Abstract
Thiovulum spp. (Campylobacterota) are large sulfur bacteria that form veil-like structures in aquatic environments. The sulfidic Movile Cave (Romania), sealed from the atmosphere for ~5 million years, has several aqueous chambers, some with low atmospheric O2 (~7%). The cave’s surface-water microbial community is dominated by bacteria we identified as Thiovulum. We show that this strain, and others from subsurface environments, are phylogenetically distinct from marine Thiovulum. We assembled a closed genome of the Movile strain and confirmed its metabolism using RNAseq. We compared the genome of this strain and one we assembled from public data from the sulfidic Frasassi caves to four marine genomes, including Candidatus Thiovulum karukerense and Ca. T. imperiosus, whose genomes we sequenced. Despite great spatial and temporal separation, the genomes of the Movile and Frasassi Thiovulum were highly similar, differing greatly from the very diverse marine strains. We concluded that cave Thiovulum represent a new species, named here Candidatus Thiovulum stygium. Based on their genomes, cave Thiovulum can switch between aerobic and anaerobic sulfide oxidation using O2 and NO3- as electron acceptors, the latter likely via dissimilatory nitrate reduction to ammonia. Thus, Thiovulum is likely important to both S and N cycles in sulfidic caves. Electron microscopy analysis suggests that at least some of the short peritrichous structures typical of Thiovulum are type IV pili, for which genes were found in all strains. These pili may play a role in veil formation, by connecting adjacent cells, and in the motility of these exceptionally fast swimmers.
Similar content being viewed by others
Introduction
Thiovulum spp. are spherical bacteria typically <25 µm in diameter [1], but some can reach up to 50 µm [2]. Thiovulum spp. are mostly known from marine environments [3], with few old descriptions from freshwater [4, 5]. Described species are sulfur-oxidizing chemolithoautotrophs [6], some of which display an extremely fast motility [7, 8]. Thiovulum cells typically form veils close to surfaces [1, 9] and can attach to these using a secreted stalk [10]. They are normally located close to the oxic-anoxic interface near sediments or microbial mats [1, 11, 12], where the planar organization of the veil and the rapid movements of the cells’ flagella produce a convective transport of O2 [13].
Movile Cave is located near the town of Mangalia, SE Romania (43°49’32“N, 28°33'38“E), 2.2 km inland from the Black Sea shore. It consists of a 200 m long upper dry passage that ends in a small lake allowing access to a 40 m long, partially submerged, lower cave level (Fig. 1). Thick and impermeable layers of clays and loess cover the limestone in which the cave is developed, preventing input of water and nutrients from the surface [14]. Sulfidic groundwater flows constantly at the bottom of Movile Cave’s lower passages. Because of the morphology of the lower cave passages (Fig. 1) and a slight difference in water temperatures, the water near the surface is practically stagnant. Riess et al. [15] determined that oxygen penetrates only the upper 0.8 mm of the water column, below which the water is anoxic.
The microbial community containing Thiovulum cells (depicted here as dots present at, and beneath the water surface) was sampled in the Lake Room, and in Air Bell 2 alongside submerged microbial mats from Air Bell 1. Thiovulum 16 S rRNA gene made up to a maximum of 5%, 0.9%, and 35% of the 16 S rRNA genes retrieved from metagenomic samples (dark gray in pies) from these cave sections, respectively. More details on community composition are presented in Fig. S1. The scale bar refers to the length of the rooms, which are all ca. 1 m in width.
Cave ecosystems are normally characterized by stable conditions and provide a window into subsurface microbiology [16] In the absence of natural light, these ecosystems are typically fueled by chemolithoautotrophy via the oxidation of reduced compounds such as H2S, Fe2+, Mn2+, NH4+, CH4, and H2. Most of the microbiological studies performed in Movile Cave, as summarized in Kumaresan et al. [17], are based on samples of microbial biofilms floating on the water surface or covering rock surfaces in the Lake Room, Air Bell 1, and in Air Bell 2 where the atmosphere is low in O2 (ca. 7%) and enriched in CO2 (ca. 2%) and CH4 (1–2%) [18]. The microbial communities at these sites were found to include sulfur oxidizing bacteria such as Beggiatoa, Sulfurospirillum, Thiobacillus, Thiomonas, Thioploca, Thiothrix, and Thiovirga [19,20,21] methylotrophs such as Methylomonas, Methylococcus and Methylocystis [22], Methylotenera, Methylophilus, and Methylovorus [19, 20]; ammonia and nitrite oxidizers such as Nitrosomonas, Nitrospira and Nitrotoga [20] and the methanogenic Archaea Methanobacterium [23] and Methanosarcina [24]. Of these, activity was confirmed only for a few taxa [20, 22,23,24,25] while for others it was inferred based on phylogenetic association.
In the lower level of Movile Cave, at and directly below the water surface (<3 mm), we observed a loose floating veil (Fig. 2 and Supplementary video 1). Using genetic and microscopic analyses, we found that this agglomeration of bacteria is dominated by a species of the genus Thiovulum (Fig. 1, S1 and results). Using a comparative genomic approach, we compare the metabolic potential of this newly identified freshwater strain and another cave Thiovulum sp. we assembled from public data to their marine counterparts, including the genomes of Candidatus Thiovulum karukerense [26] and Ca. T. imperiosus [2], whose genomes are also presented here.
Images of surface veils from the Lake Room (A and B) and Air Bell 2 (C and D) in Movile Cave. See also Supplementary Video 1. C, D are images of the same location taken above (C) and below (D) the water surface, showing the thin Thiovulum veil. The images are ca. 1 m wide.
Materials and methods
The general structure of the Movile Cave and sampling locations are depicted to scale in Fig. 1. Briefly, the lake in the Lake Room is ca. 1 m wide and 3 m long, Air Bell 1 is of similar dimensions, and Air Bell 2 is ca. 1 m wide by 5 m long. The water depth in the cave is ca. 1.5 m. The ceiling in Air Bell 2 is at about 1 m above the water surface.
Movile Cave was sampled, in most cases, from within the water with as minimal disturbance possible. Specifically, Air Bell 2 can only be reached by SCUBA diving. There, the diver stayed only so long as necessary to collect the samples and document the environment. Air bubbles ascending from the dive likely disturbed the surface resulting in the patchy appearance of the floating microbial mats (Fig. 2). Prior to disturbance by the sampling team, the microbial veils covered the entire water surface, as could be evaluated from underwater while entering Air Bell 2 and by observing side sections of the chamber that are unaffected by the divers.
Molecular analyses
Unless stated otherwise, samples of water were collected into sterile tubes from the surface (upper few mm) of the Lake Room and from the Air Bells (Fig. 1), targeting specifically the white veil.
gDNA
For total DNA extraction a total of 12 (four tubes each from the Lake Room, Air Bell 2, and submerged mats in Air Bell 1), 50-mL water samples were collected in July 2019, and preserved, on site (Lake Room) with ethanol to a final concentration of 50%. The samples were then transferred to −20 °C within several hours of fixation where they were held till further treatment.
Microorganisms from each water sample (25 mL water +25 mL ethanol) were concentrated separately using a vacuum pump and Nalgene single-use analytical filter funnels (Thermo Fisher Scientific, MA, USA), with the included filter replaced with a 0.2 µm isopore membrane filter (Millipore Sigma, MA, USA). Prior to filtration, the glass assembly components were autoclaved at 121.5 °C for 30 min wet and 20 min dry at 1.4 bar (20 psi). Filters were placed into 50 µL conical tubes with 300 µL 1× PBS, 1.5 µl 2% azide, and a sterile scalpel blade. Samples were minced using an OMNI bead ruptor elite (OMNI International, GA, USA) on a 4 ms−1 30 s program. The resulting sample was centrifuged to collect the microorganisms separated from the filters and extracted for gDNA using a modified version of the Omega Bio-Tek Universal Metagenomics kit protocol (OMEGA Bio-Tek, GA, USA). Fifteen µL of MetaPolyzyme (Millipore Sigma, MA, USA) was added to each sample and incubated at 35 °C for 13 h followed by three cycles of freeze and thaw alternating between 80 °C for 2 min and a −80 °C freezer for 10 min. Further digestion was subsequently performed by adding 35 µL Proteinase K (Omega Bio-Tek, GA, USA) and incubated at 55 °C for 1 h. Following complete enzymatic digestion, the sample was extracted using the manufacturer’s protocol (Omega Bio-Tek Universal metagenomics kit). Briefly, 500 µL ML1 buffer (CTAB) was added the digested sample and incubated at 55 °C for 15 min One volume of Tris-stabilized (pH > 7.5) phenol-chloroform-isoamyl alcohol mix (25:24:1) was used for purification and the resulting upper aqueous phase was removed and combined with RBB (guanidinium) buffer and 100% ethanol and applied to a DNA silica column supplied with the kit. Final DNA was eluted in 35 µL of elution buffer. DNA was quantified using a Qubit spectrofluorometer and Nanodrop ND-1000 (ThermoFisher Waltham MA USA).
Thiovulum aggregates
In September 2019, samples were also collected for targeted “single-cell” sequencing of Thiovulum. For this, water samples were fixed on-site (1:1 v:v) in self-made RNAlater (4 M (NH4)2SO4; 15 mM EDTA (from 0.5 M, pH 8.0 stock); 18.75 mM Na-citrate (from 1 M stock)). The samples were then transferred to 4 °C (to prevent freezing) within several hours of fixation where they were held till further treatment.
Ten individual Thiovulum aggregates were picked under a binocular at 50X magnification, after which cells lysis and DNA amplification were carried out by three initial cycles of freeze-thawing with liquid N2 and a 65 °C thermal block and subsequently the lysis and amplification protocol of the REPLI-g Single-Cell DNA amplification kit, following the manufacturer instructions (Qiagen, Hilden, Germany). Of the ten amplified aggregates, four were selected for nanopore library preparation (see below).
Total RNA
In August 2021, 8 additional tubes were collected for RNA extraction and preserved on-site with ethanol to a final concentration of 50%. The samples were then transferred to −20 °C within several hours of fixation where they were held till further treatment.
Total nucleic acids were extracted from polycarbonate filters (Millipore, 0.2 µm pore size) upon which microorganisms from 50 mL of ethanol-fixed water samples were collected. A total of six samples were extracted, three from the Lake Room and three from Air Bell 2. Extraction was done following Nercessian et al. [27] with minor modifications. In brief, a CTAB extraction buffer containing SDS and N-laurylsarcosine was added to the samples together with an equal volume of phenol/chloroform/isoamylalcohol (25:24:1) solution. The filter samples were subject to bead-beating (FastPrep-24 5 G Instrument, MP Biomedical, Eschwege, Germany), followed by centrifugation (14,000 g), a cleaning step with chloroform, and nucleic acid precipitation with PEG-6000 (Sigma-Aldrich, Taufkirchen, Germany). The precipitated nucleic acids were rinsed with 1 mL of 70% ethanol, dried and dissolved in water. DNA was digested by two sequential treatments with the TurboDNAfree Kit (Invitrogen Thermo Fisher Scientific, Dreieich, Germany) following the manufacturer’s instructions. DNA removal was evaluated using a PCR for 16 S rRNA gene. First strand cDNA was then generated using the High-Capacity cDNA Reverse Transcription Kit (Applied Biosciences, Thermo Fisher scientific), and was sent for sequencing at the Core Genomic Facility at RUSH University, Chicago, IL, USA.
Other Thiovulum spp.
Samples of Ca. T. karukerense [26] and Ca. T. imperiosus [2] were respectively obtained from veils developing above the sediment and on bones deployed in marine mangrove in Guadeloupe (Lesser Antilles). Samples were collected manually between March and May 2021 in ~1 m depth (16°16'32.7“N 61°33'28.5“W). While dense veils of Ca. T. karukerense naturally form above the mangrove sediment throughout the year, we had to deploy cleaned pig bones as described before [2] to collect live Ca. T. imperiosus individuals. The veil formation of this species depends on the anaerobic degradation of lipids from bone-marrow by marine sulfate reducing bacteria which produce high concentrations of hydrogen sulfide. Candidatus T. imperiosus veils usually appear after 8 to 10 days of deployment. Both marine veils were collected manually using distinct 60 mL syringes, brought to the laboratory. The Ca. T. karukerense sample was processed for DNA extraction within 1 h as described in the next section. The sample of Ca. T. imperiosus contained several hundreds of cells easily distinguishable under a microscope by their large size (~50 µm in diameter). Single Ca. T. imperiosus cells were isolated under a stereomicroscope and processed for single-cell genomics as described in the next section.
Shotgun metagenomic sequencing (Illumina and Oxford Nanopore)
Shotgun metagenomic sequencing of DNA from Movile Cave was accomplished using both Illumina and Oxford Nanopore sequencing technologies. For Illumina sequencing, 1 ng of genomic DNA from each sample was converted to whole-genome sequencing libraries using the Nextera XT sequencing reagents according to the manufacturer’s instructions (Illumina, San Diego CA). Final libraries were checked for library insert size using the Agilent Bioanalyser 2100 (Agilent Technologies Santa Clara, CA) and quantified using Qubit spectrofluorometry. The final sample was sequenced using paired end 2 x 150 sequencing on an MiniSeq (Illumina) system.
A first pass of Oxford Nanopore sequences was obtained using the SQK LSK109 ligation library synthesis reagents on a Rev 9.4 nanopore flow cell with the GridION X5 MK1 sequencing platform, resulting in a total of 131.8 Mbp of reads with a N50 of 1.3 kbp.
Oxford Nanopore sequencing was additionally performed on DNA amplified from individual Thiovulum aggregates. Libraries for Nanopore sequencing (four in total) were prepared using the LSK-108 kit following the manufacturer’s protocol using 1 µg of DNA but skipping the size selection step. The prepared libraries were loaded on two MIN106 R9 flow cells, generating a total of 5.7 Gbp of reads with a length N50 of ca. 3.7 kbp. Basecalling of all Oxford Nanopore reads was performed using Guppy 4.0.11.
A DNA library for Ca. Thiovulum imperiosus was prepared from a single cell, visually identified based on its large diameter. It was isolated under the stereomicroscope and washed twice in 0.1 µm filtered seawater. Seawater was removed and the cell was stored at −80 °C until further processing. The cell was thawed, and we amplified the genomic DNA by multiple displacement amplification using the REPLI-g kit (Qiagen). A DNA library was created from 200 pg of DNA from the amplified product using Nextera XT DNA library creation kit (Illumina).
A DNA library for Ca. Thiovulum karukerense was prepared from a DNA extraction performed on thousands of cells pipetted under a stereomicroscope and transferred into 0.1 um filtered seawater. Cells were then transferred to a second vial containing new 0.1 µm filtered seawater. To limit contaminations to a minimum, this step was repeated three more times. Cells were pelleted and genomic DNA was extracted with the DNeasy kit (Qiagen) following the manufacturer’s protocol. DNA libraries were created from 200 pg of DNA using Nextera XT DNA library creation kit (Illumina). Both libraries were sequenced on an Nextseq High Output (Illumina) platform.
cDNA sequencing
The provided single stranded cDNA was adjusted to 45 µL with water and sheared with the Rapid Shear gDNA shearing kit (Triangle Biotechnology, Durham, NC, USA). Briefly, 5 µL of Rapid Shear Reagent was added to the bottom of a 24-well PCR plate. cDNA (45 µL) was then added to the well and the plate was sealed and held on ice till shearing. Shearing took place for 5 min in a sonication bath. The resulting DNA of about 350 bp in length, was used directly in the Swift 1 S protocol (Accel-NGS 1 S Plus kit, Swift Biosciences, Ann Arbor, Mi, USA). The resulting sheared DNA was adjusted to 5 ng/µL, and a total of 75 ng was used for library prep, except for sample LR1 (Lake Room) that had low cDNA concentration. For this sample, the maximum volume of 15 µL was used as input. Library prep was as per the Swift Protocol with 6 cycles of PCR during indexing. Following library prep, all libraries were pooled in equal volume by combining 2 µL of each library for a final bead clean up with 0.85X AmpPure beads (Beckman Coulter Life Sciences, Indianapolis, IN, USA). This QC pool was then sequenced on an MiniSeq MO flow cell (Illumina). The resulting index distribution was used to re-pool the libraries for an SP flow cell (Illumina) sequencing run with sample LR1 pooled at maximum volume available.
Sequencing data generated in this study were deposited in NCBI Sequence Read Archive. Data from Movile Cave is available under bioproject accession number PRJNA673084. The genome of Thiovulum sp. from Frasassi is available under bioproject accession number PRJNA846597. The genomes of Ca. T. imperiosus, and Ca. T. karukerense are available under bioproject accession number PRJNA830902.
Metagenomic data analysis
To obtain information on relative Thiovulum abundance in Movile Cave, the raw short-read libraries (metagenomic) were analyzed with phyloFlash (V 3.3; [28]) that provides the relative abundance and taxonomic annotation of 16 S rRNA genes from metagenomic libraries.
To assemble the genome of the Movile Cave Thiovulum, nanopore reads were assembled using Flye 2.8.1-b1676 [29] with default parameters. The resulting assembly graph (GFA) was examined using Bandage [30], allowing to delineate a set of high-coverage (>300X) contigs against a background of low-coverage (<100X) contigs. To verify that these high-coverage contigs corresponded to Thiovulum, the published proteome of Thiovulum ES [11] was aligned on the GFA using tblastn [31] within Bandage (parameters: minimum identity 70%, minimum coverage 70%), revealing that nearly all tblastn hits were concentrated on the high-coverage contigs and vice-versa. The GFA was therefore pruned to retain only the high-coverage contigs, which were all interconnected. The remaining 31 contigs were exported as FASTA then scaffolded using SLR [32]; the nine resulting scaffolds were mapped back to the GFA to resolve most repeats, and the remaining repeats were resolved manually until obtaining a circular genome. A final polishing step was performed with unicycler-polish from Unicycler v0.4.9b [33] using the complete set of Illumina reads (for a total depth of coverage of 12X of the genome) and the subset of Nanopore reads longer than 5 kb (coverage ca. 50X). Polishing consisted of two cycles of pilon 1.23 [34], one cycle of racon 0.5.0 [35] followed by FreeBase [36], then 30 additional cycles of short-read polishing using pilon 1.23, after which the assembly reached its best ALE score [37].
Thiovulum sp. genome assembly from public databases
To obtain genomic information from additional cave-dwelling Thiovulaceae we downloaded all available metagenomic libraries from the Frasassi caves in Italy (SRR10997432, SRR1559028, SRR1559230, SRR1559353, SRR1560064, SRR1560266, SRR1560848, SRR1560849, SRR1560850, SRR8191123, SRR8194889, SRR8197024, SRR8200784, SRR8202337, SRR8203764). The short read libraries were quality-trimmed using Trimmomatic [38] and scanned for the presence of Thiovulum 16 S rRNA gene using PhyloFlash [28], revealing that library SRR1560850 contained >170,000 reads of Thiovulum sp. 16 S rRNA gene. A metagenomic assembly of library SRR1560850 was therefore conducted using Megahit [39], after which the assembly was binned using Metabat2 [40]. The obtained bins were taxonomically annotated using the GTDB-TK tool [41] resulting in one Thiovulum bin. The phylogenetic tree generated by the GTDB-TK tool from a single-copy marker gene multilocus alignment suggested that the Movile and Frasassi caves Thiovulum genomes were closely related, hence, both genomes were used to recruit all Thiovulum related reads from all Frasassi libraries. The obtained reads were re-assembled, binned and taxonomically annotated as above to obtain a more complete assembly.
Genome quality assessment, annotation, and comparison
The completeness of the Thiovulum genomes obtained was assessed using CheckM [42]. The continuity of the Movile Thiovulum genome was evaluated using the unicycler-check module in Unicycler v0.4.9b. Annotation of all genomes was performed using the command-line Prokka [43] and DRAM [44] tools as well as the KEGG [45], EggNOG 5.0 [46], PATRIC [47, 48], and RAST [49, 50] annotation servers. A COG [51] analysis was done using the ANVIO tool [52]. OperonMapper [53] was used to inspect the organization of genes into operons in the circular Movile Cave genome. CRISPRs where identified using CRISPR finder tool [54]. Metabolic models of the annotated genomes were calculated using PathwayTools (V25.5) [55] using the RAST annotation of each genome as the model basis. Components of type IV pili systems were identified using MacSyFinder [56] using precomputed models [57]. Average Nucleotide Identity (ANI) and Average Amino-acid Identity (AAI) were calculated using FastANI [58] and EzAAI [59], respectively.
16 S rRNA gene phylogenetic trees
A maximum-likelihood phylogenetic tree was calculated, including only 16 S rRNA gene sequences longer than 1200 nt, using FastTree 2 [60] using all Thiovulum sequences in the SILVA REF (V138.1) database (n = 71), three 16 S rRNA gene sequences obtained from the assembled genome of the Movile Cave Thiovulum (see below) and sequences of several different Sulfurimonas species as an outgroup. The tree was calculated using the generalized time-reversible model and the discrete gamma model with 20 rate categories. Local support values were calculated with the Shimodaira-Hasegawa test.
Transcriptomic analysis
The six libraries containing cDNA sequences (three from Air Bell 2 and three from Lake Room) were quality-trimmed using trimommatic [38] and mapped against the complete genome of the Movile Cave Thiovulum sp. using Salmon (version 1.6) [61]. Ribosomal RNA data were removed from the mapping results and TPM (Transcripts Per Kilobase Million) values were recalculated to reflect mRNA expression. Further removal of tRNA sequences did not alter the results. The RNA data was analyzed using the iDEP (v. 0.95) online tool [62] that provides an online graphical user interface for the DeSEQ2 [63] and Limma [64] packages for RNAseq analysis. Differential expression was considered significant with a 2-fold difference and a false discovery rate smaller than 0.1. Taxonomic composition of the active community was obtained by analyzing the 16 S rRNA gene from the transcriptomic read libraries using PhyloFlash as above (V 3.3; [28]). Viral transcripts were identified using VirSorter2 (v.1.1) [65], annotated against the NCBI viral refseq database [66] release 209 using BLAST and quantified using Salmon version 1.6 [61].
Water chemistry
Temperature, pH, and TDS (total dissolved solids) were measured using an Extech 341350A-P Oyster Series pH/Conductivity/TDS/ORP/Salinity Meter (FLIR System, Nashua, NH, USA) in 2019 and with a Hanna Multiparameter HI9829 (Hanna Instruments Inc., Woonsocket, RI, USA) in 2020.
Measurements of NH4+, SO42-, NO2-, NO3- were obtained as part of the cave monitoring conducted by some of the co-authors and were analyzed using ion chromatography as previously described [67, 68]
Cell enumeration and Electron microscopy and elemental analysis
For cell enumeration, two 15-mL water samples from the Lake Room were collected from outside of the water to avoid disturbance by divers and preserved on-site with formaldehyde to a final concentration of 4%. Thiovulum cell enumeration was done on a 96-grid fields 5-mL chamber microscopy slide using a light microscope (Olympus BX51, Olympus, Tokyo, Japan). Cell counts were averaged from 15-grid squares.
For transmission electron microscopy, two unpreserved 15-mL samples collected from the water surface in Movile Cave were centrifuged, and the microorganisms were resuspended and fixed for 2 h with 2.7% glutaraldehyde in phosphate buffered saline (1× PBS). The cells were then rinsed three times in 1× PBS, and finally fixed for 1 h with 2% osmic acid in 1× PBS. The cells were harvested again by centrifugation, dehydrated in graded acetone-distilled water dilutions, and embedded in epoxy resin. Sections of about 100 nm thickness were produced with a diamond knife (Diatome, Hatfield) using a Leica UC6 ultramicrotome (Leica Microsystems, Wetzlar, Germany) and were stained with lead citrate and uranyl acetate (Hayat, 2001). The grids were examined with a Jeol JEM transmission electron microscope.
Five samples were collected for scanning electron microscopy (SEM) and bright-field scanning transmission electron microscopy (BFSTEM). These were fixed on site with 2.7% glutaraldehyde in 1× PBS from concentrated stocks. The samples were transported to the microscopy center within 24 h where they were air dried in the lab on 0.22 µm mesh-sized Millipore filters, without further handling or pre-concentration step. Samples were sputter coated with 10 nm gold and examined for SEM on a JEOL JSM 5510 LV microscope (Jeol, Japan), and for BFSTEM on a Hitachi SU8230 (Tokyo, Japan).
Energy-dispersive X-ray spectroscopy (EDX) analysis was performed using an EDX analyzer (Oxford Instruments, Abingdon, UK) and with the INCA 300 software.
Results
Field observations
A pale-white veil, with a vertical thickness of 2 to 3 mm, was observed at and below the water surface in Movile Cave (Fig. 2), resembling microbial veils described for sulfur-oxidizing bacteria [7, 68]. Nevertheless, in Movile Cave, this agglomeration of cells did not form slime or a strongly cohesive aggregation. By contrast, the samples used to assemble the Frasassi caves Thiovulum genome were obtained from stream biofilms [69].
Water chemistry
Physico-chemical parameters of the Movile Cave water, as measured in the Lake Room (Table 1 and Supplementary Table 1) resembled those from previous studies of the cave [18] and revealed minimal differences between water sampled at 1 m depth (water inlet) and near-surface water.
Microscopy
A veil similar to that seen in the Lake Room (Fig. 2A, B) occurred in Air Bell 1 and was even more pronounced in Air Bell 2 (Fig. 2C, D). In Air Bell 2 the veil consisted of large, spherical to ovoid, bacterial cells (Fig. 3A, B) fitting the description of the genus Thiovulum. These cells had a diameter of 12–16 µm, contained 20–30 sulfur globules each (Fig. 3A–C), and occurred in the Lake Room at densities of approximately 5.5 × 103 cells/mL. Transmission Electron Microscopy (TEM) observations showed that these large cells were Gram-negative (Fig. 3D–F) and confirmed the existence of 20–30 irregularly shaped sulfur inclusions within each of the cells (Fig. 3D). Light and TEM imaging revealed Thiovulum-like cell divisions along the long cell axis (Fig. 3B, D). Short peritrichous filamentous appendages (Fig. 3E, F) observed on the surface of the cells resemble those noticed earlier on other Thiovulum species [6]. These filaments measured <3 µm in length (Fig. 3E) and between 20–30 nm in diameter (Fig. 3G). Closer analysis revealed that at least some of the larger structures consisted of bundles of thinner filaments (Fig. 3H) with diameters of 9–12 nm each (Fig. 3H, I). Scanning electron microscopy (SEM) revealed that some of these filaments connected the cells via multiple or single threads (Fig. 3J, K). Energy-dispersive X-ray (EDX) analysis (Fig. 3L, M) confirmed that the intracellular globules contained sulfur (20.9–26.1%), along with elements common in organic matter such as carbon (49–49.2%) and oxygen (21.1–24.6%), and a few other elements in low abundance such as sodium (2.4–3.4%) and phosphorus (1.2–2.2%).
Optical images of giant globular cells colonizing the subsurface veil from Movile Cave (A, B) including a cell undergoing division (B). Each cell carries 20 to 30 sulfur inclusions (C) and large bright spots in A, B. TEM images of Thiovulum show the cellular localization of sulfur inclusions of various shapes and sizes (D white spots). Ovoid cells divide along their long axis (B, D). The region where the cell membrane is not fully closed between dividing cells is marked with three black arrows (D). The cell is densely covered in short filamentous structures (E) protruding from the outer membrane (F). These structure with an average diameter of ca. 30 nm (G), are bundles of thinner filaments (H) with a diameter of 9–12 nm (H, I). The filaments often appear to connect Thiovulum cells one to another (J, K). EDX analysis on Thiovulum cells (L) inspected under SEM show the typical elemental composition of the cells (M) and confirm the high sulfur content of the internal globules. Note that the height of the peaks in the EDX spectra do not correlate with the element’s ratio but with the X-ray signal intensity. A, B transmitted light micrographs. C, J–L Scanning Electron Micrographs. D, F transmission electron micrographs. E, G–I scanning transmission electron micrographs.
Phylogenetic identification and relative abundance of Thiovulum sequences
Thiovulum was found in highest abundance (sequence frequency) in Air Bell 2 (<35%), followed by Lake Room (<5%) and submerged microbial mats (<0.9%) (see pie charts in Fig. 1). A detailed community composition based on 16 S rRNA genes obtained from the metagenomic libraries is presented in Supplementary Fig. 1. Thiovulum rRNA sequences made up >94% both in the Lake Room and in Air Bell 2 (Supplementary Fig. 1), revealing its dominance in the active microbial community in both cave compartments at the time of sampling.
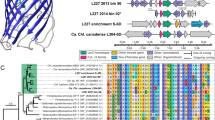
The 16 S rRNA gene sequences obtained from the closed genome of the Movile Cave Thiovulum, alongside other Thiovulum sequences obtained from Movile Cave in an earlier study [70], formed a separate clade together with other cave and subsurface, freshwater Thiovulum spp., specifically from the sulfidic Frasassi caves in Italy (Fig. 4A). This clade belonged to one of two clades, termed here 1 and 2, of otherwise marine Thiovulum spp., the other one of which included Thiovulum ES, for which a draft genome is available [11] as well as the 16 S rRNA gene sequences of Ca. T. imperiosus [2] and Ca. T. karukerense [26].
Maximum-likelihood placement of the 16 S rRNA gene of Thiovulum spp. available in the SILVA SSU database (V138.1 [103]) alongside those from the cave (Ca. Thiovulum stygium) and marine Thiovulum genomes introduced in this study (A) whose phylogenomic placement based on a concatenated protein alignment is presented in B. The protein alignment was generated using GTDB-TK [41]. The Shimodaira-Hasegawa local support values (ranging from 0 to 1) are shown next to each node. Abbreviation “st.” in figure refers to strain.
A phylogenetic tree was constructed from a multilocus alignment of single-copy marker genes from the marine Thiovulum spp.: Thiovulum ES [11], Ca. T. imperiosus, Ca. T. karukerense, and a public assembly (GCA_012963895.1) from the Tui Malila hydrothermal plume classified as Thiovulum [71], along the freshwater genome of Thiovulum from Movile Cave, a metagenome assembled genome from the Frasassi caves, and multiple groups of Campylobacterales. The tree confirms the existence of two Thiovulum clades, and the separation of the freshwater Frasassi and Movile Thiovulum genomes from the marine strains. The latter also show a higher diversity among the four marine Thiovulum spp. (Fig. 4B). This is further seen in the high Average Nucleotide Identity (ANI) and average Amino Acid Identity (AAI) between the two cave strains as compared to their dissimilarity from the marine Thiovulum spp. and the dissimilarity of those among each other. (Fig. 5A). Both AAI and ANI analyses revealed that while the Movile and Frasassi strains are highly similar (96% AAI and ANI), they are equally distant from the marine strains as those exhibit ANI values ranging between 74–78% and AAI between 62–68%.
Average Nucleotide Identity (A lower triangle), Average Amino-acid Identity (A Upper triangle) among the 6 Thiovulum strains, the relative evolutionary divergence (RED) value (A right column), and an overall comparison of the same genomes (B). The black bars represent the presence absence of a COG function in each genome. RED values were obtained from the GTDB-TK [41] tool for the newly inserted genomes. The term Candidatus was omitted from the names of Thiovulum spp. due to space considerations.
Genome analysis
Of the four genomes assembled in this study, only the assembly of metagenomic data from Movile Cave resulted in a closed circular genomic sequence (Fig. S2). The size of the assembled genomes ranged between 1.72–2.20 MBP and GC content varied between 28–37% (Table 2). Genome completeness was estimated using CheckM [42] ranging between 90–96% and evalG (as incorporated into the PATRIC server (now BV-BRC); [72]) ranging between 95.8% and 100% with the exception of Thiovulum sp. Tui Malila which was evaluated at 66% completeness. Using a Campylobacteraceae specific set of marker genes did not improve the completeness prediction. The marker genes that could not be identified are listed in Supplementary Table 2. Of these 9 genes/domains (PF00308.13, PF02464.12, PF04085.9, PF08299.6, PF08459.6, PF12344.3, TIGR00615, TIGR02124, TIGR03423) could not be found in any of the 6 genomes and additional 4 genes/domains were missing from both freshwater genomes (TIGR00196, TIGR00539, TIGR00121, PF03853.10). All genomes were analyzed using different tools with the results summarized in Supplementary Datasets 1–5.
Carbon metabolism
Similar to published Thiovulum ES, all genes required for C fixation via the reductive TCA cycle were identified. in the new freshwater and marine Thiovulum genomes. The oxidative TCA cycle was found to be complete as well in all the Thiovulum species with the citrate synthase gene (EC 2.3.3.1) replaced by ATP-citrate lyase. (EC 2.3.3.8) (Fig. 6, Supplementary Figs. 3–8, Supplementary Dataset 1–6).
Elements or reactions are colored differently whether they occur only in cave strains (purple; both genomes), marine strains (yellow; at least two of four strains), and common to both environments (peach; at least one cave and one marine, genome). Core elements or reactions are colored in brown. Green elements are unique to the Movile Cave genome. Gray arrows in the reductive TCA (rTCA) cycle show missing reactions. The sulfur/nitrogen metabolism model was drawn based on Grote et al. [104], Hamilton et al. [69], and Poser et al. [84]. SQR sulfide-quinone oxidoreductase, SDH/FR succinate dehydrogenase/fumarate reductase, FDH formate dehydrogenase, Qu quinone, Cyt b/c1 quinone cytochrome oxidoreductase, cbb3 cytochrome c oxidase, ccp cytochrome c peroxidase, NAP periplasmic nitrate reductase, NAR membrane bound nitrate reductase, Psr polysulfide reductase, εHao Epsilonproteobacterial hydroxylamine oxidoreductase, Ttr tetrathionate reductase, Rhd rhodanese-related sulfurtransferase. CRISPRs were identified in all genomes with the Movile strain data depicted here. Comparative gene expression between Thiovulum in Air Bell 2 (red) and the Lake Room (blue) are shown for each gene depicted where the TPM value was above 10. Genes were the TPM value was below 100 or above 1000 are marked with a diamond and inverse triangle, respectively. For proteins consisting of multiple subunits, the expression is the genes encoding for one of the subunits. For SQR and RhD expression is shown for both copies of the genes.
Sulfur metabolism
All annotation approaches (Supplementary Datasets 1–6) revealed only few genes involved in dissimilatory sulfur cycling, including two versions of the sulfide:quinone oxidoredutase (Type VI and IV) that oxidizes sulfide to polysulfide, and the polysulfide reductase gene nrfD that carries out the reverse process. In the Movile strain, nrfD was found in a 3-gene potential operon together with the large subunit of the assimilatory nitrate reductase (narB) and the ttrB; tetrathionate reductase subunit B. Rhodanese sulfur transferase proteins were identified in all genomes. In the Movile strain these were located in a 6-gene operon containing two other subunits of a nitrate reductase (narH and narG). As for the two other genes in this operon, one is related to cytochrome c and the other could not be annotated (G2Y-810). The tauE sulfite exporter was identified in all genomes as well, in the Movile strain genome as part of a 5-gene operon involved in the transport of molybdate (modCABD). Sulfite dehydrogenase, dissimilatory sulfite reductase (dsrAB), the sox genes or adenylyl sulfate reductase (aprAB) that carry out the sulfide oxidation to SO42- could not be found by any of the annotation tools nor by manual BLAST against all sequences available for each of those proteins in the UniProt database.
Nitrogen metabolism
The membrane-bound nitrate reductase (nar) found also in Thiovulum ES, was identified in all Thiovulum spp. genomes. The Movile and Frasassi strains possess in addition also periplasmatic nitrate reductases encoded by the nap genes encoded in one operon (napAGHB_F). A hypothetical protein encoded in this operon in the Movile genome is likely part of the napF gene (G2Y-1099) as seen by BLAST analysis in other Campylobacteraceae. The gene for hydroxylamine dehydrogenase, which is often encountered in genomes from Campylobacterota [73] (formerly referred to as Epsilonproteobacteria [74]), was also identified in the Movile strain genome as part of a 3-gene operon including two unannotated hypothetical genes.
Chemotaxis and motility
Thiovulum spp. are highly motile bacteria, therefore, we inspected motility and chemotaxis genes. All genes necessary for flagellar assembly were found in all Thiovulum genomes. The chemotaxis genes, cheA, cheW, cheD, and cheY were identified as well as additional cheY-like domains. The cheB gene was found only in the Movile strain (G2Y-1843), and cheX was missing from Thiovulum spp. strains karukerense and ES. The cetA and cetB (G2Y-174:176 cetABB’) energy taxis genes were identified in the Movile strain while all other genomes contained only cetB in 1 or 2 copies. The homolog of the Escherichia coli aerotaxis (aer) gene was found in the genomes of Ca. T. imperiosus and of the Movile strain. Additionally, multiple methyl-accepting chemotaxis proteins were identified in each genome.
Genes for type IV pili assembly and secretion were identified in all genomes. While some of these genes were identified by the classical annotation tools, other were recognized among the “hypothetical proteins” using HMM search against various families of type IV pili [57]. The various genes make up all the necessary genes for type IV pili, yet their closest relatives in the database come from different families of type IV pili, e.g., T4Pa and T4Pb.
Transcript analysis
To confirm that Thiovulum is transcriptionally active in Movile Cave, as well as to assess which genes of the predicted metabolic pathways are transcribed, samples from Air Bell 2 and from the Lake Room of Movile Cave were collected for RNA analysis. 16 S rRNA transcripts of Thiovulum dominated all samples, making up more than 94% of the active community (Fig. S1) even though Thiovulum DNA was rare in the Lake Room in our DNA samples. Despite the similarity in relative rRNA transcript abundance, the gene expression profiles differ significantly between the two sites (Fig. 7). In both the heatmap (Fig. 7A) and the principal component analysis (Fig. 7B), the samples from the different environments clustered separately, with clear clusters of genes differently expressed in the two cave compartments. Differential expression analysis (Fig. 7C; Supplementary Dataset 1) revealed that 222 genes were more expressed in the Lake Room compared to Air Bell 2, while the opposite comparison resulted in 42 genes. In contrast, the TPM normalized expression of the housekeeping genes rpoB, gyrB, and bipA was similar in both compartments, therefore, confirming that the differential expression of genes is not a methodological bias. Over half of the genes more expressed in the Lake Room encoded for hypothetical proteins to which no function could be assigned. Retron-type reverse transcriptases were the most dominant group of genes (n = 15) also exhibiting some of the highest transcription level with TPM values up to 19,000. Genes over expressed in samples from Air Bell 2 were related to energy generation including cytochromes c and b as well as F-type ATP synthase. The entire gene expression data is available in Supplementary Dataset 1 and is additionally depicted in Fig. 6 next to the displayed genes or functions.
Comparison of mRNA transcriptomic profiles of the Ca. Thiovulum stygium strain Movile obtained from triplicates samples collected in Air Bell 2 and in the Lake Room. A heatmap shows that the two environments are separated from one another with clusters of genes expressed more, or less in one of the two environments (A). The values in the heatmap are log-transformed TPM values and normalized using each gene’s standard deviation. Principle components analysis (B) demonstrates the separation between samples mainly across PC1 likely representing sample location. Differential expression analysis (C) revealed that 222 genes are significantly more expressed in the Lake Room as compared to Air Bell 2, whereas 48 genes. are significantly less expressed.
Discussion
We investigated the morphological, phylogenetic, and genomic aspects of a planktonic Thiovulum species that belongs to a newly recognized clade of freshwater Thiovulum. We were able to identify, and assemble, another genome of a freshwater Thiovulum from public metagenomic data from the Frasassi caves. We further compared these two genomes from freshwater clade to the genomes of four marine Thiovulum spp.: T. sp. ES [11] and T. sp. GCA_012963895.1 (Tui Malila) [71], retrieved from public databases, and the genomes of Ca. T. imperiosus [2] and Ca. T. karukerense [26] whose genomes are presented here for the first time. The main aspects of this comparison are depicted in Fig. 6 and in more details in Supplementary Dataset 7. Supplementary Figs. 3–8 offer a detailed, Pathway-Tools automatic- reconstruction of the pathways detected in each of the six genomes.
Phylogeny
Cave Thiovulum (Movile and Frasassi caves) are phylogenetically distinct from marine species. This is evident in both genomic and 16 S rRNA gene phylogenetic analyses for which more sequence data were available. The rRNA gene analysis also highlighted that, all sequences obtained from sulfidic caves, or subsurface environments (e.g., a drinking water well) formed a freshwater subclade in the otherwise marine Clade 2, suggesting a transition of these species from a marine to a freshwater environment. The degree of genetic diversity of the marine genomes was higher in comparison to the freshwater ones, as supported by the ANI and AAI analyses. This phenomenon could be a result of the freshwater cave Thiovulum spp. being isolated in stable subsurface environments and, therefore, diverging less from their ancestors [75]. Nevertheless, this should be further evaluated as more freshwater and marine Thiovulum, species become available for comparison. In another case of marine-freshwater transitions, it was also evidenced that freshwater Pelagibacteraceae evolved slower than their marine counterparts, revealing highly similar strains across large geographical distances [76] similar to the Movile and Frasassi Thiovulum strains.
Core marine genes (KEGG orthologs or COG) could be identified; however, these do not functionally differentiate marine from freshwater Thiovulum. Similarly, we could not detect differences in copy numbers of genes related to osmoregulation. As such, given the phylogenetic indication that the freshwater clade emerged from the marine Thiovulum, it may be that the osmoregulation genes occurring in the marine genomes, are either necessary in the cave water, or were not yet lost by the freshwater Thiovulum spp.
We propose that the two cave strains belong to the same Thiovulum species while the other 4 genomes represent 4 different Thiovulum species. In absence of cultures, to properly evaluate and define new bacterial species, we rely on molecular analyses such as ANI, AAI, and RED. Despite the great difference in genomic sequence (ANI analysis), the similarity in amino acids sequences (AAI > 62%) leads us to conservatively suggest that the investigated genomes belong all to the genus Thiovulum rather than different genera within the Thiovulaceae family [77]. The obtained RED values for the 4 new species whose genomes are presented here are between 0.83–0.88. Given the clustering of all Thiovulum genomes together, these values are in good agreement with a genus level separation as seen in Parks et al., [78] and the GTDB (v207) RED statistics report (https://gtdb.ecogenomic.org/stats/r207). This is also supported by a K/θ analysis [79,80,81] where the species separation limit was defined as K/θ > 6 (corresponding to ca. 99% species separation probability: Fig. S10). Accordingly, we propose naming the cave Thiovulum sp. as Candidatus Thiovulum stygium (sp. nov.). Stygium, referring to the mythical underworld river Styx, appropriately describes the aquatic underground biome of cave Thiovulum.
Sulfur and nitrogen metabolism
Thiovulum spp. are capable of oxidizing reduced sulfur compounds, specifically sulfide and elemental sulfur. Sulfide oxidation is evidenced by its sulfur inclusions. The amount and type of sulfur inclusions in cells is influenced by the concentrations of H2S and O2 in the environment. Typically, cells store elemental sulfur when H2S is abundant in the environment, and later use the intracellular reserves of sulfur when the sulfide source in the environment is depleted [10, 82]. Sulfur inclusions were also shown to form when the supply of O2 is limited and as a result the sulfur cannot be entirely oxidized to soluble sulfite, thiosulfate, or sulfate. Complete depletion of sulfur inclusions from cells is not likely in Movile Cave where abundant H2S (< 1 mM) [21]) is available continuously and where O2 is scarce in most habitats, and specifically in Air Bell 2 [83]. The analysis of the 6 Thiovulum genomes identified the SQR gene responsible for the oxidation of sulfide to elemental sulfur, which was also highly expressed in the Movile Cave transcripts. Nevertheless, the genes required for further oxidizing elemental sulfur to sulfate, via any of the known mechanisms, were not found. An exception to this is the possible oxidation of sulfite to sulfate via the intermediate adenylyl sulfate as suggested for Thiovulum ES, for which the gene encoding the sulfate adenylyl transferase was originally found [11] yet, according to our annotation the necessary adenylyl-sulfate reductase genes apr (EC1.8.4.9) or aprA (EC1.8.99.2) are missing from all the Thiovulum genomes.
Marshall et al. (2012) proposed that Thiovulum undergoes frequent (daily) oxic/anoxic cycles that prevent continuous accumulation of elemental sulfur in the cell. We advance three additional options by which the Thiovulum may avoid excessive sulfur accumulation. First, the presence of a polysulfide reductase (nrfD) suggests that the cells can reduce polysulfide back to sulfide (Fig. 6). In the case of the Ca. T. stygium strain Movile, activity of nrfD was confirmed by the transcriptomic data. Second, the identification of different rhodanese genes, known to be involved in thiosulfate and elemental sulfur conversion to sulfite [84], and of a sulfite exporter (tauE) in Ca. T. stygium strain Movile, all of which were found to be expressed in our samples, suggests that Thiovulum may oxidize elemental sulfur to sulfite and transport the latter out of the cell. The rhodanese genes found in the same predicted operon as the respiratory nitrate reductase genes (nar), were ca. 5-fold more expressed than a second pair of rhodanese genes. Third, we propose that cave-dwelling Thiovulum spp (Ca. T. stygium) are capable of dissimilatory nitrate reduction to ammonia (DNRA) using elemental sulfur [85] a process already shown in Campylobacterota (e.g., Sulfurospirillum deleyianum) [86]. Ca. T. stygium strain Movile and strain Frasassi contain not only the nar (narGH) genes for nitrate reductase, but also by the nap genes, coding the periplasmatic nitrate reductase known for its higher affinity and ability to function in low nitrate concentrations [87]. Additionally, they harbor the gene for the epsilonproteobacterial hydroxylamine dehydrogenase (εhao). Hydroxylamine dehydrogenase is known from other Campylobacterota (e.g., Campylobacter fetus or Nautilia profundicola) and was shown to mediate the respiratory reduction of nitrite to ammonia [73]. The hao gene was not found in the genome of any of the marine Thiovulum spp., suggesting that the hao gene may not be part of the core Thiovulum genome. Normally, Campylobacterota that utilize hydroxylamine dehydrogenase do not have formate-dependent nitrite reductase, matching the annotation of Ca. T. stygium strain Movile and strain Frasassi. Campylobacterota typically use periplasmic nitrate reductase (nap) and do not have membrane-bound narGHI system [88, 89]. All marine Thiovulum species have only nar systems while the Ca. T. stygium strain Movile and strain Frasassi have both types, suggesting that the nap genes may be a later acquisition by cave-dwelling Thiovulum. Nevertheless, while genomic information is suggestive of the presence or absence of specific enzymes and pathways, additional experiments and gene expression data are required to determine which of the genes are utilized and under which environmental conditions. Our transcriptomic analysis points out that at the time of sampling the nap and εhao genes were minimally expressed as compared to other sulfur and nitrogen metabolism genes (Fig. 6, Supplementary Dataset 1), suggesting that the DNRA pathway was not very active in the Thiovulum community at the time of sampling. In contrast, the high expression of both copies of rhodanase genes as well as the sulfite exporter (tauE) suggest that elemental sulfur may have been converted to sulfite and excreted.
We propose that, if O2 is available, sulfide is oxidized to elemental sulfur with oxygen as an electron acceptor (as may have been the case for part of the community at the time of sampling given the high expression of cytochrome c oxidase cbb3; Fig. 6). However, when cells are located below the O2 penetration depth, the Movile Cave Thiovulum may oxidize sulfide using NO3- as an electron acceptor, in a process of dissimilatory nitrate reduction to ammonium, as in other Campylobacteraceae.
Air Bell 2 vs. Lake Room
Thiovulum did not appear to be the most abundant organism in the surface water of the Lake Room either by microscopy- or DNA based observations. Yet, its rRNA made up over 94% of the transcriptomic data, similar to the RNA samples from the hypoxic Air Bell 2. Nevertheless, it is known that community profiles obtained from DNA, representing pseudo-abundance, and those from RNA, representing degree of activity, can substantially differ from each other [90, 91]. Additionally, we cannot exclude that the community composition may have shifted during the time lapse between the DNA and RNA sampling. The presence and high activity of Thiovulum at the surface of both the Lake Room and Air Bell 2, environments that differ significantly in the overlaying atmosphere (normoxic vs. hypoxic, respectively), suggest that either the Ca. T. stygium strain Movile is not sensitive to these differences in O2 concentration, or it mostly aggregates at different locations in the stagnant, oxic surface (<1 mm) of the different cave chambers.
The expression profiles differed significantly between the Lake Room and Air Bell 2, and it is evident (Fig. 6) that most genes recognizable as involved in cell metabolism had higher expression levels in Air Bell 2, though not all at statistically significant levels (Supplementary Dataset 1). More than half of the genes overexpressed by Thiovulum in the Lake Room could not be assigned any function, making it impossible to understand it’s specific metabolic activity in that compartment of the cave. However, the high expression of retron-type reverse transcriptase and Type II restriction enzymes in the Lake Room can be indicative of an ongoing phage infection [92, 93], which may explain the reduced metabolic activity and elevated expression of defense systems, though only one CRISPR associated gene was highly expressed. Quantification of viral transcripts showed an overall higher expression in the Lake Room (Fig. S9); however, at this stage it is not possible to directly connect these transcripts to Thiovulum.
Cell motility and veil formation
Thiovulum sp. often forms large veils of interconnected cells. Additionally, the cells can attach to surfaces using a slime stalk secreted by the antapical organelle located at the posterior side of the cell [1, 10]. Short peritrichous filaments (Fig. 3D) observed on the surface of the cells from Movile Cave resemble those noticed earlier in Thiovulum species and referred to as flagella [6]. While all genes necessary for flagella assembly were found in the Movile Cave and all other Thiovulum sp. genomes, so were genes for type IV pili, though with closest homologs in different types of pili systems (T4Pa, T4pB, MSH). Evaluating available electron microscopy images, we suggest that future, focused studies, should evaluate whether all these filaments are indeed flagella.
Our electron microscopy images (as well as previous ones of connected Thiovulum cells) showed that the filamentous structures that occur all around the cell are formed in bundles and some of them are connecting cells. As these filaments are not exclusively polar, they are likely not secreted from the antapical organelle [1] the fibrillar structure of which was clearly depicted in earlier studies [10, 94]. Additionally, they are much thinner than the stalk-like structure shown by de Boer et al. [10]. These peritrichous structures around the cells are commonly regarded as flagella. The length of these structures based on de Boer et al. [10], Robertson et al. [1], and our electron microscopy data, is under 3 µm. This is much shorter than flagella that are typically >10 µm in length [95], and is closer to the 1–2 µm lengths known for pili. The diameter of the Thiovulum filamentous structures see in SEM images is in the range of 20–30 nm, 2–4 times larger than typical type IV pili [96] yet in the same range as reported for pili bundles [97] or seen in images of Campylobacterota pili [98] (which may or may not be bundled). Accordingly, bright-field scanning TEM analysis on some filaments revealed that these structures consist of multiple thinner pili with diameters <10 nm (Fig. 3). In light of these observations, we propose that at least some of these structures are pili, specifically type IV pili, for which the necessary genes were found in the genomes, and that are known, among other functions, to connect cells to surfaces or other cells [99, 100].
Petroff et al. [9] investigated the physics behind the 2-dimensional plane assembly of Thiovulum veils and suggested it to be a direct result from the rotational movement attracting cells to each other. Nevertheless, our SEM images showed cells that are physically attached one to the other, suggesting that several mechanisms and steps may be involved. Type IV pili retraction can generate forces up to 150 pN known to be involved in twitching motility in bacteria [99]. Caulobacter crescentus swims at speeds of up to 100 µm s−1 using a single flagellum aided by multiple pili. Thus, if coordinated, these pili may be part of the explanation of the swimming velocity of some Thiovulum species which is, at up to ca. 615 µm s−1, 5 to 10 times higher than that of other flagellated bacteria [7]. The genomic information and re-evaluation of electron microscopy data raise new questions concerning the nature of the extracellular structures on the surface of Thiovulum and call for new targeted investigations into this topic.
Conclusions
Movile Cave is an ecosystem entirely depending on chemosynthesis. We showed that the planktonic microbial accumulations are dominated by Thiovulum, a giant bacterium typically associated with photosynthetic microbial mats. We further showed that Thiovulum dominates the active fraction in surface waters of hypoxic and normoxic compartments of the cave, suggesting an ability to adapt to different O2 concentration, by means that remain to be studied.
Our results highlight the existence of a clade of cave and subsurface Thiovulaceae that based on genomic information differs significantly from marine Thiovulum. The genomes of two conspecific cave strains from Movile and Frasassi caves, included here in the new species Candidatus Thiovulum stygium, suggest that these bacteria can perform dissimilatory reduction of nitrate to ammonium, when O2 is unavailable. Thus, Thiovulum may play a role in the nitrogen cycle of sulfidic caves, providing readily available ammonia to the surrounding microorganisms. The coupling of DNRA to sulfide oxidation provides a direct and more productive source of ammonium.
This investigation of six Thiovulum genomes, coupled with observations of current and previous microscopy images, questions the number of flagella Thiovulum cells have, suggesting that many of the peritrichous filamentous structures around the cells are in fact type IV pili that may be involved in rapid movement and cell-to-cell interactions.
The collective behavior of Thiovulum is still a puzzle and there may be more than one mechanism keeping the cells connected in clusters or in veils. Our SEM images suggest the cells are connected by thread-like structures. Petroff et al. [9] showed, on another strain, that there is no physical connection between the cells, suggesting that the swimming behavior of individual cells is what keeps them together. More research is therefore needed to understand if these different mechanisms are driven by strain variability or by different environmental conditions.
Data availability
All sequenced data from Movile Cave is available under bioproject accession number PRJNA673084. The genome of Thiovulum sp. from Frasassi is available under bioproject accession number PRJNA846597. The genomes of Ca. T. imperiosus, and Ca. T. karukerense are available under bioproject accession number PRJNA830902. All other data are provided as part of this manuscript and its supplementary material, including annotation results of all genomes that are provided as supplementary datasets.
References
Robertson LA, Gijs Kuenen J, Paster BJ, Dewhirst FE, Vandamme P Thiovulum. In: Brenner DJ, Krieg NR, Staley JT, Garrity GM (eds). Bergey’s Manual of Systematics of Archaea and Bacteria. 2015. Wiley, New York, pp 1–4.
Sylvestre M-N, Jean-Louis P, Grimonprez A, Bilas P, Collienne A, Azède C, et al. Candidatus Thiovulum sp. strain imperiosus: the largest free-living Epsilonproteobacteraeota Thiovulum strain lives in a marine mangrove environment. Can J Microbiol. 2022;68:17–30.
Robertson LA, Kuenen JG, Paster BJ, Dewhirst FE, Vandamme P Thiovulum Hinze 1913, 195AL. In: Brenner DJ, Krieg NR, Staley JT, Garrity GM (eds). Bergey’s Manual of Systematic Bacteriology. 2006. Springer, New York, pp 1189–91.
Lackey JB, Lackey EW. The habitat and description of a new genus of sulphur bacterium. J Gen Microbiol. 1961;26:29–39.
Lauterborn R. Die sapropelische Lebewelt. Ein Beitrag zur Biologie des Faulschlamms natürlicher Gewässer. Verh Naturhist Med Ver Heidelb. 1915;13:395–481.
Wirsen CO, Jannasch HW. Physiological and morphological observations on Thiovulum sp. J Bacteriol. 1978;136:765–74.
Garcia-Pichel F. Rapid bacterial swimming measured in swarming cells of Thiovulum majus. J Bacteriol. 1989;171:3560–3.
Thar R, Fenchel T. True chemotaxis in oxygen gradients of the sulfur-oxidizing bacterium Thiovulum majus. Appl Environ Microbiol. 2001;67:3299–303.
Petroff AP, Wu X-L, Libchaber A. Fast-moving bacteria self-organize into active two-dimensional crystals of rotating cells. Phys Rev Lett. 2015;114:158102.
De Boer WE, La Rivière JWM, Houwink AL. Observations on the morphology of Thiovulum majus Hinze. Antonie Van Leeuwenhoek. 1961;27:447–56.
Marshall IPG, Blainey PC, Spormann AM, Quake SR. A single-cell genome for Thiovulum sp. Appl Environ Microbiol. 2012;78:8555–63.
Jorgensen BB, Revsbech NP. Colorless sulfur bacteria, Beggiatoa spp. and Thiovulum spp., in O2 and H2S microgradients. Appl Environ Microbiol. 1983;45:1261–70.
Fenchel T, Glud RN. Veil architecture in a sulphide-oxidizing bacterium enhances countercurrent flux. Nature. 1998;394:367–9.
Lascu C, Popa R, Sarbu SM. Le karst de Movile (Dobrogea de Sud). Rev Roum de Géographie. 1994;38:85–94.
Riess W, Giere O, Kohls O, Sarbu S. Anoxic thermomineral cave waters and bacterial mats as habitat for freshwater nematodes. Aquat Micro Ecol. 1999;18:157–64.
Engel AS. Bringing microbes into focus for speleology: an introduction. In: Engel AS (ed). Microbial Life of Cave Systems. 2015. De Gruyter, Berlin, München, Boston, pp 1–22.
Kumaresan D, Wischer D, Stephenson J, Hillebrand-Voiculescu A, Murrell JC. Microbiology of Movile Cave-A chemolithoautotrophic ecosystem. Geomicrobiol J. 2014;31:186–93.
Sarbu SM. Movile Cave: A chemoautotrophically based groundwater ecosystem. In: Wilken H, Culver DC, Humphreys WF (eds). Subterranean Ecosystems. 2000. Elsevier, Amsterdam, pp 319–43.
Rohwerder T, Sand W, Lascu C. Preliminary evidence for a sulphur cycle in movile cave, Romania. Acta Biotechnol. 2003;23:101–7.
Chen Y, Wu L, Boden R, Hillebrand A, Kumaresan D, Moussard H, et al. Life without light: microbial diversity and evidence of sulfur- and ammonium-based chemolithotrophy in Movile Cave. ISME J. 2009;3:1093–104.
Flot J-F, Bauermeister J, Brad T, Hillebrand-Voiculescu A, Sarbu SM, Dattagupta S. Niphargus-Thiothrix associations may be widespread in sulphidic groundwater ecosystems: evidence from southeastern Romania. Mol Ecol. 2014;23:1405–17.
Hutchens E, Radajewski S, Dumont MG, McDonald IR, Murrell JC. Analysis of methanotrophic bacteria in Movile Cave by stable isotope probing. Environ Microbiol. 2004;6:111–20.
Schirmack J, Mangelsdorf K, Ganzert L, Sand W, Hillebrand-Voiculescu A, Wagner D. Methanobacterium movilense sp. nov., a hydrogenotrophic, secondary-alcohol-utilizing methanogen from the anoxic sediment of a subsurface lake. Int J Syst Evol Microbiol. 2014;64:522–7.
Ganzert L, Schirmack J, Alawi M, Mangelsdorf K, Sand W, Hillebrand-Voiculescu A, et al. Methanosarcina spelaei sp. nov., a methanogenic archaeon isolated from a floating biofilm of a subsurface sulphurous lake. Int J Syst Evol Microbiol. 2014;64:3478–84.
Vlasceanu L, Popa R, Kinkle BK. Characterization of Thiobacillus thioparus LV43 and its distribution in a chemoautotrophically based groundwater ecosystem. Appl Environ Microbiol. 1997;63:3123.
Gros O. First description of a new uncultured epsilon sulfur bacterium colonizing marine mangrove sediment in the caribbean: Thiovulum sp. strain karukerense. FEMS Microbiol Lett. 2017;364:fnx172.
Nercessian O, Noyes E, Kalyuzhnaya MG, Lidstrom ME, Chistoserdova L. Bacterial populations active in metabolism of C1 compounds in the sediment of Lake Washington, a freshwater lake. Appl Environ Microbiol. 2005;71:6885–99.
Gruber-Vodicka HR, Seah BKB, Pruesse E. phyloFlash: Rapid small-subunit rRNA profiling and targeted assembly from metagenomes. mSystems. 2020;5:521922.
Kolmogorov M, Yuan J, Lin Y, Pevzner PA. Assembly of long, error-prone reads using repeat graphs. Nat Biotechnol. 2019;37:540–6.
Wick RR, Schultz MB, Zobel J, Holt KE. Bandage: interactive visualization of de novo genome assemblies. Bioinformatics. 2015;31:3350–2.
Gertz EM, Yu Y-K, Agarwala R, Schäffer AA, Altschul SF. Composition-based statistics and translated nucleotide searches: improving the TBLASTN module of BLAST. BMC Biol. 2006;4:41.
Luo J, Lyu M, Chen R, Zhang X, Luo H, Yan C. SLR: a scaffolding algorithm based on long reads and contig classification. BMC Bioinforma. 2019;20:539.
Wick RR, Judd LM, Gorrie CL, Holt KE. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol. 2017;13:e1005595.
Walker BJ, Abeel T, Shea T, Priest M, Abouelliel A, Sakthikumar S, et al. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One. 2014;9:e112963.
Vaser R, Sović I, Nagarajan N, Šikić M. Fast and accurate de novo genome assembly from long uncorrected reads. Genome Res. 2017;27:737–46.
Garrison E, Marth G. Haplotype-based variant detection from short-read sequencing. ArXiv 2012;1207.
Clark SC, Egan R, Frazier PI, Wang Z. ALE: a generic assembly likelihood evaluation framework for assessing the accuracy of genome and metagenome assemblies. Bioinformatics. 2013;29:435–43.
Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–20.
Li D, Liu C-M, Luo R, Sadakane K, Lam T-W. MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics. 2015;31:1674–6.
Kang DD, Froula J, Egan R, Wang Z. MetaBAT, an efficient tool for accurately reconstructing single genomes from complex microbial communities. PeerJ. 2015;3:e1165.
Chaumeil P-A, Mussig AJ, Hugenholtz P, Parks DH. GTDB-Tk: a toolkit to classify genomes with the Genome Taxonomy Database. Bioinformatics. 2019;36:1925–7.
Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015;25:1043–55.
Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–9.
Shaffer M, Borton MA, McGivern BB, Zayed AA, La Rosa SL, Solden LM, et al. DRAM for distilling microbial metabolism to automate the curation of microbiome function. Nucleic Acids Res. 2020;48:8883–8900.
Kanehisa M, Sato Y, Kawashima M, Furumichi M, Tanabe M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2016;44:D457–62.
Huerta-Cepas J, Szklarczyk D, Heller D, Hernández-Plaza A, Forslund SK, Cook H, et al. eggNOG 5.0: a hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res. 2019;47:D309–D314.
Davis JJ, Wattam AR, Aziz RK, Brettin T, Butler R, Butler RM, et al. The PATRIC Bioinformatics Resource Center: Expanding data and analysis capabilities. Nucleic Acids Res. 2020;48:D606–12.
Brettin T, Davis JJ, Disz T, Edwards RA, Gerdes S, Olsen GJ, et al. RASTtk: a modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci Rep. 2015;5:8365.
Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, et al. The RAST Server: rapid annotations using subsystems technology. BMC Genomics. 2008;9:75.
Overbeek R, Olson R, Pusch GD, Olsen GJ, Davis JJ, Disz T, et al. The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST). Nucleic Acids Res. 2014;42:D206–14.
Tatusov RL, Galperin MY, Natale DA, Koonin EV. The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 2000;28:33–36.
Eren AM, Esen ÖC, Quince C, Vineis JH, Morrison HG, Sogin ML, et al. Anvi’o: an advanced analysis and visualization platform for 'omics data. PeerJ. 2015;3:e1319.
Taboada B, Estrada K, Ciria R, Merino E. Operon-mapper: a web server for precise operon identification in bacterial and archaeal genomes. Bioinformatics. 2018;34:4118–20.
Grissa I, Vergnaud G, Pourcel C. CRISPRFinder: a web tool to identify clustered regularly interspaced short palindromic repeats. Nucleic Acids Res. 2007;35:W52–7.
Karp PD, Midford PE, Billington R, Kothari A, Krummenacker M, Latendresse M, et al. Pathway Tools version 23.0 update: software for pathway/genome informatics and systems biology. Brief Bioinform. 2021;22:109–26.
Abby SS, Néron B, Ménager H, Touchon M, Rocha EPC. MacSyFinder: a program to mine genomes for molecular systems with an application to CRISPR-Cas systems. PLoS One. 2014;9:e110726.
Denise R, Abby SS, Rocha EPC. Diversification of the type IV filament superfamily into machines for adhesion, protein secretion, DNA uptake, and motility. PLoS Biol. 2019;17:e3000390.
Jain C, Rodriguez-R LM, Phillippy AM, Konstantinidis KT, Aluru S. High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat Commun. 2018;9:1–8. 2018 9:1
Kim D, Park S, Chun J. Introducing EzAAI: a pipeline for high throughput calculations of prokaryotic average amino acid identity. J Microbiol. 2021;59:476–80. 2021 59:5
Price MN, Dehal PS, Arkin AP. FastTree 2-approximately maximum-likelihood trees for large alignments. PLoS One. 2010;5:e9490.
Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C. Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods. 2017;14:417–9.
Ge SX, Son EW, Yao R. iDEP: an integrated web application for differential expression and pathway analysis of RNA-Seq data. BMC Bioinform. 2018;19:534.
Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550.
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47.
Guo J, Bolduc B, Zayed AA, Varsani A, Dominguez-Huerta G, Delmont TO, et al. VirSorter2: a multi-classifier, expert-guided approach to detect diverse DNA and RNA viruses. Microbiome. 2021;9:37.
O’Leary NA, Wright MW, Brister JR, Ciufo S, Haddad D, McVeigh R, et al. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016;44:D733–D745.
Brad T, Bizic M, Ionescu D, Chiriac CM, Kenesz M, Roba C, et al. Potential for natural attenuation of domestic and agricultural pollution in karst groundwater environments. Water. 2022;14:1597.
Fenchel T. Motility and chemosensory behaviour of the sulphur bacterium Thiovulum majus. Microbiol-sgm. 1994;140:3109–16.
Hamilton T, Jones DS, Schaperdoth I, Macalady J. Metagenomic insights into S(0) precipitation in a terrestrial subsurface lithoautotrophic ecosystem. Front Microbiol. 2014;5:756.
Porter M, Summers Engel A, Kane T, Kinkle B. Productivity-diversity relationships from chemolithoautotrophically based sulfidic karst systems. Int J Speleol. 2009;38:27–40.
Zhou Z, Tran PQ, Kieft K, Anantharaman K. Genome diversification in globally distributed novel marine Proteobacteria is linked to environmental adaptation. ISME J. 2020;14:2060.
Parrello B, Butler R, Chlenski P, Olson R, Overbeek J, Pusch GD, et al. A machine learning-based service for estimating quality of genomes using PATRIC. BMC Bioinform. 2019;20:1–9.
Haase D, Hermann B, Einsle O, Simon J. Epsilonproteobacterial hydroxylamine oxidoreductase (εHao): characterization of a ‘missing link’ in the multihaem cytochrome c family. Mol Microbiol. 2017;105:127–38.
Waite DW, Chuvochina MS, Hugenholtz P. Road map of the phylum Campylobacterota. Bergey’s Manual of Systematics of Archaea and Bacteria. 2019. pp 1–11.
Hoffman AA, Hercus MJ. Environmental stress as an evolutionary force. Bioscience. 2000;50:217–26.
Logares R, Brte J, Heinrich F, Shalchian-Tabrizi K, Bertilsson S. Infrequent transitions between saline and fresh waters in one of the most abundant microbial lineages (SAR11). Mol Biol Evol. 2010;27:347–57.
Rodriguez-R LM, Konstantinidis KT. Bypassing cultivation to identify bacterial species. Microbe. 2014;9:111–8.
Parks DH, Chuvochina M, Waite DW, Rinke C, Skarshewski A, Chaumeil PA, et al. A standardized bacterial taxonomy based on genome phylogeny substantially revises the tree of life. Nat Biotechnol. 2018;36:996–1004. 2018 36:10
Spöri Y, Stoch F, Dellicour S, Birky CW, Flot J-F KoT: an automatic implementation of the K/θ method for species delimitation. bioRxiv 2022; 2021.08.17.454531.
Birky CW, Adams J, Gemmel M, Perry J. Using population genetic theory and DNA sequences for species detection and identification in asexual organisms. PLoS One. 2010;5:e10609.
Birky CW, Maughan H. Evolutionary genetic species detected in prokaryotes by applying the K/θ ratio to DNA sequences. bioRxiv 2021; 2020.04.27.062828.
Volland J-M, Schintlmeister A, Zambalos H, Reipert S, Mozetič P, Espada-Hinojosa S, et al. NanoSIMS and tissue autoradiography reveal symbiont carbon fixation and organic carbon transfer to giant ciliate host. ISME J. 2018;12:714–27.
Sarbu SM, Kane TC, Kinkle BK. A chemoautotrophically based cave ecosystem. Science. 1996;272:1953–5.
Poser A, Vogt C, Knöller K, Ahlheim J, Weiss H, Kleinsteuber S, et al. Stable sulfur and oxygen isotope fractionation of anoxic sulfide oxidation by two different enzymatic pathways. Environ Sci Technol. 2014;48:9094–102.
Slobodkina GB, Mardanov AV, Ravin NV, Frolova AA, Chernyh NA, Bonch-Osmolovskaya EA, et al. Respiratory ammonification of nitrate coupled to anaerobic oxidation of elemental sulfur in deep-sea autotrophic thermophilic bacteria. Front Microbiol. 2017;8:87.
Eisenmann E, Beuerle J, Sulger K, Kroneck PMH, Schumacher W. Lithotrophic growth of Sulfurospirillum deleyianum with sulfide as electron donor coupled to respiratory reduction of nitrate to ammonia. Arch Microbiol. 1995;164:180–5.
Pandey CB, Kumar U, Kaviraj M, Minick KJ, Mishra AK, Singh JS. DNRA: A short-circuit in biological N-cycling to conserve nitrogen in terrestrial ecosystems. Sci Total Environ. 2020;738:139710.
Kern M, Simon J. Electron transport chains and bioenergetics of respiratory nitrogen metabolism in Wolinella succinogenes and other Epsilonproteobacteria. Biochimica et Biophysica Acta (BBA) - Bioenerg. 2009;1787:646–56.
Meyer JL, Huber JA. Strain-level genomic variation in natural populations of Lebetimonas from an erupting deep-sea volcano. ISME J. 2014;8:867–80.
Shu D, Guo J, Zhang B, He Y, Wei G. rDNA- and rRNA-derived communities present divergent assemblage patterns and functional traits throughout full-scale landfill leachate treatment process trains. Sci Tot Environ. 2019;646:1069–79.
Bižic-Ionescu M, Ionescu D, Grossart HP. Organic particles: Heterogeneous hubs for microbial interactions in aquatic ecosystems. Front Microbiol. 2018;9:2569.
Millman A, Bernheim A, Stokar-Avihail A, Fedorenko T, Voichek M, Leavitt A, et al. Bacterial retrons function in anti-phage defense. Cell. 2020;183:1551–.e12.
Pingoud A, Wilson GG, Wende W. Type II restriction endonucleases—a historical perspective and more. Nucleic Acids Res. 2014;42:7489–527.
Fauré-Fremiet E, Rouiller CH. Étude au microscope électronique d’une bactérie sulfureuse, Thiovulum majus Hinze. Exp Cell Res. 1958;14:29–46.
Renault TT, Abraham AO, Bergmiller T, Paradis G, Rainville S, Charpentier E, et al. Bacterial flagella grow through an injection-diffusion mechanism. Elife. 2017;6:e23136.
Karami Y, López-Castilla A, Ori A, Thomassin JL, Bardiaux B, Malliavin T, et al. Computational and biochemical analysis of type IV pilus dynamics and stability. Structure. 2021;29:1397–.e6.
Biais N, Ladoux B, Higashi D, So M, Sheetz M. Cooperative retraction of bundled type IV pili enables nanonewton force generation. PLoS Biol. 2008;6:e87.
Higashi DL, Biais N, Weyand NJ, Agellon A, Sisko JL, Brown LM, et al. N. elongata produces type IV pili that mediate interspecies gene transfer with N. gonorrhoeae. PLoS One. 2011;6:e21373.
Craig L, Forest KT, Maier B. Type IV pili: dynamics, biophysics and functional consequences. Nat Rev Microbiol. 2019;17:429–40.
Gao Y, Neubauer M, Yang A, Johnson N, Morse M, Li G, et al. Altered motility of Caulobacter crescentus in viscous and viscoelastic media. BMC Microbiol. 2014;14:322.
Sarbu SM, Popa R. A unique chemoautotrophically based cave ecosystem. In: Camacho AI (ed). The natural history of biospeleology. 1992. Mus. Nat. de Hist. Naturales, Madrid, pp 637–66.
Brad T, Iepure S, Sarbu SM. The chemoautotrophically based movile cave groundwater ecosystem, a hotspot of subterranean biodiversity. Diversity. 2021;13:128.
Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, et al. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013;41:D590–6.
Grote J, Schott T, Bruckner CG, Glöckner FO, Jost G, Teeling H, et al. Genome and physiology of a model Epsilonproteobacterium responsible for sulfide detoxification in marine oxygen depletion zones. Proc Natl Acad Sci USA. 2012;109:506–10.
Acknowledgements
The authors thank GESS team for logistics with sampling in the cave. We would also like to thank Pheobe Laaguiby at the University of Vermont for performing the outstanding Oxford Nanopore sequencing and Bo Barker Jørgensen, Emily Fleming, Carl Wirsen, Tom Fenchel and the anonymous reviewers for their valuable suggestions that improved the quality of this manuscript. We would also like to thank the Extreme Microbiome Project (XMP) for providing the DNA extraction reagents and methods as well as Laura Gray and Mehdi Keddache at Illumina Corp for providing partial sequencing reagents through its partnership with XMP. TB was supported by a grant of Ministry of Research and Innovation, project number PN-III-P4-ID-PCCF-2016–0016 (DARKFOOD). SMS was supported by a grant of Ministry of Research and Innovation (UEFISCDI) projects number PN-III-P4-ID-PCE-2020–2843 (EVO-DEVO-CAVE). JWA acknowledges the support from a grant from the User Support Program Space Research (grant ALW-GO/13-09) of the Netherlands Organization for Scientific Research (NWO). MB was funded through the German Research Foundation (DFG) Eigene Stelle project BI 1987/2-1. The computational resources for the assembly of the Thiovulum genome were provided to J.-F. Flot by the Consortium des Équipements de Calcul Intensif (CÉCI) funded by the Fonds de la Recherche Scientifique de Belgique (F.R.S.-FNRS) under Grant No. 2.5020.11; DI was funded through the German Research Foundation (DFG) Eigene Stelle project IO 98/3-1. JM Volland was supported by Gordon and Betty Moore Foundation grant GBMF7617. The work (proposal: 10.46936/10.25585/60001074) conducted by the U.S. Department of Energy Joint Genome Institute (https://ror.org/04xm1d337), a DOE Office of Science User Facility, is supported by the Office of Science of the U.S. Department of Energy operated under Contract No. DE-AC02-05CH11231.
Funding
Open Access funding enabled and organized by Projekt DEAL.
Author information
Authors and Affiliations
Contributions
TB, SMS, JS, MB, DI, and RP collected and processed samples from Movile Cave; P-EC, J-MV, and OG collected and processed samples from Guadeloupe; TB and SMS conducted light microscopical analysis; LBT conducted electron microscopy analysis; ST, DV, MB, DI, sequenced the samples from Movile Cave; MB, DI, LZ, JA, JO, J-FF conducted bioinformatic analysis on sequences from Movile Cave; P-EC, J-MV, DI, MB, LZ conducted bioinformatic analysis on sequences from Guadeloupe; All authors contributed to the writing of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Bizic, M., Brad, T., Ionescu, D. et al. Cave Thiovulum (Candidatus Thiovulum stygium) differs metabolically and genomically from marine species. ISME J 17, 340–353 (2023). https://doi.org/10.1038/s41396-022-01350-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41396-022-01350-4