, New York, NY, 1981.
Elliot Murray, D. J. & Pearson, L. S. C. D. The British Ability Scales. (NFER, Slough, UK., 1979).
Acknowledgements
A.G. and T.F.M.A. were supported by the Munich Cluster for Systems Neurology (SyNergy). S.P. is a Royal Society University research Fellow. B.M.M., C.F., B.S.P., and S.E.F. are supported by the Max Planck Society. A.P.M. is a Wellcome Senior Fellow in Basic Biomedical Science (WT098017). F.R. is supported by Agence Nationale de la Recherche (ANR-06-NEURO-019-01, ANR-10-LABX-0087 IEC, ANR-10-IDEX-0001-02 PSL, ANR-11-BSV4-014-01), European Commission (LSHM-CT-2005-018696), Ville de Paris. We would also like to acknowledge our project partners Catherine Billard, Caroline Bogliotti, Vanessa Bongiovanni, Laure Bricout, Camille Chabernaud, Yves Chaix, Isabelle Comte-Gervais, Florence Delteil-Pinton, Jean-François Démonet, Florence George, Christophe-Loïc Gérard, Stéphanie Iannuzzi, Marie Lageat, Marie-France Leheuzey, Marie-Thérèse Lenormand, Marion Liébert, Emilie Longeras, Emilie Racaud, Isabelle Soares-Boucaud, Sylviane Valdois, Nadège Villiermet, Johannes Ziegler, Dr Else Eising, Dr Holger Kirsten, and Dr Bent Müller.
Author information
Authors and Affiliations
Contributions
A.G., T.F.M.A., N.M.S., D.C., and K.M. contributed to genotype QC and imputation, and to phenotype QC. A.G. carried out statistical analyses. A.G., T.F.M.A., K.M., B.M.M., and G.S.K. wrote the paper, with additional contributions by all the co-authors. All the co-authors contributed to collection, phenotypic assessment, and genotyping of the datasets included in the present study. B.M.M. and G.S.K. supervised the present work.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Gialluisi, A., Andlauer, T.F.M., Mirza-Schreiber, N. et al. Genome-wide association scan identifies new variants associated with a cognitive predictor of dyslexia. Transl Psychiatry 9, 77 (2019). https://doi.org/10.1038/s41398-019-0402-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41398-019-0402-0
This article is cited by
-
A genome-wide association study of Chinese and English language phenotypes in Hong Kong Chinese children
npj Science of Learning (2024)
-
Towards a dynamic, comprehensive conceptualization of dyslexia
Annals of Dyslexia (2024)
-
Cumulative genetic score of KIAA0319 affects reading ability in Chinese children: moderation by parental education and mediation by rapid automatized naming
Behavioral and Brain Functions (2023)
-
Properties of white matter tract diffusivity in children with developmental dyslexia and comorbid attention deficit/hyperactivity disorder
Journal of Neurodevelopmental Disorders (2023)
-
Associative gene networks reveal novel candidates important for ADHD and dyslexia comorbidity
BMC Medical Genomics (2023)