Abstract
Nearly 95% of susceptibility SNPs identified by genome-wide association studies (GWASs) are located in non-coding regions, which causes a lot of difficulty in deciphering their biological functions on disease pathogenesis. Here, we aimed to conduct a comprehensive functional annotation for all the schizophrenia susceptibility loci obtained from GWASs. Considering varieties of epigenomic regulatory elements, we annotated all 22,688 acquired susceptibility SNPs according to their genomic positions to obtain functional SNPs. The comprehensive annotation indicated that these functional SNPs are broadly involved in diverse biological processes. Histone modification enrichment showed that H3K27ac, H3K36me3, H3K4me1, and H3K4me3 were related to the development of schizophrenia. Transcription factors (TFs) prediction, methylation quantitative trait loci (meQTL) analyses, expression quantitative trait loci (eQTL) analyses, and proteomic quantitative trait loci analyses (pQTL) identified 447 target protein-coding genes. Subsequently, differential expression analyses between schizophrenia cases and controls, nervous system phenotypes from mouse models, and protein–protein interaction with known schizophrenia-related pathways and genes were carried out with our target genes. We finaly prioritized 10 target genes for schizophrenia (CACNA1C, CLU, CSNK2B, GABBR1, GRIN2A, MAPK3, NOTCH4, SRR, TNF, and SYNGAP1). Our results may serve as an encyclopedia of schizophrenia susceptibility SNPs and offer holistic guides for post-GWAS functional experiments.
Similar content being viewed by others
Introduction
Schizophrenia is one of the most mysterious and costliest mental disorders with a lifetime risk about 1% [1, 2]. Patients with a diagnosis of schizophrenia live 12–15 years shorter than normal people, and this mortality difference increases in recent decades1. Published twin studies found that the heritability of schizophrenia was 73–90% and environmental influence was estimated as 3–19%2,3. Basing on the high heritability of schizophrenia, genetic susceptibility factor decipherment would lead us to a better understanding of the genetic basis of schizophrenia.
To date, genome-wide association studies (GWASs) have identified many schizophrenia susceptibility loci. However, most of the disease-associated variants locate in intronic or intergenic regions4, which causes difficulties in clarifying their effects on diseases pathogenesis. Also, till now, the majority of functional SNPs remain unrevealed in schizophrenia studies.
Recent years, the increasing epigenomic datasets, including the Encyclopedia of DNA Elements (ENCODE)5 and Roadmap Epigenomics Project6, make it possible to understand the function of non-coding variants from epigenomic level. It has been reported that the non-coding regions could indirectly participate in the regulation of proximal or distal genes expression by functioning as regulatory elements. Typically, these include the DNA methylation sites, histone modification sites, DNase I hypersensitive sites, transcription factor (TF)-binding sites, as well as the proved enhancer and promoter regions, which are related to the cleavage, transcription, and translation of genes4,7. Given these, some functional annotations of GWASs reported SNPs, and the epigenomic contribution have identified a bunch of functional SNPs and extended our understanding in genetic regulation mechanisms8,9,10. For schizophrenia, a comprehensive annotation of GWASs results is needed.
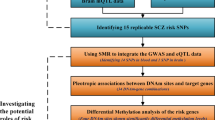
In this study, we obtained schizophrenia-associated SNPs from GWASs and aimed to perform comprehensive functional annotation for all susceptibility loci. At first, we acquired known schizophrenia susceptibility SNPs (referred to as index SNPs) and SNPs in high linkage disequilibrium (LD) with the index SNPs. We also acquired schizophrenia susceptibility SNPs from GWAS summary studies. For SNPs in coding DNA sequences (CDS) regions, we predicted the potential effect of missense SNPs on protein functions. For SNPs in untranslated regions (UTRs), we predicted their potential effects on microRNA binding. For SNPs located in promoters, we predicted whether they have effects on TF binding. For SNPs located in intronic or intergenic region, we detected whether they might regulate enhancer activity through affecting TF-binding ability. For all annotated functional SNPs, histone modification enrichment, methylation quantitative trait loci (meQTL), expression quantitative trait loci (eQTL), and proteomic quantitative trait loci (pQTL) analyses were subsequently performed. In addition, we used multiple ways to validate the correlation of target genes with schizophrenia. Our results may serve as an encyclopedia of schizophrenia susceptibility SNPs and offer guides for post-GWAS functional experiments.
Materials and methods
Acquisition of schizophrenia-associated SNPs
Figure 1 shows the analysis strategy of this study. SNPs associated with schizophrenia were obtained from two common resources of SNP–trait associations: GWAS-catalog11 and phenotype–genotype integrator (http://www.ncbi.nlm.nih.gov/gap/phegeni). SNPs reported to be associated with schizophrenia with P < 5 × 10−8 were selected. These were further referred to as index SNPs. Due to the low genomic coverage of microarrays, those true causal variants may not be detected12. Therefore, we also obtained the SNPs in strong LD (r2 ≥ 0.8) with the index SNPs using the 1000 Genomes Phase III data. These were further referred to as LD SNPs. In addition, we also acquired schizophrenia susceptibility SNPs from GWAS summary studies13,14 with P < 5 × 10−8, which were called summary SNPs hereafter. All of the index, LD and summary SNPs were considered as schizophrenia susceptibility SNPs and were subjected to subsequently analyses. Using GencodeV19 reference genome (http://www.gencodegenes.org/releases/), the susceptibility SNPs were annotated with ANNOVAR15 to get their genomic region information.
Schizophrenia-associated index SNPs were obtained from public GWASs databases. LD analyses were carried out using the 1000 genome data and GWASs summary data were used to obtain all schizophrenia susceptibility SNPs. Functional annotation was carried out according to the genomic regions of all susceptibility SNPs. For annotated functional SNPs, histone modification enrichment, meQTL, eQTL, and pQTL analyses then were carried out. Various methods were used to validate target genes
Functional annotation of schizophrenia-associated SNPs
For SNPs in CDS, we focused on nonsynonymous SNPs. We predicted the potential effect of missense SNPs on protein functions using PROVEAN16, SIFT17, Polyphen2 (ref. 18), and CADD19. For SNPs located in 3′-UTR, we predicted the miRNA-binding affinity with the UTR sequences using MirSNP database20.
For SNPs located within 1 kb region upstream of transcription start site (TSS), we considered that these SNPs are located in promoters. For SNPs located in intronic or intergenic regions, we used the chromatin 15-states data in brain and blood tissues (Supplementary Table S1) from the Roadmap project (http://www.roadmapepigenomics.org/) to check whether they are located in enhancers). Subsequently, we detected whether the promoter and enhancer SNPs would affect TF binding using the SNP2TFBS database21. We also investigated which TFs they were enriched for disruption. The GEO dataset GSE69838 (ref. 22) was used to check whether these enriched TFs were differentially expressed during neuronal differentiation of SH-SY5Y cell (0 day vs 11 day).
Histone modification enrichment
We used histone modification data (including 11 histone marks H3K27ac, H3K27me3, H3K36me3, H3K4me1, H3K4me2, H3K4me3, H3K79me2, H3K9ac, H3K9me, H3K9me3, and H4K20me1) in brain and blood tissues from the Roadmap project (supplementary Table S1) to predict these schizophrenia functional SNPs were enriched in what kind of histone marks using Variant Set Enrichment (VSE)23. In the VSE method, a disjoint list of associated variant set (AVS) had be constructed at the first step, in which only one SNP is present in one LD block to avoid inflating test statistics.
meQTL analyses
We used the meQTL data in the prefrontal cortex from 335 controls and 191 schizophrenia patients reported by Jaffe et al.24 to investigate whether the annotated functional SNPs could affect DNA methylation levels.
eQTL analyses
eQTL results derived from (1) the dorsolateral prefrontal cortex of schizophrenia patients (N = 258) and control subjects (N = 279) from CommonMind Consortium (CMC)25 and (2) whole blood from 3841 samples reported by Bonder et al.26 were used to investigate whether the annotated functional SNPs could affect gene expression levels in schizophrenia-related tissues. For functional enhancer SNPs, genes with TSS flanking ±1 Mb around the SNPs were used for investigating eQTL signals in all related tissues. For other functional SNPs, their located genes were used. The Benjamini–Hochberg procedure was executed to account for the multiple testing problems. We also applied independent integrative analysis (Summary-data-based Mendelian Randomization, SMR) approach developed by Zhu et al.27 to the GWASs (summary-level data from GWASs summary studies previously mentioned13,14) and eQTL data (summary-level data from the Genotype-Tissue Expression (GTEx) v7 project28, after integrating 13 brain subregions, and whole blood). After applying a conservative threshold (PHEIDI > 0.05 and PeQTL < 0.05), genes with FDR q < 0.05 in both eQTL and SMR results in brain or blood separately were obtained.
pQTL analyses
We used the data from blood plasma proteome reported by Suhre et al.29 to investigate whether the annotated functional SNPs in blood could affect protein levels. The expression levels of 1124 proteins were quantified in 1000 individuals of the population-based KORA study. For functional enhancer SNPs, all proteins associated with susceptibility SNPs were used. For other functional SNPs, proteins coded by their located genes were used. The Benjamini–Hochberg procedure was executed, and the threshold of FDR q < 0.05 was used to filter out non-significant signals.
Differential expression analyses
For all target genes (obtained from meQTL, eQTL, and pQTL analyses, and enriched TFs) which encode proteins (GencodeV19 annotation data), we further checked whether they were differentially expressed between schizophrenia case and control samples. Meta-analysis using sample-size weighted model in the METAL software30 was carried out to combine the results of differential expression analyses from GSE53987 (prefrontal cortex, striatum, and hippocampus from 48 schizophrenia patients and 55 control subjects)31, GSE93987 (dorsolateral prefrontal cortex from 102 patients and 106 controls)32, and GSE38484 (whole blood from 106 patients and 96 controls)33). Effect direction was taken into account in this model. The Benjamini–Hochberg procedure was executed, and the threshold was same as mentioned before.
Associated genes from mouse models
We used phenotype data in mouse models from two projects, Mouse Genome Database (MGD)34 and International Mouse Phenotyping Consortium (IMPC)35, to verify whether these target genes deficiency could lead to disorders in nervous system.
Protein–protein interaction
We also compare our target genes with known schizophrenia-related pathways and genes information reported by MalaCards database (http://www.malacards.org/). For those genes not included, we checked whether they could be related to known schizophrenia pathways or genes through gene–gene interaction using data from MyProteinNet database (http://netbio.bgu.ac.il/myproteinnet/).
Pathway enrichment
For target genes which had at least two hits in differential expression analyses, mouse models and Malacards, we conducted KEGG pathway enrichment analyses with these genes to detect their related pathways (http://amp.pharm.mssm.edu/Enrichr/).
Targeted drugs
We used data of targeted drugs from DrugBank (https://www.drugbank.ca/) to investigate whether there were any connection between target genes and approved biotech drugs.
Results
Acquisition of schizophrenia-associated SNPs
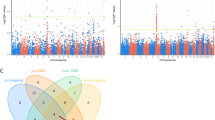
There were 347 schizophrenia-associated SNPs obtained from GWAS-Catalog and PheGenI in total (Fig. 2a, Supplementary Table S2). Among the index SNPs, 248 SNPs were from European-specific GWAS sample. Subsequently, we acquired 6926 SNPs in high LD with the index SNPs. We also obtained 20,573 susceptibility SNPs from GWAS summary studies (Fig. 2b). All susceptibility SNPs (22,688) were annotated with ANNOVAR and their classifications are shown in Fig. 2c.
Functional annotation of associated SNPs
We identified 338 susceptibility SNPs located in CDS. Of these SNPs, 161 encoded synonymous substitutions, 172 encoded missense substitutions, 1 frameshift substitution, and 4 still unknown. We explored the potential effects of the missense SNPs using PROVEAN, SIFT, Polyphen2, and CADD. Sixty-six SNPs were predicted to be damaging by at least one algorithm (Supplementary Table S3), including five index SNPs, rs1051061, rs13107325, rs16897515, rs2247870, and rs950169. Seven SNPs were predicted to be damaging by all four algorithms, including rs9257834, rs678, rs4584886, rs34788973, rs13195402, rs13195401, and rs1029871. Moreover, rs13195401, rs13195402, and rs34788973 were all in high LD with index SNPs rs13194053, rs55834529, and rs6932590, which may cause damaging changes in BTN2A1 and OR2B2.
We identified 357 SNPs in 3′-UTR, and 119 SNPs were predicted to be involved in microRNA targets (Supplementary Table S4), including four index SNPs (rs10786736, rs13205911, rs3735025, and rs4702). In addition, another two SNPs, rs1376607 and rs3735026, were in complete LD with the index SNP rs3735025.
Of all 388 promoter SNPs, 89 SNPs might disrupt TF binding, including index SNP rs796364. For example, the substitution from C to A of rs796364 would affect the binding affinity of NFIC in the upstream of FTCDNL1 gene. For SNPs in intronic or intergenic region, we identified 2820 and 4694 SNPs located in the enhancer regions of brain and blood cells, respectively. Among them, 659/1028 SNPs in brain/blood might regulate enhancer activity through affecting TF binding. These promoter SNPs were enriched for disruption of eight TFs binding (Fig. 3a) and enhancer SNPs were found to potentially disrupt 12/21 TFs binding in brain/blood (Fig. 3b, c), 34 in total (supplementary Table S5). Comparisons of the gene expression levels during SH-SY5Y cell differentiation detected significant expression changes for 13 TFs (MYC, MAFG, JUND, NFKB1, EGR1, ETS1, KLF4, NFE2L1, RXRA, SOX2, SP2, TCF12, and ZFX; Fig. 3d), suggesting these TFs may have significant functions on nerve cells differentiation or proliferation (Supplementary Table S5).
a Transcription factor enrichment results for the promoter SNPs. b TF enrichment results for the enhancer SNPs in brain tissues. c TF enrichment results for the enhancer SNPs in blood tissues. d Differential expression analyses results (0 day vs 11 day) of the promoter and enhancer SNPs enriched TFs in human SH-SY5Y cells. *adjust P < 0.05, **adjust P < 0.01, ***adjust P < 0.001. e The results of histone modification enrichment for all functional SNPs in brain cell lines. f The results of histone modification enrichment for all functional SNPs in blood cell lines. The shallow of red represents the value of −log10 (adjust P)
Finally, we identified 934 functional SNPs in brain and 1300 (rs134885, rs1800629, and rs2747054 were identified in both enhancer and promoter regions and were only counted once) in blood, including 1 splicing SNP, 66 missense SNPs which may damage protein functions, 119 UTR SNPs involved in potential microRNA targets, 89 promoter, and 659/1028 enhancer SNPs which might disrupt TF binding in brain/blood.
Histone modification enrichment
Histone modification enrichment showed that these functional SNPs were enriched in H3K27ac (active enhancer), H3K36me3 (active elongation), H3K4me1 (active enhancer), and H3K4me3 (active promoter) in most of the brain (Fig. 3e) and blood cell lines (Fig. 3f). All of these histone marks would activate gene expression.
QTL analyses
According to meQTL results, 516 of 934 annotated functional SNPs in brain were associated with DNA methylation alterations of 224 genes (Supplementary Table S6). For all annotated functional SNPs, we performed SMR and eQTL analyses to find their target genes. One thousand six hundred and one genes in brain tissues and 552 genes in blood tissue were identified in SMR analyses. One hundred and seventy-five genes in CMC and 568 genes in data from Bonder et al. were identified in eQTL analyses (Supplementary Table S6). As a result, 299 genes in blood and 147 genes in brain were overlapped in SMR and eQTL analyses, respectively, 380 genes in total. We also carried out pQTL analyses in blood tissue. Due to the low genotyping density of the current available proteomic data, only 41 functional SNPs were identified and were associated with 19 proteins (supplementary Table S6). Besides, C2, C4A, C4B, MICA, and MICB were identified as the targets of the functional SNPs from both gene and protein expression levels.
Validation of target genes
Finally, we acquired 538 target genes (Fig. 4a, including 34 enriched TFs, 224 meQTL, 380 eQTL, and 19 pQTL target genes), including 447 protein-coding genes. For these protein-coding genes, 168 genes (37.58%) were differentially expressed between schizophrenia cases and controls (Supplementary Table S7). We also confirmed our results in mouse models and 103 (23.04%) genes were related to nervous system phenotype (Supplementary Table S8). Using the schizophrenia-related pathways data from MalaCards, we found 33 protein-coding genes could be assigned to several signal pathways of schizophrenia (Fig. 4b). Thirteen genes (C4A, C4B, CACNA1C, CYP2D6, DRD2, GABBR1, GRIN2A, ITIH3, KCTD13, MICB, NOTCH4, SRR, and TNF) had been reported related to schizophrenia. Among them, CACNA1C, CYP2D6, DRD2, GABBR1, GRIN2A, NOTCH4, and SRR were also assigned to related pathways. However, there were still some target genes could not be explained. Using gene–gene interaction data, we found that 221 of the 408 unexplained genes (54.17%) could be related to known schizophrenia pathways through gene–gene interaction (Supplementary Figure S1A) and 65 genes could be related to known schizophrenia-related genes (including 18 genes related to 6 schizophrenia elite genes, Supplementary Figure S1B). Subsequently, we conducted KEGG pathway enrichment analysis for 59 target genes which had at least two hits in above methods (Fig. 4c), and they were enriched in 13 related signal pathways (Fig. 4d). In addition, there were 12 approved drugs targeted to functional genes or schizophrenia-related genes which interacted with functional genes (Fig. 4e).
a The Venn diagram of the target genes from QTLs analyses. b The target protein-coding genes (ordinates) assigned to known schizophrenia-related pathways (abscissa). c The Venn diagram of the validation of target genes. d The enriched schizophrenia-related pathways of 59 target genes which had at least two hits in validations. e Connections between target genes (dark blue), genes from PPI (blue), and drugs (green) indicated for other diseases (purple)
Notably, we prioritized 10 target genes, CACNA1C, CLU, CSNK2B, GABBR1, GRIN2A, MAPK3, NOTCH4, SRR, TNF, and SYNGAP1, which were differentially expressed between schizophrenia cases and controls, validated in mouse models and MalaCards directly (Fig. 4c, Table 1, Supplementary Table S9). The correlation with schizophrenia of six among them were already identified. CLU and MAPK3 were only identified as new schizophrenia susceptibility loci by GWASs13, but the other two genes (CSNK2B and SYNGAP1) were not identified yet.
Discussion
In this study, we obtained SNPs associated with schizophrenia from the current results of GWASs and conducted functional annotation comprehensively for all susceptibility loci. Different from previous studies, not only the SNPs in exon, UTR, and promoter regions, we also focused on the annotation in intronic and intergenic regions using epigenomic data.
We found 538 target genes, including 447 protein-coding genes. One hundred and sixty-eight (37.58%) of them were differentially expressed between schizophrenia cases and controls, and 103 of them were confirmed related to nervous system phenotype in mouse models, both indicating their important involvement in the development of schizophrenia. Only 33 protein-coding genes could be directly assigned to schizophrenia-related pathways, and 13 were confirmed as schizophrenia-related genes. Furthermore, 54.67% of the rest target protein-coding genes could be related to known schizophrenia-related genes or pathways through gene–gene interaction, which confirmed gene regulatory networks are sufficient to interconnect these genes with disease-related genes36.
C2, C4A, C4B, MICA, and MICB were identified as the targets of the functional SNPs from both gene and protein expression levels. MICB was found differentially expressed between schizophrenia cases and controls. C4A37, C4B37, and MICB38,39 were previously reported to be associated with schizophrenia. Hakobyan et al.40 identified that the hemolytic activity of the C2 complement components was significantly lower in the serum of the schizophrenic patients.
We prioritized 10 target genes for schizophrenia. They were all differentially expressed between schizophrenia cases and controls, validated in mouse models and MalaCards directly. The correlation with schizophrenia of six among them were already identified. CACNA1C41,42, GRIN2A43 (interacted with elite gene AKT1), TNF44, and aberrant SRR45 may contribute to schizophrenia pathogenesis. GABBR146 and NOTCH447 may confer susceptibility to the development of schizophrenia. Interaction between COMT (elite gene) and NOTCH4 genotypes may predict the treatment response to typical neuroleptics in schizophrenia patients48. CLU and MAPK3 were indentified as new loci in GWAS study13.
Besides, the other two genes (SYNGAP1 and CSNK2B) were most likely to be schizophrenia susceptibility genes. SYNGAP1 was assigned to Neuroscience pathway directly, and CSNK2B in beta-adrenergic signaling pathway. However, SYNGAP1 and CSNK2B are located in the extended MHC complex on chromosome 6, so it is doubtful those are truly novel associations with schizophrenia. We performed Pearson correlation analyses between CSNK2B (or SYNGAP1) and C4A, C4B37 or the other gene SYNGAP1 (or CSNK2B), and did conditional eQTL analyses between each eQTL top SNPs and corresponding target genes (CSNK2B and SYNGAP1) by adjusting the residual effect of the C4A and C4B using genotype and expression data (after covariates) of 13 brain subregions and whole blood from the GTEx-v7 project. (1) Pearson correlation analyses results showed that there was no significant correlations between CSNK2B and C4A or C4B in 13/14 or 11/14 tissues, respectively, and there was no significant correlations between SYNGAP1 and C4A or C4B in 11/14 or all 14 tissues, respectively. Moreover, we explored the correlations between CSNK2B and SYNGAP1 and found that there was no significant correlations for CSNK2B and SYNGAP1 in 12/14 tissues (supplementary Table S10). (2) In the conditional eQTL analyses, we used the top SNPs obtained in the SMR analyses. As a result, 10 SNPs were used for CSNK2B, and 3 SNPs for SYNGAP1. We checked if the eQTL association remained significant after adjusting the residual effect of C4A and C4B (Supplementary Table S10). For CSNK2B, conditional eQTL association remained significant (P < 0.05) in 21 SNP-tissues pairs (8 SNPs and 10 tissues, respectively, and there were 30 pairs before adjusting the C4 signal). SYNGAP1 remained significant in 2/3 SNP-tissues pairs. Thus, the observed eQTL associations between corresponding SNPs and CSNK2B, SYNGAP1 were independent of C4 signal. These analyses results suggested us that CSNK2B and SYNGAP1 were independent from the C4 signal and each other.
There were 12 approved drugs from biotech targeted to functional genes, while Pegademase bovine is used for the treatment of adenosine deaminase, which is involved in the pathophysiology of schizophrenia49. Botulinum Toxin Type B is used for dystonia, which is one of the symptoms of schizophrenia. The other 10 drugs were all non-psychiatric medications. We searched for the potential clinical relevance of those non-psychiatric medications in context of schizophrenia. (1) There are four drugs (Tenecteplase, Antihemophilic factor, human recombinant, and Oxytocin) approved for use in blood disorders. Patients with schizophrenia have excess cardiovascular morbidity and mortality50. There was high prevalence of psychopathology in children with blood disorders51. (2) Four drugs (Ibalizumab, Pegaptanib, Palifermin, and SGN-30) are applied to autoimmune diseases or inflammation. It was found that autoimmune diseases were associated with an increased risk of developing schizophrenia52. An important role of peripheral immune-to-brain communication pathways was suggested in schizophrenia, and there was an association between elevated levels of circulating inflammatory cytokines and subsequent risk of psychosis53. (3) Three3 medications (SGN-30, LErafAON, and Capromab pendetide) are used for various forms of cancers. Patients with schizophrenia have been found with a reduced risk of cancer54. It has been found that patients with schizophrenia have a hyper-opaminergic system and dopamine has the ability to inhibit tumor angiogenesis55. Consequently, these non-psychiatric medications may have chance to be applied to schizophrenia, as well to a better understanding of the schizophrenia pathogenesis.
There are still some limitations of this study. Because of the limitation of current available pQTL data, only blood tissue was used in this study. Thus, we may miss many pQTL target genes. Therefore, more pQTL target genes may be obtained if more large-scale data from schizophrenia relevant tissues are available. In addition, we mainly focused on protein-coding genes. Other target genes which encode non-coding RNAs may also play roles in the pathogenesis of schizophrenia. Besides, we did meta-analyses across distinct tissue types firstly because blood was used as a surrogate for brain tissues in schizophrenia studies56,57,58. We aimed to find the genes expressed consistently in these schizophrenia-related tissues. Secondly, meta-analysis applied to different tissues to identify non-tissue specific genes was commonly used13,59. This did have some limitations since different tissues have different gene expression patterns, which may introduce heterogeneity.
In conclusion, we acquired SNPs associated with schizophrenia and provided a comprehensive annotation for all susceptibility loci. We identified 934 functional SNPs in brain and 1300 in blood, including 66 missense SNPs, 1 splicing SNP, 119 UTR SNPs, 89 promoter, and 659/1028 enhancer SNPs. These promoter and enhancer SNPs were enriched for disruption of 34 TFs. H3K27ac, H3K36me3, H3K4me1, and H3K4me3 were more likely to be related to the development of schizophrenia. meQTL analyses for the functional SNPs showed that 516 functional SNPs in brain would affect DNA methylation levels of 224 genes. eQTL analyses for the functional SNPs identified 380 target genes. pQTL analyses showed that 19 proteins might be affected by functional SNPs. Finally, we acquired 447 target protein-coding genes. One hundred and sixty-eight of these genes (37.58%) were differentially expressed between schizophrenia cases and controls, and 103 (23.04%) were related to nervous system phenotype in mouse models. Besides, 260 (58.17%) could be correlated with schizophrenia-related pathways or related genes directly or through gene–gene interaction. We prioritized 10 target genes for schizophrenia, CLU and MAPK3 were identified by GWASs, but the other two genes (CSNK2B and SYNGAP1) were novel. Our results may offer holistic guides for post-GWAS functional experiments.
References
Saha, S., Chant, D. & McGrath, J. A systematic review of mortality in schizophrenia—is the differential mortality gap worsening over time? Arch. Gen. Psychiatry 64, 1123–1131 (2007).
Sullivan, P. F., Kendler, K. S. & Neale, M. C. Schizophrenia as a complex trait—evidence from a meta-analysis of twin studies. Arch. Gen. Psychiatry 60, 1187–1192 (2003).
Brennand, K. J. et al. Modelling schizophrenia using human induced pluripotent stem cells. Nature 473, 221–225 (2011).
Maurano, M. T. et al. Systematic localization of common disease-associated variation in regulatory DNA. Science 337, 1190–1195 (2012).
Dunham, I. et al. An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 (2012).
Kundaje, A. et al. Integrative analysis of 111 reference human epigenomes. Nature 518, 317–330 (2015).
Wang, G. J., Yang, P. & Xie, H. G. Gene variants in noncoding regions and their possible consequences. Pharmacogenomics 7, 203–209 (2006).
Hazelett, D. J. et al. Comprehensive functional annotation of 77 prostate cancer risk loci. PLoS Genet. 10, e1004102 (2014).
Yao, L. J., Tak, Y. G., Berman, B. P. & Farnham, P. J. Functional annotation of colon cancer risk SNPs. Nat. Commun. 5, 5114 (2014).
Coetzee, S. G. et al. Enrichment of risk SNPs in regulatory regions implicate diverse tissues in Parkinson’s disease etiology. Sci. Rep. 6, 30509 (2016).
Welter, D. et al. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 42, D1001–D1006 (2014).
McClellan, J. & King, M. C. Genetic heterogeneity in human disease. Cell 141, 210–217 (2010).
Ripke, S. et al. Biological insights from 108 schizophrenia-associated genetic loci. Nature 511, 421–427 (2014).
Pardinas, A. F. et al. Common schizophrenia alleles are enriched in mutation-intolerant genes and in regions under strong background selection. Nat. Genet. 50, 381–389 (2018).
Wang, K., Li, M. Y. & Hakonarson, H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 38, e164–e164 (2010).
Choi, Y. & Chan, A. P. PROVEAN web server: a tool to predict the functional effect of amino acid substitutions and indels. Bioinformatics 31, 2745–2747 (2015).
Kumar, P., Henikoff, S. & Ng, P. C. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat. Protoc. 4, 1073–1082 (2009).
Adzhubei, I. A. et al. A method and server for predicting damaging missense mutations. Nat. Methods 7, 248–249 (2010).
Kircher, M. et al. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 46, 310–315 (2014).
Liu, C. X. et al. MirSNP, a database of polymorphisms altering miRNA target sites, identifies miRNA-related SNPs in GWAS SNPs and eQTLs. BMC Genomics 13, 661 (2012).
Kumar, S., Ambrosini, G. & Bucher, P. SNP2TFBS—a database of regulatory SNPs affecting predicted transcription factor binding site affinity. Nucleic Acids Res. 45, D139–D144 (2017).
Chiocchetti, A. G. et al. Transcriptomic signatures of neuronal differentiation and their association with risk genes for autism spectrum and related neuropsychiatric disorders. Transl. Psychiatry 6, e864 (2016).
Cowper-Sal-lari, R. et al. Breast cancer risk-associated SNPs modulate the affinity of chromatin for FOXA1 and alter gene expression. Nat. Genet. 44, 1191–1198 (2012).
Jaffe, A. E. et al. Mapping DNA methylation across development, genotype and schizophrenia in the human frontal cortex. Nat. Neurosci. 19, 40–47 (2016).
Fromer, M. et al. Gene expression elucidates functional impact of polygenic risk for schizophrenia. Nat. Neurosci. 19, 1442–1453 (2016).
Bonder, M. J. et al. Disease variants alter transcription factor levels and methylation of their binding sites. Nat. Genet. 49, 131–138 (2017).
Zhu, Z. H. et al. Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets. Nat. Genet. 48, 481–487 (2016).
Lonsdale, J. et al. The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 45, 580–585 (2013).
Suhre, K. et al. Connecting genetic risk to disease end points through the human blood plasma proteome. Nat. Commun. 8, 14357 (2017).
Willer, C. J., Li, Y. & Abecasis, G. R. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 26, 2190–2191 (2010).
Lanz, T. A. et al. STEP levels are unchanged in pre-frontal cortex and associative striatum in post-mortem human brain samples from subjects with schizophrenia, bipolar disorder and major depressive disorder. PLoS ONE 10, e0121744 (2015).
Arion, D. et al. Distinctive transcriptome alterations of prefrontal pyramidal neurons in schizophrenia and schizoaffective disorder. Mol. Psychiatry 20, 1397–1405 (2015).
de Jong, S. et al. A gene co-expression network in whole blood of schizophrenia patients is independent of antipsychotic-use and enriched for brain-expressed genes. PLoS ONE 7, e39498 (2012).
Blake, J. A. et al. Mouse Genome Database (MGD)-2017: community knowledge resource for the laboratory mouse. Nucleic Acids Res. 45, D723–D729 (2017).
Dickinson, M. E. et al. High-throughput discovery of novel developmental phenotypes. Nature 537, 508–514 (2016).
Boyle, E. A., Li, Y. I. & Pritchard, J. K. An expanded view of complex traits: from polygenic to omnigenic. Cell 169, 1177–1186 (2017).
Sekar, A. et al. Schizophrenia risk from complex variation of complement component 4. Nature 530, 177–183 (2016).
Carter, C. J. Schizophrenia susceptibility genes directly implicated in the life cycles of pathogens: cytomegalovirus, influenza, herpes simplex, rubella, and Toxoplasma gondii. Schizophr. Bull. 35, 1163–1182 (2009).
Shirts, B. H. et al. Polymorphisms in MICB are associated with human herpes virus seropositivity and schizophrenia risk. Schizophr. Res. 94, 342–353 (2007).
Hakobyan, S., Boyajyan, A. & Sim, R. B. Classical pathway complement activity in schizophrenia. Neurosci. Lett. 374, 35–37 (2005).
Zhang, S. Y. et al. Role of CACNA1C gene polymorphisms and protein expressions in the pathogenesis of schizophrenia: a case-control study in a Chinese population. Neurol. Sci. 38, 1393–1403 (2017).
Ripke, S. et al. Genome-wide association study identifies five new schizophrenia loci. Nat. Genet. 43, 969–976 (2011).
Javitt, D. C. & Zukin, S. R. Recent advances in the phencyclidine model of schizophrenia. Am. J. Psychiatry 148, 1301–1308 (1991).
Hoseth, E. Z. et al. A study of TNF pathway activation in schizophrenia and bipolar disorder in plasma and brain tissue. Schizophr. Bull. 43, 881–890 (2017).
Labrie, V. et al. Serine racemase is associated with schizophrenia susceptibility in humans and in a mouse model. Human. Mol. Genet. 18, 3227–3243 (2009).
Zhao, X. et al. Systematic study of association of four GABAergic genes: glutamic acid decarboxylase 1 gene, glutamic acid decarboxylase 2 gene, GABA(B) receptor 1 gene and GABA(A) receptor subunit beta 2 gene, with schizophrenia using a universal DNA microarray. Schizophr. Res. 93, 374–384 (2007).
Luo, X. G. et al. NOTCH4 gene haplotype is associated with schizophrenia in African Americans. Biol. Psychiatry 55, 112–117 (2004).
Anttila, S. et al. Interaction between NOTCH4 and catechol-O-methyltransferase genotypes in schizophrenia patients with poor response to typical neuroleptics. Pharmacogenetics 14, 303–307 (2004).
Seibt, K. J., Oliveira, R. D., Bogo, M. R., Senger, M. R. & Bonan, C. D. Investigation into effects of antipsychotics on ectonucleotidase and adenosine deaminase in zebrafish brain. Fish Physiol. Biochem. 41, 1383–1392 (2015).
Kugathasan, P. et al. Association of secondary preventive cardiovascular treatment after myocardial infarction with mortality among patients with schizophrenia. JAMA Psychiatry 75, 1234–1240 (2018).
Sadowski, H. et al. Psychopathology in children from families with blood disorders: a cross-national study. Eur. Child Adolesc. Psychiatry 11, 151–161 (2002).
Wang, L.-Y., Chen, S.-F., Chiang, J.-H., Hsu, C.-Y. & Shen, Y.-C. Autoimmune diseases are associated with an increased risk of schizophrenia: a nationwide population-based cohort study. Schizophr. Res. 202, 297–302 (2018).
Khandaker, G. M. & Dantzer, R. Is there a role for immune-to-brain communication in schizophrenia? Psychopharmacology 233, 1559–1573 (2016).
Barak, Y., Achiron, A., Mandel, M., Mirecki, I. & Aizenberg, D. Reduced cancer incidence among patients with schizophrenia. Cancer 104, 2817–2821 (2005).
Asada, M. et al. Reduced tumor growth in a mouse model of schizophrenia, lacking the dopamine transporter. Int. J. Cancer 123, 511–518 (2008).
Glatt, S. J. et al. Comparative gene expression analysis of blood and brain provides concurrent validation of SELENBP1 up-regulation in schizophrenia. Proc. Natl. Acad. Sci. USA 102, 15533–15538 (2005).
Sullivan, P. F., Fan, C. & Perou, C. M. Evaluating the comparability of gene expression in blood and brain. Am. J. Med. Genet. B Neuropsychiatr. Genet. 141B, 261–268 (2006).
Tsuang, M. T. et al. Assessing the validity of blood-based gene expression profiles for the classification of schizophrenia and bipolar disorder: a preliminary report. Am. J. Med. Genet. B Neuropsychiatr. Genet. 133B, 1–5 (2005).
Mei, H. et al. Tissue non-specific genes and pathways associated with diabetes: an expression meta-analysis. Genes 8, E44 (2017).
Acknowledgements
Data were generated as part of the CommonMind Consortium supported by funding from Takeda Pharmaceuticals Company Limited, F. Hoffman-La Roche Ltd and NIH grants R01MH085542, R01MH093725, P50MH066392, P50MH080405, R01MH097276, RO1-MH-075916, P50M096891, P50MH084053S1, R37MH057881, R37MH057881S1, HHSN271201300031C, AG02219, AG05138, and MH06692. Brain tissue for the study was obtained from the following brain bank collections: the Mount Sinai NIH Brain and Tissue Repository, the University of Pennsylvania Alzheimer’s Disease Core Center, the University of Pittsburgh NeuroBioBank and Brain and Tissue Repositories, and the NIMH Human Brain Collection Core. This study is supported by National Natural Science Foundation of China (81573241, 31701095, and 31771399); China Postdoctoral Science Foundation (2016M602797, 2018T111037); Innovative Talent Promotion Plan of Shaanxi Province for Young Sci-Tech New Star (2018KJXX-010); Zhejiang Provincial Natural Science Foundation of China (LGF18C060002); the Fundamental Research Funds for the Central Universities; and China Scholarship Council.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Niu, HM., Yang, P., Chen, HH. et al. Comprehensive functional annotation of susceptibility SNPs prioritized 10 genes for schizophrenia. Transl Psychiatry 9, 56 (2019). https://doi.org/10.1038/s41398-019-0398-5
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41398-019-0398-5
This article is cited by
-
G protein-coupled receptors in neurodegenerative diseases and psychiatric disorders
Signal Transduction and Targeted Therapy (2023)
-
Functional connectivity signatures of NMDAR dysfunction in schizophrenia—integrating findings from imaging genetics and pharmaco-fMRI
Translational Psychiatry (2023)
-
Integrative omics of schizophrenia: from genetic determinants to clinical classification and risk prediction
Molecular Psychiatry (2022)
-
A preliminary genetic association study of GAD1 and GABAB receptor genes in patients with treatment-resistant schizophrenia
Molecular Biology Reports (2022)