Abstract
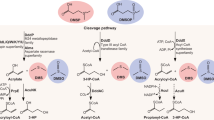
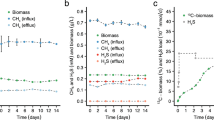
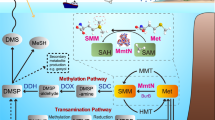
Dimethylsulfide (DMS) is the major biosulfur source emitted to the atmosphere with key roles in global sulfur cycling and potentially climate regulation. The main precursor of DMS is thought to be dimethylsulfoniopropionate. However, hydrogen sulfide (H2S), a widely distributed and abundant volatile in natural environments, can be methylated to DMS. The microorganisms and the enzymes that convert H2S to DMS, and their importance in global sulfur cycling were unknown. Here we demonstrate that the bacterial MddA enzyme, previously known as a methanethiol S-methyltransferase, could methylate inorganic H2S to DMS. We determine key residues involved in MddA catalysis and propose the mechanism for H2S S-methylation. These results enabled subsequent identification of functional MddA enzymes in abundant haloarchaea and a diverse range of algae, thus expanding the significance of MddA mediated H2S methylation to other domains of life. Furthermore, we provide evidence for H2S S-methylation being a detoxification strategy in microorganisms. The mddA gene was abundant in diverse environments including marine sediments, lake sediments, hydrothermal vents and soils. Thus, the significance of MddA-driven methylation of inorganic H2S to global DMS production and sulfur cycling has likely been considerably underestimated.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
We are sorry, but there is no personal subscription option available for your country.
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The predicted structure can be obtained from the AlphaFold Protein Structure Database (https://alphafold.ebi.ac.uk/) with accession code A0A0F6P9C0. The sequences of MddA and other proteins can be found in NCBI database as well as Supplementary Information files.
References
Shemi A, Alcolombri U, Schatz D, Farstey V, Vincent F, Rotkopf R, et al. Dimethyl sulfide mediates microbial predator-prey interactions between zooplankton and algae in the ocean. Nat Microbiol. 2021;6:1357–66.
De Zwart JMM, Kuenen JG. C1-cycle of sulfur compounds. Biodegradation. 1992;3:37–59.
Charlson RJ, Lovelock JE, Andreae MO, Warren SG. Oceanic phytoplankton, atmospheric sulfur, cloud albedo and climate. Nature. 1987;326:655–61.
Quinn PK, Bates TS. The case against climate regulation via oceanic phytoplankton sulphur emissions. Nature. 2011;480:51–6.
Kettle AJ, Andreae MO. Flux of dimethylsulfide from the oceans: a comparison of updated data seas and flux models. J Geophys Res Atmos. 2000;105:26793–808.
Watts SF. The mass budgets of carbonyl sulfide, dimethyl sulfide, carbon disulfide and hydrogen sulfide. Atmos Environ. 2000;34:761–79.
Vallina SM, Simo R. Strong relationship between DMS and the solar radiation dose over the global surface ocean. Science. 2007;315:506–8.
Curson ARJ, Liu J, Martinez AB, Green RT, Chan YH, Carrion O, et al. Dimethylsulfoniopropionate biosynthesis in marine bacteria and identification of the key gene in this process. Nat Microbiol. 2017;2:17009.
Curson AR, Todd JD, Sullivan MJ, Johnston AW. Catabolism of dimethylsulphoniopropionate: microorganisms, enzymes and genes. Nat Rev Microbiol. 2011;9:849–59.
Yoch DC. Dimethylsulfoniopropionate: Its sources, role in the marine food web, and biological degradation to dimethylsulfide. Appl Environ Microbiol. 2002;68:5804–15.
Alcolombri U, Ben-Dor S, Feldmesser E, Levin Y, Tawfik DS, Vardi A. Identification of the algal dimethyl sulfide-releasing enzyme: A missing link in the marine sulfur cycle. Science. 2015;348:1466–9.
Li CY, Wang XJ, Chen XL, Sheng Q, Zhang S, Wang P, et al. A novel ATP dependent dimethylsulfoniopropionate lyase in bacteria that releases dimethyl sulfide and acryloyl-CoA. eLife. 2021;10:e64045.
Kiene RP, Hines ME. Microbial formation of dimethyl sulfide in anoxic sphagnum peat. Appl Environ Microbiol. 1995;61:2720–6.
Zinder SH, Brock TD. Dimethyl sulphoxide reduction by micro-organisms. J Gen Microbiol. 1978;105:335–42.
Spiese CE, Kieber DJ, Nomura CT, Kiene RP. Reduction of dimethylsulfoxide to dimethylsulfide by marine phytoplankton. Limnol Oceanogr. 2009;54:560–70.
Stets EG, Hines ME, Kiene RP. Thiol methylation potential in anoxic, low-pH wetland sediments and its relationship with dimethylsulfide production and organic carbon cycling. FEMS Microbiol Ecol. 2004;47:1–11.
Carrion O, Pratscher J, Curson ARJ, Williams BT, Rostant WG, Murrell JC, et al. Methanethiol-dependent dimethylsulfide production in soil environments. ISME J. 2017;11:2379–90.
Carrion O, Curson AR, Kumaresan D, Fu Y, Lang AS, Mercade E, et al. A novel pathway producing dimethylsulphide in bacteria is widespread in soil environments. Nat Commun. 2015;6:6579.
Reisch CR, Stoudemayer MJ, Varaljay VA, Amster IJ, Moran MA, Whitman WB. Novel pathway for assimilation of dimethylsulphoniopropionate widespread in marine bacteria. Nature. 2011;473:208–11.
Lomans BP, Smolders AJP, Intven LM, Pol A, denCamp HJMO, vanderDrift C. Formation of dimethyl sulfide and methanethiol in anoxic freshwater sediments. Appl Environ Microbiol. 1997;63:4741–7.
Bak F, Finster K, Rothfuss F. Formation of dimethylsulfide and methanethiol from methoxylated aromatic compounds and inorganic sulfide by newly isolated anaerobic bacteria. Arch Microbiol. 1992;157:529–34.
Carrion O, Pratscher J, Richa K, Rostant WG, Ul Haque MF, Murrell JC, et al. Methanethiol and dimethylsulfide cycling in stiffkey saltmarsh. Front Microbiol. 2019;10:1040.
Andreae MO. Ocean-atmosphere interactions in the global biogeochemical sulfur cycle. Mar Chem. 1990;30:1–29.
Bagarinao T. Sulfide as an environmental factor and toxicant: tolerance and adaptations in aquatic organisms. Aquat Toxicol. 1992;24:21–62.
Johnson KS, Beehler CL, Sakamoto-Arnold CM, Childress JJ. In situ measurements of chemical distributions in a deep-sea hydrothermal vent field. Science 1986;231:1139–41.
Thompson BE, Bay SM, Anderson JW, Laughlin JD, Greenstein DJ, Tsukada DT. Chronic effects of contaminated sediments on the urchin Lytechinus pictus. Environ Toxicol Chem. 1989;8:629–37.
Malone Rubright SL, Pearce LL, Peterson J. Environmental toxicology of hydrogen sulfide. Nitric Oxide. 2017;71:1–13.
Cuevasanta E, Moller MN, Alvarez B. Biological chemistry of hydrogen sulfide and persulfides. Arch Biochem Biophys. 2017;617:9–25.
Weisiger RA, Pinkus LM, Jakoby WB. Thiol S-methyltransferase: suggested role in detoxication of intestinal hydrogen sulfide. Biochem Pharm. 1980;29:2885–7.
Sun Y, Wang M, Zhong Z, Chen H, Wang H, Zhou L, et al. Adaption to hydrogen sulfide-rich environments: Strategies for active detoxification in deep-sea symbiotic mussels, Gigantidas platifrons. Sci Total Environ. 2022;804:150054.
Itoh N, Toda H, Matsuda M, Negishi T, Taniguchi T, Ohsawa N. Involvement of S-adenosylmethionine-dependent halide/thiol methyltransferase (HTMT) in methyl halide emissions from agricultural plants: isolation and characterization of an HTMT-coding gene from Raphanus sativus (daikon radish). BMC Plant Biol. 2009;9:116.
Maldonato BJ, Russell DA, Totah RA. Human METTL7B is an alkyl thiol methyltransferase that metabolizes hydrogen sulfide and captopril. Sci Rep. 2021;11:4857.
Sambrook J, Russell DW. Molecular cloning, a laboratory manual. 3rd ed. New York, USA: Cold Spring Harbor Laboratory Press; 2001.
Wang PX, Yu ZC, Li BY, Cai XS, Zeng ZS, Chen XL, et al. Development of an efficient conjugation-based genetic manipulation system for Pseudoalteromonas. Micro Cell Fact. 2015;14:11.
Kovach ME, Elzer PH, Hill DS, Robertson GT, Farris MA, Roop RM, et al. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene. 1995;166:175–6.
Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596:583–9.
Varadi M, Anyango S, Deshpande M, Nair S, Natassia C, Yordanova G, et al. AlphaFold Protein Structure Database: massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 2022;50:D439–D44.
Fu LM, Niu BF, Zhu ZW, Wu ST, Li WZ. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics 2012;28:3150–2.
Katoh K, Rozewicki J, Yamada KD. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform. 2019;20:1160–6.
Capella-Gutierrez S, Silla-Martinez JM, Gabaldon T. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 2009;25:1972–3.
Price MN, Dehal PS, Arkin AP. FastTree 2-approximately maximum-likelihood trees for large alignments. PLoS One. 2010;5:e9490.
Letunic I, Bork P. Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021;49:W293–W6.
Vernette C, Lecubin J, Sanchez P, Tara Oceans C, Sunagawa S, Delmont TO, et al. The Ocean Gene Atlas v2.0: online exploration of the biogeography and phylogeny of plankton genes. Nucleic Acids Res. 2022;50:W516–26.
Chen IA, Chu K, Palaniappan K, Ratner A, Huang J, Huntemann M, et al. The IMG/M data management and analysis system v.6.0: new tools and advanced capabilities. Nucleic Acids Res. 2021;49:D751–D63.
Zhang YJ, Liu XF, Kuang BZ, Zhang XY, Zhou MY, Chen S. Neptunicoccus sediminis gen. nov., sp. nov., a member of the family Rhodobacteraceae isolated from the Yellow Sea. Int J Syst Evol Microbiol. 2018;68:1702–6.
Toda H, Itoh N. Isolation and characterization of a gene encoding a S-adenosyl-L-methionine-dependent halide/thiol methyltransferase (HTMT) from the marine diatom Phaeodactylum tricornutum: Biogenic mechanism of CH3I emissions in oceans. Phytochemistry 2011;72:337–43.
Furne J, Springfield J, Koenig T, DeMaster E, Levitt MD. Oxidation of hydrogen sulfide and methanethiol to thiosulfate by rat tissues: a specialized function of the colonic mucosa. Biochem Pharm. 2001;62:255–9.
Wirth JS, Wang T, Huang QY, White RH, Whitman WB. Dimethylsulfoniopropionate sulfur and methyl carbon assimilation in Ruegeria species. mBio. 2020;11:e00329-20.
Liscombe DK, Louie GV, Noel JP. Architectures, mechanisms and molecular evolution of natural product methyltransferases. Nat Prod Rep. 2012;29:1238–50.
Sun Q, Huang MY, Wei YQ. Diversity of the reaction mechanisms of SAM-dependent enzymes. Acta Pharmaceutica Sin B. 2021;11:632–50.
Tobias H, Christian B. Oxidation and incorporation of hydrogen sulfide by dissolved organic matter. Chem Geol. 2006;235:12–20.
Kiene RP, Bates TS. Biological removal of dimethyl sulfide from sea-water. Nature. 1990;345:702–5.
Kappler U, Schafer H. Transformations of dimethylsulfide. Met Ions Life Sci. 2014;14:279–313.
Mathai JC, Missner A, Kugler P, Saparov SM, Zeidel ML, Lee JK, et al. No facilitator required for membrane transport of hydrogen sulfide. Proc Natl Acad Sci USA. 2009;106:16633–8.
Riahi S, Rowley CN. Why can hydrogen sulfide permeate cell membranes? J Am Chem Soc. 2014;136:15111–3.
Cuevasanta E, Denicola A, Alvarez B, Moller MN. Solubility and permeation of hydrogen sulfide in lipid membranes. PLoS One. 2012;7:e34562.
Horinouchi M, Kasuga K, Nojiri H, Yamane H, Omori T. Cloning and characterization of genes encoding an enzyme which oxidizes dimethyl sulfide in Acinetobacter sp. strain 20B. FEMS Microbiol Lett. 1997;155:99–105.
Fuse H, Takimura O, Murakami K, Yamaoka Y, Omori T. Utilization of dimethyl sulfide as a sulfur source with the aid of light by Marinobacterium sp. strain DMS-S1. Appl Environ Microbiol. 2000;66:5527–32.
Boden R, Murrell JC, Schafer H. Dimethylsulfide is an energy source for the heterotrophic marine bacterium Sagittula stellata. FEMS Microbiol Lett. 2011;322:188–93.
Lidbury I, Krober E, Zhang ZD, Zhu YJ, Murrell JC, Chen Y, et al. A mechanism for bacterial transformation of dimethylsulfide to dimethylsulfoxide: a missing link in the marine organic sulfur cycle. Environ Microbiol. 2016;18:2754–66.
Sunda W, Kieber DJ, Kiene RP, Huntsman S. An antioxidant function for DMSP and DMS in marine algae. Nature. 2002;418:317–20.
Teng ZJ, Wang P, Chen XL, Guillonneau R, Li CY, Zou SB, et al. Acrylate protects a marine bacterium from grazing by a ciliate predator. Nat Microbiol. 2021;6:1351–6.
Acknowledgements
We thank Terry McGenity from University of Essex for providing the Haladaptatus sp. W1 strain, and Emese Bartha from University of East Anglia for providing advice on how to culture it. This work was supported by the Marine S&T Fund of Shandong Province for Qingdao Marine Science and Technology Center (No. 2022QNLM030004-3), the National Key Research and Development Program of China (2022YFC2807500), the National Science Foundation of China (grants 42276102, 92251303, 42076229, 31961133016), the Fundamental Research Funds for the Central Universities (202172002, 202041011), the Major Scientific and Technological Innovation Project (MSTIP) of Shandong Province (2019JZZY010817), the Program of Shandong for Taishan Scholars (tspd20181203), the Biotechnology and Biological Sciences Research Council, UK, grant (BB/X005968), Natural Environment Research Council, UK, Standard grants (NE/X000990, NE/V000756 and NE/S001352) and the Leverhulme Trust research grant (RPG-2020-413).
Author information
Authors and Affiliations
Contributions
CYL and YZZ designed and directed the research. JDT designed some experiments. HYC, CYL, QW, OC, XZ and JM performed the experiments. PW, XZ and XLC helped in data analysis. HYC, CYL, JDT and YZZ wrote the manuscript. XLC edited the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Li, CY., Cao, HY., Wang, Q. et al. Aerobic methylation of hydrogen sulfide to dimethylsulfide in diverse microorganisms and environments. ISME J 17, 1184–1193 (2023). https://doi.org/10.1038/s41396-023-01430-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41396-023-01430-z