Abstract
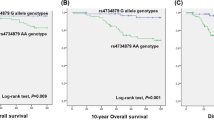
The identification of predictive biomarkers for the first-line treatment of epithelial ovarian cancer (EOC) remains a challenge. Although genome-wide association studies (GWAS) have identified several genetic polymorphisms as predictors of EOC clinical outcome, the subsequent validation has not yet been performed. This study aims to validate the influence of Neuregulin 3 (NRG3) rs1649942 and Brain and reproductive organ-expressed (TNFRSF1A modulator) (BRE) rs7572644 GWAS-identified variants in an independent cohort of EOC patients from the North region of Portugal (n = 339) submitted to first-line treatment. Polymorphism genotypes were determined by real-time PCR using validated assays. Patients carrying the NRG3 rs1649942 A allele presented a significantly longer overall survival (OS) when compared to GG-genotype patients (log-rank test, P = 0.011) in the FIGO IV stage subgroup. No impact was observed for early-stage patients or considering disease-free survival (DFS) as an outcome. For FIGO I/II stage patients, BRE rs7572644 C allele carriers exhibit a decreased OS (P = 0.014) and DFS (P = 0.032) when compared to TT-homozygous patients. Furthermore, a Multivariate Cox regression analysis revealed a three-fold increase in the risk of death (HR, 3.09; P = 0.015) and recurrence (HR, 3.33; P = 0.009) for FIGO I/II C allele carriers. No significant impact was observed for late-stage patients. The BRE rs7572644 and NRG3 rs1649942 genetic variants were validated in an independent cohort of EOC Portuguese patients, particularly in specific subgroups considering FIGO staging. Further functional post-GWAS analyses are indispensable to understand the biological mechanisms underlying the observed results.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 6 print issues and online access
$259.00 per year
only $43.17 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer . 2015;136:E359–386.
Pereira D, Assis J, Gomes M, Nogueira A, Medeiros R. Improvement of a predictive model in ovarian cancer patients submitted to platinum-based chemotherapy: implications of a GST activity profile. Eur J Clin Pharmacol. 2016;72:545–53.
Diaz-Padilla I, Amir E, Marsh S, Liu G, Mackay H. Genetic polymorphisms as predictive and prognostic biomarkers in gynecological cancers: a systematic review. Gynecol Oncol. 2012;124:354–65.
Caiola E, Broggini M, Marabese M. Genetic markers for prediction of treatment outcomes in ovarian cancer. Pharm J. 2014;14:401–10.
Assis J, Pereira D, Gomes M, Marques D, Marques I, Nogueira A, et al. Influence of CYP3A4 genotypes in the outcome of serous ovarian cancer patients treated with first-line chemotherapy: implication of a CYP3A4 activity profile. Int J Clin Exp Med. 2013;6:552–61.
Huang RS, Duan S, Shukla SJ, Kistner EO, Clark TA, Chen TX, et al. Identification of genetic variants contributing to cisplatin-induced cytotoxicity by use of a genomewide approach. Am J Hum Genet. 2007;81:427–37.
Pinto R, Assis J, Nogueira A, Pereira C, Pereira D, Medeiros R. Rethinking ovarian cancer genomics: where genome-wide association studies stand? Pharmacogenomics. 2017;18:1611–25.
Prat J, Oncology FCoG. FIGO’s staging classification for cancer of the ovary, fallopian tube, and peritoneum: abridged republication. J Gynecol Oncol. 2015;26:87–89.
Rustin GJ, Vergote I, Eisenhauer E, Pujade-Lauraine E, Quinn M, Thigpen T, et al. Definitions for response and progression in ovarian cancer clinical trials incorporating RECIST 1.1 and CA 125 agreed by the Gynecological Cancer Intergroup (GCIG). Int J Gynecol Cancer. 2011;21:419–23.
MacArthur J, Bowler E, Cerezo M, Gil L, Hall P, Hastings E, et al. The new NHGRI-EBI Catalog of published genome-wide association studies (GWAS Catalog). Nucleic Acids Res. 2017;45(D1):D896–901.
Saccone SF, Bolze R, Thomas P, Quan J, Mehta G, Deelman E, et al. SPOT: a web-based tool for using biological databases to prioritize SNPs after a genome-wide association study. Nucleic Acids Res. 2010;38(Web Server issue):W201–9.
Desmet FO, Hamroun D, Lalande M, Collod-Beroud G, Claustres M, Beroud C. Human Splicing Finder: an online bioinformatics tool to predict splicing signals. Nucleic Acids Res. 2009;37:e67.
Coetzee SG, Coetzee GA, Hazelett DJ. motifbreakR: an R/Bioconductor package for predicting variant effects at transcription factor binding sites. Bioinformatics. 2015;31:3847–9.
Eisenhauer EA, Therasse P, Bogaerts J, Schwartz LH, Sargent D, Ford R, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur J Cancer. 2009;45:228–47.
Manolio TA. Genomewide association studies and assessment of the risk of disease. N Engl J Med. 2010;363:166–76.
Patel JN, McLeod HL, Innocenti F. Implications of genome-wide association studies in cancer therapeutics. Br J Clin Pharmacol. 2013;76:370–80.
Manolio TA. Bringing genome-wide association findings into clinical use. Nat Rev Genet. 2013;14:549–58.
Chen K, Ma H, Li L, Zang R, Wang C, Song F, et al. Genome-wide association study identifies new susceptibility loci for epithelial ovarian cancer in Han Chinese women. Nat Commun. 2014;5:4682.
Huang RS, Johnatty SE, Gamazon ER, Im HK, Ziliak D, Duan S, et al. Platinum sensitivity-related germline polymorphism discovered via a cell-based approach and analysis of its association with outcome in ovarian cancer patients. Clin Cancer Res. 2011;17:5490–500.
Lu Y, Chen X, Beesley J, Johnatty SE, Defazio A, Lambrechts S, et al. Genome-wide association study for ovarian cancer susceptibility using pooled DNA. Twin Res Human Genet. 2012;15:615–23.
Earp MA, Kelemen LE, Magliocco AM, Swenerton KD, Chenevix-Trench G, Lu Y, et al. Genome-wide association study of subtype-specific epithelial ovarian cancer risk alleles using pooled DNA. Hum Genet. 2014;133:481–97.
Mostowska A, Sajdak S, Pawlik P, Markowska J, Pawalowska M, Lianeri M, et al. Replication study for the association of seven genome- GWAS-identified loci with susceptibility to ovarian cancer in the Polish population. Pathol Oncol Res. 2015;21:307–13.
Zhang D, Sliwkowski MX, Mark M, Frantz G, Akita R, Sun Y, et al. Neuregulin-3 (NRG3): a novel neural tissue-enriched protein that binds and activates ErbB4. Proc Natl Acad Sci USA. 1997;94:9562–7.
Falls DL. Neuregulins: functions, forms, and signaling strategies. Exp Cell Res. 2003;284:14–30.
Maihle NJ, Baron AT, Barrette BA, Boardman CH, Christensen TA, Cora EM, et al. EGF/ErbB receptor family in ovarian cancer. Cancer Treat Res. 2002;107:247–58.
Sheng Q, Liu J. The therapeutic potential of targeting the EGFR family in epithelial ovarian cancer. Br J Cancer. 2011;104:1241–5.
National Center for Biotechnology Information. Gene. Available from: https://www.ncbi.nlm.nih.gov/gene/?term= [Accessed Feb 2018].
Fane M, Harris L, Smith AG, Piper M. Nuclear factor one transcription factors as epigenetic regulators in cancer. Int J Cancer. 2017;140:2634–41.
Tak YG, Farnham PJ. Making sense of GWAS: using epigenomics and genome engineering to understand the functional relevance of SNPs in non-coding regions of the human genome. Epigenetics Chromatin. 2015;8:57.
Yeung TL, Leung CS, Yip KP, Au Yeung CL, Wong ST, Mok SC. Cellular and molecular processes in ovarian cancer metastasis. A review in the theme: cell and molecular processes in cancer metastasis. Am J Physiol Cell Physiol. 2015;309:C444–56.
De Smet F, Pochet NL, Engelen K, Van Gorp T, Van Hummelen P, Marchal K, et al. Predicting the clinical behavior of ovarian cancer from gene expression profiles. Int J Gynecol Cancer. 2006;16(Suppl 1):147–51.
Breuleux M. Role of heregulin in human cancer. Cell Mol life Sci. 2007;64:2358–77.
Pradeep S, Kim SW, Wu SY, Nishimura M, Chaluvally-Raghavan P, Miyake T, et al. Hematogenous metastasis of ovarian cancer: rethinking mode of spread. Cancer Cell. 2014;26:77–91.
Fridley BL, Ghosh TM, Wang A, Raghavan R, Dai J, Goode EL, et al. Genome-wide study of response to platinum, taxane, and combination therapy in ovarian cancer: in vitro phenotypes, inherited variation, and disease recurrence. Front Genet. 2016;7:37.
Rebbeck TR, Mitra N, Domchek SM, Wan F, Friebel TM, Tran TV, et al. Modification of BRCA1-associated breast and ovarian cancer risk by BRCA1-interacting genes. Cancer Res. 2011;71:5792–805.
Ho DV, Chan JY. Induction of Herpud1 expression by ER stress is regulated by Nrf1. FEBS Lett. 2015;589:615–20.
Huang H, Tindall DJ. Dynamic FoxO transcription factors. J Cell Sci. 2007;120(Pt 15):2479–87.
Goto T, Takano M, Hirata J, Tsuda H. The involvement of FOXO1 in cytotoxic stress and drug-resistance induced by paclitaxel in ovarian cancers. Br J Cancer. 2008;98:1068–75.
Shi W, Tang MK, Yao Y, Tang C, Chui YL, Lee KK. BRE plays an essential role in preventing replicative and DNA damage-induced premature senescence. Sci Rep. 2016;6:23506.
Li Y, Qi K, Zu L, Wang M, Wang Y, Zhou Q. Anti-apoptotic brain and reproductive organ-expressed proteins enhance cisplatin resistance in lung cancer cells via the protein kinase B signaling pathway. Thorac Cancer. 2016;7:190–8.
Chan BC, Li Q, Chow SK, Ching AK, Liew CT, Lim PL, et al. BRE enhances in vivo growth of tumor cells. Biochem Biophys Res Commun. 2005;326:268–73.
Konig IR. Validation in genetic association studies. Brief Bioinform. 2011;12:253–8.
McCarthy MI, Abecasis GR, Cardon LR, Goldstein DB, Little J, Ioannidis JP, et al. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat Rev Genet. 2008;9:356–69.
Freedman ML, Monteiro AN, Gayther SA, Coetzee GA, Risch A, Plass C, et al. Principles for the post-GWAS functional characterization of cancer risk loci. Nat Genet. 2011;43:513–8.
Acknowledgements
We would like to thank the Liga Portuguesa Contra o Cancro-Centro Regional do Norte, Ministério da Saúde de Portugal (CFICS-45/2007), IPO-Porto (CI-IPOP-22-2015), and Fundação para a Ciência e Tecnologia (FCT). Joana Assis (SFRH/BD/98536/2013), Augusto Nogueira (SFRH/BD/124155/2016), and Carina Pereira (SFRH/BPD/114803/2016) are grant holders from FCT.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Pinto, R., Assis, J., Nogueira, A. et al. Pharmacogenomics in epithelial ovarian cancer first-line treatment outcome: validation of GWAS-associated NRG3 rs1649942 and BRE rs7572644 variants in an independent cohort. Pharmacogenomics J 19, 25–32 (2019). https://doi.org/10.1038/s41397-018-0056-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41397-018-0056-y
This article is cited by
-
Implications of venous thromboembolism GWAS reported genetic makeup in the clinical outcome of ovarian cancer patients
The Pharmacogenomics Journal (2021)