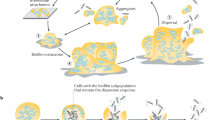
Abstract
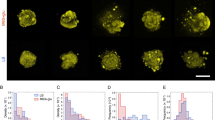
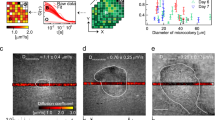
Microbial communities that form surface-attached biofilms must release and disperse their constituent cells into the environment to colonize fresh sites for continued survival of their species. For pathogens, biofilm dispersal is crucial for microbial transmission from environmental reservoirs to hosts, cross-host transmission, and dissemination of infections across tissues within the host. However, research on biofilm dispersal and its consequences in colonization of fresh sites remain poorly understood. Bacterial cells can depart from biofilms via stimuli-induced dispersal or disassembly due to direct degradation of the biofilm matrix, but the complex heterogeneity of bacterial populations released from biofilms rendered their study difficult. Using a novel 3D-bacterial “biofilm-dispersal-then-recolonization” (BDR) microfluidic model, we demonstrated that Pseudomonas aeruginosa biofilms undergo distinct spatiotemporal dynamics during chemical-induced dispersal (CID) and enzymatic disassembly (EDA), with contrasting consequences in recolonization and disease dissemination. Active CID required bacteria to employ bdlA dispersal gene and flagella to depart from biofilms as single cells at consistent velocities but could not recolonize fresh surfaces. This prevented the disseminated bacteria cells from infecting lung spheroids and Caenorhabditis elegans in on-chip coculture experiments. In contrast, EDA by degradation of a major biofilm exopolysaccharide (Psl) released immotile aggregates at high initial velocities, enabling the bacteria to recolonize fresh surfaces and cause infections in the hosts efficiently. Hence, biofilm dispersal is more complex than previously thought, where bacterial populations adopting distinct behavior after biofilm departure may be the key to survival of bacterial species and dissemination of diseases.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
We are sorry, but there is no personal subscription option available for your country.
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
Hengge R. Principles of c-di-GMP signalling in bacteria. Nat Rev Microbiol. 2009;7:263–73.
McDougald D, Rice SA, Barraud N, Steinberg PD, Kjelleberg S. Should we stay or should we go: mechanisms and ecological consequences for biofilm dispersal. Nat Rev Microbiol. 2012;10:39–50.
Fleming D, Rumbaugh K. The consequences of biofilm dispersal on the host. Sci Rep. 2018;8:10738.
Wolcott R. Biofilm and catheter-related bloodstream infections. Br J Nurs. 2021;30:S4–s9.
Minasyan H. Sepsis: mechanisms of bacterial injury to the patient. Scand J Trauma Resusc Emerg Med. 2019;27:19.
Qin B, Fei C, Bridges AA, Mashruwala AA, Stone HA, Wingreen NS, et al. Cell position fates and collective fountain flow in bacterial biofilms revealed by light-sheet microscopy. Science 2020;369:71–7.
Paula AJ, Hwang G, Koo H. Dynamics of bacterial population growth in biofilms resemble spatial and structural aspects of urbanization. Nat Commun. 2020;11:1354.
Li Y, Petrova OE, Su S, Lau GW, Panmanee W, Na R, et al. BdlA, DipA and Induced Dispersion Contribute to Acute Virulence and Chronic Persistence of Pseudomonas aeruginosa. PLOS Pathog. 2014;10:e1004168.
Barraud N, Hassett DJ, Hwang SH, Rice SA, Kjelleberg S, Webb JS. Involvement of nitric oxide in biofilm dispersal of Pseudomonas aeruginosa. J Bacteriol. 2006;188:7344–53.
Rumbaugh KP, Sauer K. Biofilm dispersion. Nat Rev Microbiol. 2020;18:571–86.
Yu M, Chua SL. Demolishing the great wall of biofilms in Gram-negative bacteria: To disrupt or disperse? Med Res Rev. 2020;40:1103–16.
Guilhen C, Forestier C, Balestrino D. Biofilm dispersal: multiple elaborate strategies for dissemination of bacteria with unique properties. Mol Microbiol. 2017;105:188–210.
Chua SL, Liu Y, Yam JKH, Chen Y, Vejborg RM, Tan BGC, et al. Dispersed cells represent a distinct stage in the transition from bacterial biofilm to planktonic lifestyles. Nat Commun. 2014;5:4462.
Goodwine J, Gil J, Doiron A, Valdes J, Solis M, Higa A, et al. Pyruvate-depleting conditions induce biofilm dispersion and enhance the efficacy of antibiotics in killing biofilms in vitro and in vivo. Sci Rep. 2019;9:3763.
Yu S, Su T, Wu H, Liu S, Wang D, Zhao T, et al. PslG, a self-produced glycosyl hydrolase, triggers biofilm disassembly by disrupting exopolysaccharide matrix. Cell Res. 2015;25:1352–67.
Martí M, Trotonda MP, Tormo-Más MÁ, Vergara-Irigaray M, Cheung AL, Lasa I, et al. Extracellular proteases inhibit protein-dependent biofilm formation in Staphylococcus aureus. Microbes Infect. 2010;12:55–64.
Hall-Stoodley L, Nistico L, Sambanthamoorthy K, Dice B, Nguyen D, Mershon WJ, et al. Characterization of biofilm matrix, degradation by DNase treatment and evidence of capsule downregulation in Streptococcus pneumoniae clinical isolates. BMC Microbiol. 2008;8:173.
Reffuveille F, de la Fuente-Núñez C, Mansour S, Hancock RE. A broad-spectrum antibiofilm peptide enhances antibiotic action against bacterial biofilms. Antimicrob Agents Chemother. 2014;58:5363–71.
Crone S, Vives-Flórez M, Kvich L, Saunders AM, Malone M, Nicolaisen MH, et al. The environmental occurrence of Pseudomonas aeruginosa. APMIS 2020;128:220–31.
Wheatley RM, Caballero JD, van der Schalk TE, De Winter FH, Kapel N, Recanatini C, et al. Assessing the contribution of gut-to-lung translocation to bacterial colonization and antibiotic resistance in an ICU patient. medRxiv. 2022, https://www.medrxiv.org/content/10.1101/2022.01.17.22269403v1.
Chuang C-H, Wang Y-H, Chang H-J, Chen H-L, Huang Y-C, Lin T-Y, et al. Shanghai fever: a distinct Pseudomonas aeruginosa enteric disease. Gut. 2014;63:736–43.
Cerioli M, Batailler C, Conrad A, Roux S, Perpoint T, Becker A, et al. Pseudomonas aeruginosa implant-associated bone and joint infections: experience in a regional reference center in France. Front Med. 2020;7:513242.
Shah NB, Osmon DR, Steckelberg JM, Sierra RJ, Walker RC, Tande AJ, et al. Pseudomonas prosthetic joint infections: a review of 102 episodes. J Bone Jt Infect. 2016;1:25–30.
van Delden C. Pseudomonas aeruginosa bloodstream infections: how should we treat them? Int J Antimicrobial Agents. 2007;30:71–5.
Jugulete G, Luminos M, Visan A, Draganescu A, Merisescu M, Vasile M, et al. Severe sepsis with multiple organ dysfunctions caused by Pseudomonas aeruginosa in an immunocompetent child. Crit Care. 2012;16:P69.
Stoner SN, Baty JJ, Scoffield JA. Pseudomonas aeruginosa polysaccharide Psl supports airway microbial community development. ISME J. 2022;16:1730–9.
Jennings LK, Dreifus JE, Reichhardt C, Storek KM, Secor PR, Wozniak DJ, et al. Pseudomonas aeruginosa aggregates in cystic fibrosis sputum produce exopolysaccharides that likely impede current therapies. Cell Rep. 2021;34:108782.
Roberts AEL, Kragh KN, Bjarnsholt T, Diggle SP. The limitations of in vitro experimentation in understanding biofilms and chronic infection. J Mol Biol. 2015;427:3646–61.
Sternberg C, Tolker-Nielsen T. Growing and analyzing biofilms in flow cells. Curr Protoc Microbiol. 2006;Chapter 1:Unit 1B.2. https://doi.org/10.1002/9780471729259.mc01b02s00.
Khoo BL, Shang M, Ng CH, Lim CT, Chng WJ, Han J. Liquid biopsy for minimal residual disease detection in leukemia using a portable blast cell biochip. NPJ Precis Oncol. 2019;3:30.
Khoo BL, Grenci G, Jing T, Lim YB, Lee SC, Thiery JP, et al. Liquid biopsy and therapeutic response: Circulating tumor cell cultures for evaluation of anticancer treatment. Sci Adv. 2016;2:e1600274.
Deng Y, Fu Y, Chua SL, Khoo BL. Biofilm Potentiates Cancer‐Promoting Effects of Tumor‐Associated Macrophages in a 3D Multi‐Faceted Tumor Model. Small 2023;19:2205904.
Liu YS, Deng Y, Chen CK, Khoo BL, Chua SL. Rapid detection of microorganisms in a fish infection microfluidics platform. J Hazard Mater. 2022;431:128572.
Shigematsu M, Meno Y, Misumi H, Amako K. The measurement of swimming velocity of Vibrio cholerae and Pseudomonas aeruginosa using the video tracking methods. Microbiol Immunol. 1995;39:741–4.
Chua SL, Tan SY, Rybtke MT, Chen Y, Rice SA, Kjelleberg S, et al. Bis-(3’-5’)-cyclic dimeric GMP regulates antimicrobial peptide resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2013;57:2066–75.
Morgan R, Kohn S, Hwang SH, Hassett DJ, Sauer K. BdlA, a chemotaxis regulator essential for biofilm dispersion in Pseudomonas aeruginosa. J Bacteriol. 2006;188:7335–43.
Petrova OE, Sauer K. Dispersion by Pseudomonas aeruginosa requires an unusual posttranslational modification of BdlA. Proc Natl Acad Sci. 2012;109:16690–5.
Chua SL, Ding Y, Liu Y, Cai Z, Zhou J, Swarup S, et al. Reactive oxygen species drive evolution of pro-biofilm variants in pathogens by modulating cyclic-di-GMP levels. Open Biol. 2016;6:160162.
Colvin KM, Irie Y, Tart CS, Urbano R, Whitney JC, Ryder C, et al. The Pel and Psl polysaccharides provide Pseudomonas aeruginosa structural redundancy within the biofilm matrix. Environ Microbiol. 2012;14:1913–28.
Chimenti I, Pagano F, Angelini F, Siciliano C, Mangino G, Picchio V, et al. Human lung spheroids as in vitro niches of lung progenitor cells with distinctive paracrine and plasticity properties. Stem Cells Transl Med. 2017;6:767–77.
Fong ELS, Toh TB, Yu H, Chow EK-H. 3D culture as a clinically relevant model for personalized medicine. SLAS Technol. 2017;22:245–53.
Chan SY, Liu SY, Seng Z, Chua SL. Biofilm matrix disrupts nematode motility and predatory behavior. ISME J. 2021;15:260–9.
Li S, Liu SY, Chan SY, Chua SL. Biofilm matrix cloaks bacterial quorum sensing chemoattractants from predator detection. ISME J. 2022;16:1388–96.
Clark AS, Huayta J, Arulalan KS, San-Miguel A. Chapter 13 - Microfluidic devices for imaging and manipulation of C. elegans. In: Liu X, Sun Y, editors. Micro and nano systems for biophysical studies of cells and small organisms. Academic Press; 2021. p. 295–321.
Letizia MC, Cornaglia M, Trouillon R, Sorrentino V, Mouchiroud L, Bou Sleiman MS, et al. Microfluidics-enabled phenotyping of a whole population of C. elegans worms over their embryonic and post-embryonic development at single-organism resolution. Microsyst Nanoengineering. 2018;4:6.
Deng Y, Liu SY, Chua SL, Khoo BL. The effects of biofilms on tumor progression in a 3D cancer-biofilm microfluidic model. Biosens Bioelectron. 2021;180:113113.
Straub H, Eberl L, Zinn M, Rossi RM, Maniura-Weber K, Ren Q. A microfluidic platform for in situ investigation of biofilm formation and its treatment under controlled conditions. J Nanobiotechnology. 2020;18:166.
Christensen LD, Gennip MV, Rybtke MT, Wu H, Chiang W-C, Alhede M, et al. Clearance of Pseudomonas aeruginosa Foreign-Body Biofilm Infections through Reduction of the Cyclic Di-GMP Level in the Bacteria. Infect Immun. 2013;81:2705–13.
Dang H, Lovell CR. Microbial surface colonization and biofilm development in marine environments. Microbiol Mol Biol Rev. 2016;80:91–138.
Qin Z, Yang X, Yang L, Jiang J, Ou Y, Molin S, et al. Formation and properties of in vitro biofilms of ica-negative Staphylococcus epidermidis clinical isolates. J Med. Microbiol. 2007;56:83–93.
Weaver WM, Milisavljevic V, Miller JF, Di Carlo D. Fluid flow induces biofilm formation in Staphylococcus epidermidis polysaccharide intracellular adhesin-positive clinical isolates. Appl Environ Microbiol. 2012;78:5890–6.
Zhao T, Liu Y. N-acetylcysteine inhibit biofilms produced by Pseudomonas aeruginosa. BMC Microbiol. 2010;10:140.
Lu TK, Collins JJ. Dispersing biofilms with engineered enzymatic bacteriophage. Proc Natl Acad Sci. 2007;104:11197–202.
Rao Y, Wang W, Tan F, Cai Y, Lu J, Qiao X. Influence of different ions doping on the antibacterial properties of MgO nanopowders. Appl Surf Sci. 2013;284:726–31.
Zhang T, Deng Y, Liu YS, Chua SL, Tang BZ, Khoo BL. Bacterial targeted AIE photosensitizers synergistically promote chemotherapy for the treatment of inflammatory cancer. Chem Eng J. 2022;447:137579.
Chen CK, Zhang J, Bhingarde A, Matotek T, Barrett J, Hardesty BD, et al. A portable purification system for the rapid removal of microplastics from environmental samples. Biochem Eng J. 2022;428:132614.
Liao JC, Zou SJ, Deng YL, Jiang Y, Chua SL, Khoo BL. Multivariate analysis of liquid biopsies for real-time detection of patients with biofilm-associated infections (BAI). Biochem Eng J. 2023:453;139595.
Liu SY, Leung MM-L, Fang JK-H, Chua SL. Engineering a microbial ‘trap and release’ mechanism for microplastics removal. Chem Eng J. 2021;404:127079.
Masuko T, Minami A, Iwasaki N, Majima T, Nishimura S, Lee YC. Carbohydrate analysis by a phenol-sulfuric acid method in microplate format. Anal Biochem. 2005;339:69–72.
Wang S, Chan SY, Deng Y, Khoo BL, Chua SL. Oxidative stress induced by Etoposide anti-cancer chemotherapy drives the emergence of tumor-associated bacteria resistance to fluoroquinolones. J Adv Res. 2023;S2090-1232(23)00062-0. https://doi.org/10.1016/j.jare.2023.02.011.
Acknowledgements
This research is supported by The Hong Kong Polytechnic University, Department of Applied Biology and Chemical Technology Startup Grant (BE2B), Departmental General Research Fund (UALB), One-line account (ZVVV), Environmental and Conservation Fund (ECF-48/2019 and ECF-84/2021), Health and Medical Research Fund (HMRF-20190302), and State Key Laboratory of Chemical Biology and Drug Discovery Fund (1-BBX8). This work was also supported by the City University of Hong Kong [7005208,7005464,7020002,9610430,9667220]; Hong Kong Center for Cerebro- Cardiovascular Health Engineering (COCHE); Research Grants Council of the Hong Kong Special Administrative Region [21200921]; Pneumoconiosis Compensation Fund Board [9211276]; and the Hetao Shenzhen-Hong Kong Science and Technology Innovation Cooperation Zone Shenzhen Park Project (HZQB-KCZYZ-2021017).
Author information
Authors and Affiliations
Contributions
SLC and BLK designed methods and experiments. YM and YD performed laboratory experiments and analyzed the data. YM, YD, BLK and SLC interpreted the results and wrote the paper. All authors have contributed to, seen, and approved the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare the following financial interests/personal relationships, which may be considered as potential competing interests: One or more authors have a pending patent related to this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Ma, Y., Deng, Y., Hua, H. et al. Distinct bacterial population dynamics and disease dissemination after biofilm dispersal and disassembly. ISME J 17, 1290–1302 (2023). https://doi.org/10.1038/s41396-023-01446-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41396-023-01446-5
This article is cited by
-
Staphylococcus aureus and biofilms: transmission, threats, and promising strategies in animal husbandry
Journal of Animal Science and Biotechnology (2024)