Abstract
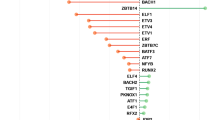
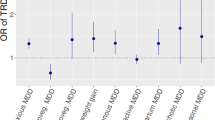
In clinical practice, an antidepressant prescription is a trial and error approach, which is time consuming and discomforting for patients. This study investigated an in silico approach for ranking antidepressants based on their hypothetical likelihood of efficacy. We predicted the transcriptomic profile of citalopram remitters by performing an in silico transcriptomic-wide association study on STAR*D GWAS data (N = 1163). The transcriptional profile of remitters was compared with 21 antidepressant-induced gene expression profiles in five human cell lines available in the connectivity-map database. Spearman correlation, Pearson correlation, and the Kolmogorov–Smirnov test were used to determine the similarity between antidepressant-induced profiles and remitter profiles, subsequently calculating the average rank of antidepressants across the three methods and a p value for each rank by using a permutation procedure. The drugs with the top ranks were those having a high positive correlation with the expression profiles of remitters and that may have higher chances of efficacy in the tested patients. In MCF7 (breast cancer cell line), escitalopram had the highest average rank, with an average rank higher than expected by chance (p = 0.0014). In A375 (human melanoma) and PC3 (prostate cancer) cell lines, escitalopram and citalopram emerged as the second-highest ranked antidepressants, respectively (p = 0.0310 and 0.0276, respectively). In HA1E (kidney) and HT29 (colon cancer) cell types, citalopram and escitalopram did not fall among top antidepressants. The correlation between citalopram remitters’ and (es)citalopram-induced expression profiles in three cell lines suggests that our approach may be useful and with future improvements, it can be applicable at the individual level to tailor treatment prescription.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 6 print issues and online access
$259.00 per year
only $43.17 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout

Similar content being viewed by others
References
James SL, Abate D, Abate KH, Abay SM, Abbafati C, Abbasi N, et al. Global, regional, and national incidence, prevalence, and years lived with disability for 354 diseases and injuries for 195 countries and territories, 1990-2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet. 2018;392:1789–858.
Fabbri C, Tansey KE, Perlis RH, Hauser J, Henigsberg N, Maier W, et al. New insights into the pharmacogenomics of antidepressant response from the GENDEP and STAR∗D studies: rare variant analysis and high-density imputation. Pharmacogenomics J. 2018;18:413–21.
Leuchter AF, Cook IA, Hamilton SP, Narr KL, Toga A, Hunter AM, et al. Biomarkers to predict antidepressant response. Curr Psychiatry Rep. 2010;12:553–62.
Tansey KE, Guipponi M, Hu X, Domenici E, Lewis G, Malafosse A, et al. Contribution of common genetic variants to antidepressant response. Biol Psychiatry. 2013;73:679–82.
Gandal MJ, Leppa V, Won H, Parikshak NN, Geschwind DH. The road to precision psychiatry: translating genetics into disease mechanisms. Nat Neurosci. 2016;19:1397–407.
Uher R, Tansey KE, Rietschel M, Henigsberg N, Maier W, Mors O, et al. Common genetic variation and antidepressant efficacy in major depressive disorder: a meta-analysis of three genome-wide pharmacogenetic studies. Am J Psychiatry. 2013;170:207–17.
Wigmore EM, Hafferty JD, Hall LS, Howard DM, Clarke TK, Fabbri C, et al. Genome-wide association study of antidepressant treatment resistance in a population-based cohort using health service prescription data and meta-analysis with GENDEP. Pharmacogenomics J. 2020;20:329–41.
Lamb J, Crawford ED, Peck D, Modell JW, Blat IC, Wrobel MJ, et al. The connectivity map: using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313:1929–35.
Tsuchimine S, Ochi S, Tajiri M, Suzuki Y, Sugawara N, Inoue Y, et al. Effects of cytochrome P450 (CYP) 2C19 genotypes on steady-state plasma concentrations of escitalopram and its desmethyl metabolite in Japanese patients with depression. Ther Drug Monit. 2018;40:356–61.
Fava M, Rush AJ, Trivedi MH, Nierenberg AA, Thase ME, Sackeim HA, et al. Background and rationale for the sequenced treatment alternatives to relieve depression (STAR*D) study. Psychiatr Clin N Am. 2003;26:457–94.
Garriock HA, Kraft JB, Shyn SI, Peters EJ, Yokoyama JS, Jenkins GD, et al. A genomewide association study of citalopram response in major depressive disorder. Biol Psychiatry. 2010;67:133–8.
Novick D, Hong J, Montgomery W, Dueñas H, Gado M, Haro JM. Predictors of remission in the treatment of major depressive disorder: Real-world evidence from a 6-month prospective observational study. Neuropsychiatr Dis Treat. 2015;11:197–205.
Rush AJ, Trivedi MH, Ibrahim HM, Carmody TJ, Arnow B, Klein DN, et al. The 16-item quick inventory of depressive symptomatology (QIDS), clinician rating (QIDS-C), and self-report (QIDS-SR): a psychometric evaluation in patients with chronic major depression. Biol Psychiatry. 2003;54:573–83.
Gaynes BN, Warden D, Trivedi MH, Wisniewski SR, Fava M, Rush AJ. What did STAR* D teach us? Results from a large-scale, practical, clinical trial for patients with depression. Psychiatric services. 2009;60:1439–45.
Lam M, Awasthi S, Watson HJ, Goldstein J, Panagiotaropoulou G, Trubetskoy V, et al. RICOPILI: Rapid Imputation for COnsortias PIpeLIne. Bioinformatics. 2020;36:930–3.
Gusev A, Ko A, Shi H, Bhatia G, Chung W, Penninx BWJH, et al. Integrative approaches for large-scale transcriptome-wide association studies. Nat Genet. 2016;48:245–52.
Gusev A, Mancuso N, Won H, Kousi M, Finucane HK, Reshef Y, et al. Transcriptome-wide association study of schizophrenia and chromatin activity yields mechanistic disease insights. Nat Genet. 2018;50:538–48.
Mancuso N, Gayther S, Gusev A, Zheng W, Penney KL, Kote-Jarai Z, et al. Large-scale transcriptome-wide association study identifies new prostate cancer risk regions. Nat Commun. 2018;9:1–11.
Stegle O, Parts L, Durbin R, Winn J. A bayesian framework to account for complex non-genetic factors in gene expression levels greatly increases power in eQTL studies. PLoS Comput Biol. 2010;6:1–11.
The GTEx Consortium. Genetic effects on gene expression across human tissues. Nature. 2017;7675:204–13.
Pain O, Pocklington AJ, Holmans PA, Bray NJ, O’Brien HE, Hall LS, et al. Novel insight into the etiology of autism spectrum disorder gained by integrating expression data with genome-wide association statistics. Biol Psychiatry. 2019;86:265–73.
Subramanian A, Narayan R, Corsello SM, Peck DD, Natoli TE, Lu X, et al. A next generation connectivity map: l1000 platform and the first 1,000,000 profiles. Cell. 2017;171:1437–52.e17.
So HC, Chau CKL, Chiu WT, Ho KS, Lo CP, Yim SHY, et al. Analysis of genome-wide association data highlights candidates for drug repositioning in psychiatry. Nat Neurosci. 2017;20:1342–9.
Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–50.
Wise LH, Lanchbury JS, Lewis CM. Meta-analysis of genome searches. Ann Hum Genet. 1999;63:263–72.
Hicks JK, Sangkuhl K, Swen JJ, Ellingrod VL, Müller DJ, Shimoda K, et al. Clinical pharmacogenetics implementation consortium guideline (CPIC) for CYP2D6 and CYP2C19 genotypes and dosing of tricyclic antidepressants: 2016 update. Clin Pharmacol Ther. 2017;102:37–44.
Fabbri C, Tansey KE, Perlis RH, Hauser J, Henigsberg N, Maier W, et al. Effect of cytochrome CYP2C19 metabolizing activity on antidepressant response and side effects: Meta-analysis of data from genome-wide association studies. Eur Neuropsychopharmacol. 2018;28:945–54.
Sirota M, Dudley JT, Kim J, Chiang AP, Morgan AA, Sweet-Cordero A, et al. Discovery and preclinical validation of drug indications using compendia of public gene expression data. Sci Transl Med. 2011;3:96ra77.
Dudley JT, Sirota M, Shenoy M, Pai RK, Roedder S, Chiang AP, et al. Computational repositioning of the anticonvulsant topiramate for inflammatory bowel disease. Sci Transl Med. 2011;3:96ra76.
Jiménez-Rojo L, Granchi Z, Graf D, Mitsiadis TA. Stem cell fate determination during development and regeneration of ectodermal organs. Front Physiol. 2012;3:1–11.
Sakka L, Delétage N, Chalus M, Aissouni Y, Sylvain-Vidal V, Gobron S, et al. Assessment of citalopram and escitalopram on neuroblastoma cell lines. Cell toxicity and gene modulation. Oncotarget. 2017;8:42789–807.
Jacobsen JPR, Plenge P, Sachs BD, Pehrson AL, Cajina M, Du Y, et al. The interaction of escitalopram and R-citalopram at the human serotonin transporter investigated in the mouse. Psychopharmacology. 2014;231:4527–40.
Musa A, Tripathi S, Kandhavelu M, Dehmer M, Emmert-streib F. Harnessing the biological complexity of Big Data from LINCS gene expression signatures. PLoS ONE. 2018;13:1–16.
Acknowledgements
We thank the NIMH for providing the opportunity of analyzing their data on the STAR*D sample. We would like to thank the CMap team for making their data available for community research use. This paper represents independent research part-funded by the National Institute for Health Research (NIHR) Biomedical Research Center at Oxford, South London, Maudsley NHS Foundation Trust, and King’s College London. The views expressed are those of the authors and not necessarily those of the NHS, the NIHR, or the Department of Health and Social Care. We acknowledge the use of research computing facility at King’s College London, Rosalind (https://rosalind.kcl.ac.uk), which is delivered in partnership with the National Institute for Health Research (NIHR) Biomedical Research Center at South London & Maudsley and Guy’s & St. Thomas’ NHS Foundation Trusts, and part-funded by capital equipment grants from the Maudsley Charity (award 980) and Guy’s & St. Thomas’ Charity (TR130505).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
CML is a member of the Scientific Advisory Board of Myriad Neurosciences. The other authors declare no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
41397_2020_186_MOESM1_ESM.doc
Supplementary Material for ‘Investigating an in silico approach for prioritizing antidepressant drug prescription based on drug-induced expression profiles and predicted gene expression’
Rights and permissions
About this article
Cite this article
Shoaib, M., Giacopuzzi, E., Pain, O. et al. Investigating an in silico approach for prioritizing antidepressant drug prescription based on drug-induced expression profiles and predicted gene expression. Pharmacogenomics J 21, 85–93 (2021). https://doi.org/10.1038/s41397-020-00186-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41397-020-00186-5