Abstract
Comparative and pan-genomic analyses of the endophytic fungus Pezicula neosporulosa (Helotiales, Ascomycota) from needles of the relict fir, Abies beshanzuensis, showed expansions of carbohydrate metabolism and secondary metabolite biosynthetic genes characteristic for unrelated plant-beneficial helotialean, such as dark septate endophytes and ericoid mycorrhizal fungi. The current species within the relatively young Pliocene genus Pezicula are predominantly saprotrophic, while P. neosporulosa lacks such features. To understand the genomic background of this putatively convergent evolution, we performed population analyses of 77 P. neosporulosa isolates. This revealed a mosaic structure of a dozen non-recombining and highly genetically polymorphic subpopulations with a unique mating system structure. We found that one idiomorph of a probably duplicated mat1-2 gene was found in putatively heterothallic isolates, while the other co-occurred with mat1-1 locus suggesting homothallic reproduction for these strains. Moreover, 24 and 81 genes implicated in plant cell-wall degradation and secondary metabolite biosynthesis, respectively, showed signatures of the balancing selection. These findings highlight the evolutionary pattern of the two gene families for allowing the fungus a rapid adaptation towards endophytism and facilitating diverse symbiotic interactions.
Similar content being viewed by others
Introduction
One of the main tasks of evolutionary biology is to reveal how organisms adapt to different ecological niches and lifestyles. Trophic lability may shape the specific genomic landscape. To tackle such questions, a genomic prediction that uses both populations- and individual-level data could provide complementary insights into the mechanisms underlying trophic adaptations [1]. Fungi represent some of the primary drivers of ecosystem processes and are the ideal model for addressing these issues. Fungi have evolved diversified nutrition strategies (biotrophic, necrotrophic, and saprotrophic lifestyles), which likely exert evolutionary pressures contributing to genome evolution [2]. For several groups of plant symbiotic fungi, such as the ectomycorrhizal basidiomycetes (e.g. Laccaria bicolor), ericoid mycorrhizal (ERM) fungi, sebacinaceous fungi, and root dark septate endophytes (DSEs), the analyses of genome architecture uncovered the mechanisms of adaptations to two main trophic statuses: primary symbiotrophy and transient saprotrophy [3,4,5,6].
Foliar endophytes (FEs) are fungi that colonize the healthy photosynthetic tissues of plants without causing diseases [7]. Unlike the aforementioned fungal symbionts, FEs are characterized by highly localized infection and hyper-diversity within leaf tissues [7]. There is a growing consensus that horizontally transmitted FEs serve as defensive inducible mutualists to protect plants through the production of bioactive compounds [8,9,10,11]. This is contrasted to vertically transmitted grass endophytes which refer to constitutive mutualists. The “Foraging Ascomycete” hypothesis [12] predicts that saprotrophs may shift life-history strategies to endophytism to better disperse and persist [13,14,15]. Despite the ubiquitous distribution of FEs and their central role in plant-environmental interactions, the common adaptation strategy that enables an endophytic lifestyle at the evolutionary timescale remains elusive. Furthermore, there has been no concerted effort to explore the intraspecific genome-level polymorphism in FEs [16], hampering our understanding of trophic mode-associated genetic variations within populations.
We seek to bridge this gap by focusing on a model endophyte system on the relict fir species Abies beshanzuensis (Pinaceae), which is a critically endangered tree species left from the Quaternary glacial period (only three individual plants are known in the wild and all within 100 m of one another) and endemic in Qingyuan, Zhejiang Province (China) (Table S1) [17]. Earlier, we recorded a high level of endophytic fungal species diversity on this relict fir [18]. Among these species, Pezicula neosporulosa (Helotiales, Ascomycota) – recently recognized as a new species – dominates the needle tissues [19]. Besides, P. neosporulosa is known to be a rare species that has otherwise only been sampled twice in the Netherlands [19]. Because the helotialean ancestors of Pinaceae endophytes have been co-evolving with their hosts for more than 300 million years [20], it can be considered that P. neosporulosa has already been shifted to endophytic habitat. Research on P. neosporulosa is beginning to gather pace in part due to its broad spectrum of phenotypic and physiological polymorphism (observed quantitatively different litter decomposition activity and production of antibiotic compounds) over their very restricted geographic range [19], indicating intriguing population biology. In fungi, such cryptic morpho-physiological variants may be an underappreciated source of intraspecific ecologically relevant phenotypic diversification [21]. The prevalence of this phenomenon calls for a holistic understanding of its evolutionary benefits. For these reasons, P. neosporulosa provides an excellent opportunity to understand how it adapts to endophytic lifestyle at both individual and population levels.
In this work, we hypothesize that the evolution of P. neosporulosa could have shaped a set of common genomic features and evolutionary mechanisms facilitating its endophytic phase. We first focus on describing the evolutionary history of carbohydrate-active enzymes repertoire (CAZymes) and secondary metabolite gene clusters (SMGCs) gene families (expansions or contractions) of this endophyte by comparing genomes of individuals, two closely-related saprotrophic Pezicula species, and other plants beneficial helotialean fungi. We also performed a pan-genomic strategy to identify changes in gene family size and pan-genome dynamics based on 11 representative individuals. We point to population intra-specific polymorphism within an evolutionary context to identify the natural selection signatures. We further ask a related question: do recombination events likely occur within this population? Given that sexual recombination contributes greatly to genetic variations and adaptation of fungi, we intend to search for sexual recombination footprints and examine the reproduction system in P. neosporulosa.
Materials and methods
Pezicula neosporulosa isolates
In total, 75 endophytic P. neosporulosa isolates, and two P. neosporulosa isolates (CBS 101.96 and CBS 102.96) from the CBS-KNAW Culture Collection were used in this work. Detailed information on their origin and cultivation is provided in Supplementary Information Text S1.
Annotation of CAZymes and biosynthetic gene clusters
Genomes from 35 fungal species related to P. neosporulosa were downloaded from the JGI MycoCosm database (http://jgi.doe.gov/fungi) [22]. Two genomes from saprotrophic P. sporulosa (CBS 225.96) and P. cinnamomea (CBS 240.96) were sequenced in this study. Our comparative analyses focused on CAZomes (www.cazy.org) [23] and gene clusters involved in secondary metabolite biosynthesis. Three tools were used in combination for CAZomes annotation in dbCAN2 (http://cys.bios.niu.edu/dbCAN2, v2.0.1) [24]. The genes and gene clusters involved in secondary metabolism in 38 fungal species were predicted using antiSMASH v4.0.2 [25].
Phylogenomic reconstruction and molecular dating
Gene families or orthologous groups of these species were determined by OrthoFinder v2.3.8 with the default inflation parameter of 1.5 [26]. We identified 1497 homologous single copy gene families. After filtering short low-quality genes (encoding proteins with <200 amino acids), a total of 1,169 single-copy genes were used for constructing a phylogenomic tree based on a concatenated sequence alignment. The single-copy orthologous protein-coding sequences (CDS) were aligned using MUSCLE [27]. The unambiguously-aligned conserved blocks were extracted using Gblocks 0.91b with default parameters [28]. The concatenated alignment was used to create a Bayesian inference of phylogeny using MrBayes v3.1.2. The phylogeny was calibrated using two calibration points as follows: the most recent common ancestor (MRCA) of Leotiomycetes and ERM were estimated to have occurred c. 148 and 118 Mya, respectively [6]. Divergence time of each tree node was inferred using Bayesian Markov-chain Monte Carlo (MCMC) tree (MCMCTree) package in PAML v4.9 with the GTR nucleotide substitution model.
Gene family evolution
To monitor the dynamic changes during the evolution of gene families, we estimated gene family size expansion and contraction on each branch of the phylogenomic tree. We clustered the protein sequences into families using the Markov Cluster Algorithm (MCL) [29]. Possible expansions or contractions of gene family were estimated using the Computational Analysis of gene Family Evolution (CAFÉ) software package v4.2.1 [30]. Expansions or contractions of gene families with a p value ≤ 0.05 were considered significant.
Metabolomic analysis using high-performance liquid chromatography (HPLC)
While previous work has shown significant variation in antimicrobial activity among isolates, we proceeded to analyze the secondary metabolites produced by six representative isolates of endophytic P. neosporulosa through the HPLC technique. The metabolite extraction procedure and HPLC condition are described in Supplementary Information Text S1.
Pan-genome construction
To further infer intra-genomic diversity and trophic mode-specific markers of evolution and adaptation, we constructed the P. neosporulosa pan-genome. Illumina-based platform often yield highly fragmented genome assembly, which will bias the genome annotation. In our case, however, we still can obtain assembled genomes with relative high quality for several isolates when mapping reads to the reference genome M44 (Table S2). The pan-genome is pieced together using 10 short-read sequenced isolates (including all strains in HPLC analysis) covering the different physiological traits, and mating types (discussed below). This set of isolates provides us the opportunity to conduct a pilot study to estimate the pan-genome structure, which was constructed iteratively. Briefly, the list was started by including all the genes from M44, then, genes from other genomes were compared to the current pan-genome with BLASTn and added if the E-value was >1 × 10−10 according to the methods described by Walkowiak et al. [31]. The calculation approach of pan-genome is available in Supplementary Information Text S1.
Variants (SNPs and indels) calling and quality filtering
Whole-genome re-sequencing of 77 individuals was performed (see Supplementary Information Text S1). High-quality reads were aligned to the reference genome using BWA v0.7.17 with the MEM module [32]. SAM alignment files were sorted and converted into BAM files with SAMtools v1.3 [33]. Then HaplotypeCaller, CombineGVCFs, GenotypeGVCFs, SelectVariants, and VariantFiltration, implemented in the GATK package [34], were applied to call, select, and filter SNPsusing criteria “QD < 2.0 || FS > 80.0 || MQ < 20.0 || SOR > 3.0 || MQRankSum < −12.5 || ReadPosRankSum < −8.0 || QUAL < 40.0”. All SNP sets were filtered for a maximum of 50% missing data per site and a minimum minor allele frequency (MAF) of 0.02.
Genome-wide scan of natural selection signatures
We tested for deviations from neutrality using Tajima’s D [35] and Fu and Li’s D/D* and F/F* [36] statistics to search for signals of natural selection using VariScan v2.0.2 [37]. In addition, non-synonymous to synonymous substitution (pN/pS) ratios (measure the efficacy of selection to purge slightly deleterious mutations) [38], nucleotide diversities (θπ), and MAFs were obtained for all CAZymes and SMGCs to further confirm the signal for natural selection. Full details on statistical hypothesis and genome-wide scans for selection are provided in Supplementary Information Text S1.
Recombination detection and identification of MAT loci
We estimated the recombination events of the population using SplitsTree v4.14.4 [39]. We identified MAT loci in P. neosporulosa to understand which mating system (homothallic and heterothallic) is predominant, and check if mating type allele frequencies deviated from 1:1 ratio, which would indicate frequent asexual reproduction. The method for accurate identification of MAT loci is given in Supplementary Information Text S1. To validate these identified MAT-encoding genes, we designed several pairs of degenerate primers using Primer Premier 6.0 to amplify the target regions with PCR and Sanger sequencing (Table S3). As a result, 11 isolates were randomly selected for validation of their different mating types.
Results
The genome of P. neosporulosa resembles that of unrelated DSEs or other plant-associated helotialean fungi
The M44 reference genome for P. neosporulosa was assembled into 58 scaffolds with a total length of 54.3 Mbp and an N50 of 1.53 Mbp. A 95% completeness was confirmed, with 720 complete (718 single-copy BUSCOs and two duplicated BUSCOs), five fragmented, and 33 missing BUSCO orthologs (a total of 758 BUSCO groups were searched, https://busco.ezlab.org/frames/fungi.htm) [40]. More details on the genomic features of P. neosporulosa are presented in Table S4.
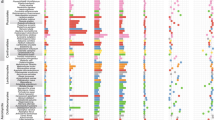
To better understand how evolution has acted on key regulatory gene families over evolutionary time, we investigated the gene family expansion/contraction events that took place on the Pezicula lineage. Results indicated that the branch to the Pezicula lineage showed a history of substantial family contractions and expansions, greatly exceeding that observed on other branches (Fig. 1A). In total, we identified 169 gene families exhibiting significant expansions (97, a total of 661 genes) and contractions (72, a total of 85 genes) in the P. neosporulosa compared to 35 reference genomes (Mann–Whitney test, p value < 0.05). We detected a significant enrichment of GO terms that were over-represented in the gene family expansion including monooxygenase (GO:0004497; adjusted p value = 1.10 × 10−5), oxidoreductase (GO:0016705; adjusted p value = 0.003), hydrolase (GO:0004553; adjusted p value = 0.0009), and chitinase (GO:0004568; adjusted p value = 0.0008) (Fig. 2), which were implicated in saprotrophic ability. Only one significantly enriched GO term in contracted families was identified for molecular functions: protein binding (GO:0005515; adjusted p value = 4.50 × 10−7).
A Phylogenomic species tree constructed with 1169 core orthologous single-copy genes using the GTR model with gamma-distributed rate variation. Two important groups of mutualistic helotialean fungi, including dark septate endophytes and ericoid mycorrhizal fungi, are indicated in dark blue and purple, respectively. The arrows indicate two calibration points. The numbers of expanded (orange) and contracted (blue) gene families at each internal node corresponded to the size of the bubbles. On the Pezicula clade and P. neosporulosa branch, the numbers of expansions and contractions that were statistically significant are shown in brackets. For better visualization, bubbles at terminal nodes or nodes with no clear gene family size changes are not shown. Gene number and genome size of all species are shown on the right side. Two Eurotiomycetes (Aspergillus spp.) and Xylona heveae (Xylonomycete) are the outgroup taxa. Bayesian posterior p values of 1.00 are indicated by a black square. Text with gray shading indicates three Pezicula species. B Heat map and dendrogram of all detected SMBCs. Text with gray shading indicates that P. neosporulosa is close to two DSE species in relation to their SMGCs patterns. C Comparison of the CAZymes repertoires identified in genomes of three Pezicula species. CAZomes categories include Glycoside Hydrolases (GHs), GlycosylTransferases (GTs), Carbohydrate Esterases (CEs), Polysaccharide Lyases (PL), Carbohydrate-Binding Modules (CBMs), and the Auxiliary Activities enzymes (AAs).
Description of enriched GO terms associated with significantly expanded gene families through a two-dimensional scatterplot space derived by applying multidimensional scaling to a matrix of the GO terms’ semantic similarities through REVIGO (http://revigo.irb.hr/). The bubble color indicates significance (−log10 p value), and size indicates the frequency of the GO terms in the underlying gene ontology database.
To determine the evolutionary relationships between P. neosporulosa and other members of the Leotiomycetes class, we constructed a phylogenomic tree based on 1169 single-copy core orthologous genes in 38 Pezizomycotina genomes (Fig. 1A). The three Pezicula species were monophyletic and occupied the basal position in the order Helotiales. The MRCA of Pezicula and other helotialean fungi was estimated to have occurred c. 127 Ma (95% CI, 120.1 to 136.0). The subsequent prediction that P. sporulosa and P. neosporulosa diverged c. 3.4 Mya (95% CI, 2.2 to 4.9), suggesting that these species diverged very recently compared to many other fungal genera. The phylogenomic analysis reveals that P. neosporulosa is distantly related to other helotialean plant mutualists, such as ERM and DSEs (Fig. 1A). However, the plant pathogenic species included in this study are also separated by considerable evolutional distance.
Considering the important roles of secondary metabolites and plant cell wall degradation ability in determining fungal lifestyles, we focused on identifying gene repertoire related to the CAZome and secondary metabolite biosynthesis. In total, we detected 84 secondary metabolites biosynthetic gene (SMGC) clusters in P. neosporulosa (supplementary material dataset), of which 29 belonged to the type I polyketide synthases (T1PKS), 16 to the non-ribosomal peptide synthases (NRPS), seven were the NRPS/PKS hybrids, ten belonged to the terpene synthase group, and one to the type III polyketide synthases (T3PKS). The remaining SMGCs were assigned to the unknown group (Fig. 1B). Unexpectedly, the total number of SMGCs of P. neosporulosa was higher than that of most other helotialean fungi (Table S5) and exceeded an average of 48 clusters for the order. In particular, the expansion of NRPS, T1PKS, and currently unknown gene clusters was detected. The cluster analysis of the SMGCs revealed that the endophytic P. neosporulosa is sharply different from two sister Pezicula species that are putatively saprotrophic but are very similar to the two other DSE species, such as Acephala macrosclerotiorum and Phialocephala scapiformis. The above two species, together with P. neosporulosa, share nearly the same number of genes involved in the production of phosphonates and have a reduced number of NRPS and beta-lactone biosynthetic genes compared to P. sporulosa and P. cinnamomea. Considering the very short evolutionary distance between Pezicula species (Fig. 1A), this observation points to the potential role of SMGCs in the trophic adaptation of P. neosporulosa to foliar endophytism. In line with this observation, Looney et al. [41] also speculated that a reduction of NRPS may be required for adopting a symbiotic lifestyle for ectomycorrhizal fungi.
The CAZome assembly responsible for plant cell wall degradation is also highly variable in this order (Fig. 1C). The P. neosporulosa genome is rich in predicted CAZymes (595 genes), which is similar or only slightly less than a few other DSEs and ERM in Leotiomycetes, such as Acephala macrosclerotiorum, Oidiodendron maius, Meliniomyces bicolor, and Phialocephala scopiformis. We further compared the composition of auxiliary activities (AAs) gene families. Among these, laccases (AA1) and cellobiose dehydrogenases (AA3)-encoding genes are directly involved in lignin degradation, and we found that the number of AA1 and AA3 genes exceed what is reported for most other fungal species but are comparable to mycorrhizal species, such as M. bicolor, and the DSEs P. scopiformis, and Phialocephala subalpine. In addition, cellobiose dehydrogenases (AA3), glucooligosaccharide oxidases (AA7), cellulose-degrading auxiliary activity family 9 (AA9), and β-1,4 endoglucanase (glycoside hydrolase family 5, GH5) also showed a similar pattern. Interestingly, the comparison of the three Pezicula species revealed that P. neosporulosa had a considerably richer CAZome than the two saprotrophic species (Fig. 1C), which also points to the unique ecological adaptation of this species. The principal component analysis revealed that the content of P. neosporulosa CAZome is unique with a sign of proximity to A. macrosclerotiorum and Meliniomyces bicolor that both are beneficial associates of plants (Fig. S1).
Thus, the phylogenomic analysis revealed the genetic separation and the recent speciation in the genus Pezicula and the unique genomic feature of the endophytic P. neosporulosa that resembles unrelated helotialean plant-beneficial fungi by the composition of CAZomes and the richness of SMGCs.
Gene family expansions in accessory genome highlight the adaptation of P. neosporulosa to foliar endophytism
As assessing the physiology of a species based on a single isolate is not sufficient, we have characterized and curated the pan-genome using 11 isolates with a sufficiently good quality of assembly to account for intraspecific variability (Fig. 3). We found that the pan-genome of P. neosporulosa remains open as the curve showing the relation of the pan-genome size and the number of isolates did not reach saturation and is also evidenced by the estimated exponent γ (= 0.037 ± 0.0004) > 0 (Fig. 3A). In this dataset, 13,271 and 16,881 genes constituted core- and pan-genomes, respectively (Figs. 3B and 3D). More specifically, only very few orphan genes (a total of 648 genes private to all isolates) were present in a range of 8 to 218 genes per individual. The plot for estimating the number of new genes added by each genomic sequence fitted a decaying exponential, and mathematical extrapolation predicts that an average of 74 new genes will be identified for every new genome sequenced (Fig. 3C).
A Estimation of P. neosporulosa pan-genome size. The curve is a least-squares fit of the power law to medians. B Estimation of P. neosporulosa core genome. The parameter κc is the amplitude of the exponential decay, τc is the decay constant, and Ω measures the best-fit value of the core genome. C Estimation of new genes; the number of individual-specific genes is plotted as a function of the number of genomes sequentially added. The parameters κs and τs are equivalent to κc and τc, and tg(θ) measures the best-fit number of specific genes. D The proportion of genes in the core, dispensable, and private genomes. E Distribution of COG categories between the core genome, accessory genome, and variable genomes.
In the context of nutritional adaptation and possible switch from saprotrophy to symbiotrophy, we aimed to identify if mutualistic and saprotrophic potentials are fueled by dispensable genes, which might have adaptive values. The outcomes of the attribution of the KOG functional categories to the core, pan, and accessory gene pools of 11 P. neosporulosa isolates are depicted in Fig. 3. Accessory genomes are enriched in genes encoding for the proteins assigned to category Q (secondary metabolism) and category G (carbohydrate transport and metabolism) (Fig. 3E).
We also provided direct chemical and molecular evidence from six isolates to demonstrate the variable secondary metabolite productions and diverged SMGC clusters organization (Fig. 4). Though several peaks were identical, most were different in peak areas, and some were unique among HPLC profiles. Similar differences were also observed in their SM gene clusters. These gene content polymorphisms were most likely generated through deletion or insertion. One prime example involves the NRPS-T1PKS hybrid gene cluster (Fig. 4D). We discovered that this cluster differed in the number and type of transport and additional modification genes associated with them, thus adding further potential to produce structurally diverse compounds from these isolates. Intriguingly, the M86 isolate lacks transport-related and other modification genes compared to remaining isolates; this could in part contribute to its poor metabolite production, evidenced by fewer peaks and peak areas in their metabolome profiles (Fig. 4C), although the chemical structures have not yet been identified.
A A schematic outline showing the endophytic lifestyle of P. neosporulosa determined by a diverse repertoire of genes related to CAZymes and SMGCs. AA9 and GH5 are the potential genetic determinants of endophytism. The Foraging Ascomycete (FA) hypothesis is presented to elucidate the benefits of endophytism. B Semiquantitative plate assays were performed to determine intraspecific variations in antibiotic activity, cellulase, laccase, and peroxidase production from six representative P. neosporulosa isolates. Fusarium oxysporum was used as a pathogen for the confrontation test. Congo red, syringaldazine, and Azure-B were used to detect cellulase, laccase, and peroxidase activities, respectively. This result supports the notion that the individuals did indeed display a remarkable degree of a physiological diversity. C Comparison of HPLC profiling of secondary metabolites produced by six isolates. The compounds were detected using a UV detector at a wavelength of 210 nm. D Comparison of micro-synteny and sequence similarity between SMGC clusters in six isolates investigated. Left: a Bayesian phylogenetic tree inference from 10,956 core orthologous single-copy genes. The approximately-maximum-likelihood phylogenetic tree was constructed using FastTree v2.1.7 under a JTT substitution model. Differences in gene content in the NRPS-T1PKS hybrid gene cluster in six isolates are shown. All predicted genetic components of this cluster are located at one scaffold.
A high level of standing genetic variation within the P. neosporulosa population
A total of 19.3 billion paired-end reads (comprising 198 Gb of high-quality raw nucleotide data) were generated FROM sequenced 77 P. neosporulosa strains. The average sequence depth per individual was 67.8× coverage (ranging from 41.3 to 122.6×) (Table S6). We surveyed genome-wide patterns of genetic variation in non-overlapping 1 kbp sliding windows and generated a Circos plot for visualization (Fig. 5A). Totally, we recovered 1,912,822 quality-filtered SNPs after singleton exclusion and the average SNP density of 35 SNPs/kbp, corresponding to diversity θW and θπ of 0.0073 and 0.0093, respectively. It suggested a relatively high level of total genetic diversity. A total of 967,088 non- transposable elements (TE) SNPs (50.56%) were located at intergenic regions. SNPs in genic regions, excluding TEs, included 375,450 synonymous and 200,988 nonsynonymous SNPs (Fig. 5B, C). The site-frequency spectrum for the various functional SNP classes indicated that both nonsynonymous and synonymous polymorphisms were skewed toward lower frequencies, suggesting that they were enriched for slightly deleterious mutations (Fig. 5D). An extensive genomic distribution of small insertions and deletions (indels, 1–40 bp) were located throughout the genome at an average density of 2.94 indel/kbp A total of 70,231 insertions and 89,481 deletions were identified, with average lengths of 4.06 and 4.72 bp, respectively (Fig. 5E). We annotated 15,333 (9.60%) indels in coding regions of the genome.
A Genome-wide averages of summary statistics were visualized using Circos. The data tracks are organized concentrically from the outer circle to the inner circle: distribution of SMGC clusters, gene density, TE density, SNP density, indel distribution, Tajima’s D, Watterson’s theta (θW, nucleotide polymorphism based on segregating sites), and Circos plot of the 58 scaffolds scaffold. The graph was plotted using Circos with a consecutive, non-overlapping 1 kbp sliding window. B, C Distribution of SNPs among functional effect classes compared to the proportion of sites in the reference genome. D Minor allele site-frequency spectrum. E Statistics of short indel (insertions and deletions) distribution. F Recombination pattern based on SplitsTree neighbor-net network analysis. Branch lengths represent pairwise Hamming distances (uncorrected P distances). The un-rooted radial phylogram showed a star-like topology without clear reticulations, suggestive of a low degree of recombination. The branches are shadowed in light blue indicate the isolates carrying mat1-2-1 (b) idiomorph (see more details in Fig. 6).
Restricted recombination and loss of sex in P. neosporulosa
To ascertain if the frequent sexual recombination will result in increased intraspecific genetic diversity, we next set out to search for population-level signs of recombination in the P. neosporulosa population. We assessed the extent of network structure among branches. A striking feature of the non-recombining network is the obtained combinations of 27 extremely long branches that have several terminal short-branched networks each. We speculated that this pattern might represent the typical pattern of diversified clonal expansions. Overall, the star-shaped topology with no geographic structure showed no evidence of recombination, indicating that adaptive radiation had occurred in P. neosporulosa.
The extremely low level of recombination is a sign of clonal, strongly inbreeding, or selfing population structure. We, therefore, ask whether P. neosporulosa undergoes sexual reproduction in either a heterothallic (self-sterile) or homothallic (self-fertile) fashion. We analyzed the structure and allele distribution of the mating-type loci determined by searching the P. neosporulosa genome for homologous MAT-locus sequences from known helotialean fungi. Intriguingly, we identified a putative unique reproductive system in this fungus, which contained two divergent mat1-2 idiomorphs in a ratio (30:47) that did not deviate from 1:1 (see Fig. 5F; χ2-test, p = 0.174) (Fig. 6A). The former individuals carry genes from mat1-2 and mat1-1 located at a single locus, with gene arrangement (mat1-1-1, mat1-1-5, and mat1-2-1) consistent with primary homothallism, while the latter lacked opposite mat1-1 idiomorphs (Fig. 6B). Furthermore, we revealed the presence of two types of mat1-2-1 idiomorphs with dissimilar sequences of different sizes (342 and 298 amino acids for mating types 1 and 2, respectively), but each allele contained the conserved PRkXseXrrR sequence in the C-terminal of the high-mobility group (HMG-box) domain. The mat1-2-1 cladogram revealed the genetic divergence of the mat1-2-1 idiomorphs, and one of them was clustered with two other Pezicula species (Fig. 6A). This suggests that the duplication of mat1-2-1 is quite old and that the two alleles have different evolutionary histories. The structure of mating-type 2 loci was similar to that of the homothallic Hymenoscyphus albidus [42], of which mat1-2-1 clusters with the heterothallic form mating type 1, suggesting an independent origin of this structure. We did not find isolates carrying only mat1-1 idiomorph, which suggests the absence of a heterothallic sexual system. The organization of the mating-type loci and flanking regions (apn2 and cox13) is shown in Fig. 6B. PCR and Sanger sequencing validated the occurrence of these mating-type genes in different groups (Fig. 6C).
A mat1-2-1-based phylogeny among helotialean-related fungi via the maximum likelihood (ML) method using PhyML v3.1 with the default model of HKY85. Sequences were derived from our work and ML bootstrap values/Bayesian posterior probabilities above 85% and 0.99, respectively, are indicated as black filled circles on nodes; ML bootstrap value below 80% while Bayesian posterior probabilities above 0.99 are indicated as gray filled circles on nodes. B Organization of the two types of the mating locus in each genome. C PCR validation of mat1-1-1, mat1-1-5, and two mat1-2-1 idiomorphs from representative isolates; in each sample, we found a single band corresponding to one idiomorph.
A set of CAZymes and SMGCs show signatures of selection
The high density of SNPs around SMGCs (Fig. 5A) implies that there may be selective maintenance of diversity in specific regions due to balanced polymorphisms what would be an alternative explanation of the above observations. To explore this possibility, we estimated genome-wide distribution plots for neutrality test statistics to see whether there are patterns in DNA sequence variation that fit the expectations of the hypothesis of neutrality. Data showed that both empirical and permuted null distribution of all neutrality test statistics calculated from SNP data in 50-kbp intervals is skewed toward positive statistics (Fig. S2). The average windowed Tajima’s D, Fu, and Li’s D*, F*, D, and F in the population were 0.58, 1.57, 1.39, 1.73, and 1.54, respectively, indicative of a genome-wide excess of intermediate frequency variants.
To look for potential outliers, we compared the distribution of diversity and test statistics over the average of the whole genome and gene families. The results indicated that the GH, PL, NRPS, and T1PKS families had higher average Tajima’s D values than the observed genome-wide distribution data (right-tailed Mann–Whitney U-test, p values = 6.23 × 10−5, 0.0041, 0.00013, and 0.0078, respectively) (Fig. 7A). A one-sided test for an excess of rare variants is justified here, as we are interested in sequences showing extremely positive Tajima’s D.
A Comparison of Tajima’s D statistics among the four gene families (GH, PL, T1PKS, and NRPS) and the whole-genome average at the population level. A 1-kbp sliding window was used to calculate the test statistics values. Significance was assessed by comparing the observed sliding-window values of each metric against a null genome-wide distribution derived through random permutation (right-tailed Mann–Whitney U-test). ** and *** indicate significance at the p < 0.01 and p < 0.001 levels, respectively. B Scatterplots of the Tajima’s D with θπ, Fu and Li’s D*, and pN/pS. First, we chose 24 and 81 candidate genes encoding CAZymes and secondary metabolite biosynthetic enzymes, respectively, which showed positive outlier values of at least one neutrality test statistic. Second, we randomly selected 100 genes as the control (genome average). The sample () function was used to obtain the random sampling in R v3.6.2. Third, we compared the θπ, Fu and Li’s D* (as an example), and pN/pS for plotting to detect the strong selection signatures. We set the thresholds as follows: Tajima’s D > 95% percentile of null genome-wide distribution and values of θπ, Fu and Li’s D* were set above the genome average, and pN/pS > 1. Gray lines are indicated in each plot as thresholds. The shaded dots in each scatterplot represent the genes under strong balancing selection. A right-tailed Mann–Whitney U-test is performed to compare whether the average θπ, Fu, and Li’s D* and pN/pS values of the two groups of candidate genes are higher than those of the reference gene set. The one-sided test for an excess of rare variants is justified here because we are interested in sequences showing extreme positive Tajima’s D, Fu, and Li’s D*, and pN/pS values. C local regions of diversity and patterns of selection. Tajima’s D and θπ values are plotted for a 1-kbp sliding window across four representative genes belonging to GH15, T1PKS, NRPS, and terpene synthase, showing a strong signal for balancing selection with outstanding peaks of elevated Tajima’s D and θπ. The x-axis represents the position on the scaffold. D The distribution of minor allele frequencies spectrum of corresponding genes compared to the null distribution of minor allele frequencies and to the whole genomic average.
We next sought to determine how many genes residing in each family show selection signals. We assessed the significance of Tajima’s D and Fu and Li’s D/D* and F/F* values against the genome-wide distribution. To avoid false positives, we report significance for statistical values that exceeded the positive 5% thresholds of the genome-wide non-parametric permutation distributions. Taking Tajima’s D statistics as an example, this resulted in assigning a cut-off value of ≥2.41 to separate outliers.
Our data revealed that many genes showed positive outlier values. In brief, values of 24 CAZymes and 81 SMGCs exceeded the 95% tails of the genome-wide distributions of at least one test statistic, suggesting that selection has acted on these genes. Among these, genes within the GHs and T1PKS families recorded the highest numbers of outliers (10 and 31 genes, respectively, Table S7). We further created stringent criteria to confirm their strong selection signatures by correlating Tajima’s D values with θπ, Fu and Li’s D* (as an example), and pN/pS (three scatterplots are shown in Fig. 7B). As a control, we looked at 100 randomly selected genes from the entire genome dataset. The higher pN/pS (>1) values obtained suggested that these were among the fastest evolving regions in the genome and considered to be under balancing selection [38]. Subsequently, we compared the values for test statistics between the average of two gene sets and reference genes. Except for pN/pS, the observed significant differences in other statistics (right-tailed Mann–Whitney U-test, all p values < 0.01) exceeded random fluctuations and were likely influenced by selection forces (Fig. 7B). One CAZyme and eight SMGCs showed outlier values for Tajima’s D and pN/pS values > 1. Notably, the gene found to have the highest pN/pS (estimated to be 2.88) was t1pks, with 23 nonsynonymous and eight synonymous mutations, indicating an enrichment for nonsynonymous mutations. We further showed four candidate genes that exhibited a particularly high level of nucleotide diversity (Fig. 7C). Within each genic region, at least one or two significant peaks exceeded the upper 5.0% tail of the genome-wide diversity distribution, with low diversity in the surrounding regions, suggesting these are among the fastest evolving regions in the genome, and evolving under very low evolutionary constraints.
To explore the extent to which allele frequency distributions in the candidate loci differed from those of the whole genome, we compared the minor allele frequency (MAF) spectra. The MAF for genome-wide SNPs exhibited the expected frequency of rare alleles, consistent with the idea that most polymorphisms are neutral or nearly neutral (Fig. S3); however, the overall MAFs for many candidate genes’ SNPs was not consistent with neutrality (Fig. 7D). The effect of balancing selection was to shift the selected loci toward a distinguishable enrichment of MAF of 0.3–0.5 (right-skewed).
Discussion
This study reveals the genomic hallmarks of foliar endophytism of a relict fir-associated endophytic fungus and uncovers its evolving narrative at multiple individual and population levels. Helotialean mutualistic fungi improve the fitness of many plants, including those forming mycorrhiza [6, 43, 44]. Intriguingly, unlike genomes of specialized or obligate plant-associated ectomycorrhizal, arbuscular mycorrhizal fungi and some endophytes, in which the CAZome sizes are remarkably reduced [45, 46], helotialean mycorrhizal and endophytic fungi have a rich repertoire of genes encoding lignocellulolytic enzymes of carbohydrate metabolism [5, 44, 47], reflecting their facultative biotrophy or incomplete saprotrophy to biotrophy transition. This pattern suggests that they have similar genetic repertoires to both plants pathogenic or saprotrophic species. However, large-scale comparative genomics and functional verifications could provide some new clues on the identification of a core set of CAZymes genes contributing to endophytic lifestyle [48,49,50]. In our case, we found two potential genetic determinants of endophytism: GH5 and AA9, which are richer in plant beneficial helotialean fungi (gene numbers ranging from 9–33 and 1–24, with an average of 24 and 15, respectively) than closely-related pathogenic or saprotrophic relatives (gene number ranging from 4–22 and 0–27, with an average of 15 and 11, respectively). GH5 and AA9 are thought to be vital for establishing ectomycorrhizal symbiosis and endophytism adaptation [49, 50].
As mentioned above, instead of improving plant nutrient uptake provided by the helotialean mycorrhizal and root endophytic fungi, helotialean FEs most likely act as defensive mutualists of trees by functioning as antagonists of pathogens or pests. This process is mainly driven by producing a series of biologically active secondary metabolites [9, 20]. Schulz et al. [51], Tanney et al. [52], and Yue et al. [53] revealed that many endophytic Pezicula species constituted an immense reservoir of antimicrobial compounds, which are in line with our findings. The amount of identified SMGCs in the P. neosporulosa genome is essentially higher than in other helotialean fungi, suggesting a vast and still uncharacterized metabolic potential. More interestingly, the evolutionary distance between the three Pezicula species is very small, but they are genomically and ecologically distinct. P. neosporulosa genome differs from the two other species and resembles those from unrelated plant beneficial helotialean fungi enabled by subtle yet highly specific changes in the gene content of a couple of SMGCs. It thus clearly suggests that within symbiotic Helotiales, mutualistic endophytism can be achieved among unrelated taxa using highly similar genomic toolkits, indicating the possibility of convergent evolution.
It could be speculated that gene family expansions or contractions play a major role in shaping organismal adaptation and the emergence of evolutionary innovations [54]. The specialized expansions of several lignocellulose-degrading gene families could be envisaged to adapt to environmental challenges [55]. We also attempted to construct the P. neosporulosa pan-genome to see if there are some expanded unique genome contents contributing to endophytic trophic advantage. We posit that enrichment in accessory genes related to CAZomes and secondary metabolite biosynthesis may be involved in maintaining endophytism, which is consistent with previous investigations [56, 57].
A key result in the present study is the pronounced standing diversity in P. neosporulosa population, suggesting sufficient variation for this fungus to evolve in response to environments. Intraspecific genetic variation in fungi is pervasive, and its role in trait evolution is increasingly appreciated [21, 58,59,60]. Indeed, genetic polymorphisms have been characterized in populations of the endophytic fungal Rhabdocline spp. (Helotiales) and Lophodermium spp. (Rhytismatales) from Pinaceae and mycorrhizal O. maius forming ericoid mycorrhizae with plants [16, 61,62,63,64]. However, less attention has been directed to their ecological and evolutionary implications. In this study, we assessed the magnitude of the polymorphism based on whole-genome sequence comparisons. It is reasonable that both SNPs and indels contribute to the extensive intra-genomic variations in the P. neosporulosa genome. The average SNP density (35 SNPs/kbp) is much higher than other reported fungal populations [65, 66]. This, however, leads to an as yet unanswered question on why the extremely low genetic diversity of the relic host tree still hosts a highly diverse population of a fungal endophyte. This suggests that a predominantly clonal population structure has not eroded genotypic diversity. Thus, we conclude that the potential sources of such astonishing diversity within a small temporal scale may be related to large effective population size [16] or high de novo mutations [67]. Unexpectedly, we found that two Dutch isolates are included within the diversity of Chinese isolates, suggesting no geographical clustering and long-distance dispersal of P. neosporulosa, although the avenues of its dispersal remain unknown. Thus, we cautiously advocate that selection could have led to the evolution of phenotypic variability, which might have prevailed over genetic differentiation to allow rapid adaptation to hosts.
Subsequently, we searched for signatures of selection using a genome-scanning approach. It has been suggested that the signals of balancing selection in highly self-fertilizing and clonal populations could be stronger in the case of peaks of variability in genomic regions [68, 69]. Our findings support this claim. A dozen outlier loci showing pervasive signature of selection in genes associated with T1PKS, NRPS, PLs, and GHs families indicate these genes experience rapid evolutionary change. Carbone et al. [70] and Drott et al. [71] confirmed that the costs of aflatoxins in the absence of insects and soil microbes are outweighed by benefits in their presence, resulting in balancing selection on the aflatoxin gene cluster of Aspergillus parasiticus. Our findings may fit this scenario as balancing selection will retain the chemotype polymorphisms of secondary metabolites in P. neosporulosa for protecting trees against herbivorous insects or needle pathogens. It is thus easy to envision that the diverse alleles may display variable activities of CAZomes and SMGCs, thus increasing the evolutionary potential and resilience of a symbiont population over long periods.
Another contribution of our study is the discovery of a unique mating system. To the best of our knowledge, this work represents the first documentation of two divergent mat1-2 alleles taking place simultaneously in a population that has both homothallic and heterothallic MAT loci arrangements. The two mat1-2-1 alleles differ in length and sequence at the nucleotide and amino acid levels, which fits with the observation of rapid evolutions in the mat1-2 sequences [72]. Furthermore, the apparent absence of isolates carrying only the opposite mat1-1 locus is surprising. We observed 30 isolates in total with a single mating type (mat1-2-1), and according to the typical bipolar mating system of all ascomycete fungi, both opposite mating types (mat1-1 and mat1-2) are needed for sexual recombination. Thus, we speculate that this is likely a sign of a severe bottleneck that generates genetic drift. It is noteworthy that the occurrence of two types that do not deviate from a balanced ratio and the age of the two mat1-2-1 idiomorphs might also indicate either the existence of heterothallism in which mat1-2-1 (a) acts as a mat1-1 mating type, a mixed system with outcrossing and haploid selfing, or a transition from heterothallism to homothallism. To distinguish between these hypotheses would require observation of the mating system in crossing assays.
Conclusion
Our results reveal a new example of convergent genomic adaptation to endophytism in P. neosporulosa, mostly determined by enzymes involved in carbohydrate metabolism and secondary metabolite biosynthesis. It seems clear that different plant-beneficial helotialean fungi, including P. neosporulosa, use similar genomic toolkits to function as symbiotrophs. Notably, a hallmark of P. neosporulosa lies in its highly intra-genomic polymorphism despite the signature of clonality. We acknowledge that it is not the first attempt to estimate the incidence of intra-specific variations in FEs. However, our focus on P. neosporulosa from the relict Abies is justified because the studied population experiences increased selection pressure to diversify CAZomes and SMGCs. This evolutionary force could render the Pezicula fungi have more endophytic niche fitness and serve as a considerable “ecological regulator” in forest ecosystems.
Data availability
M44 genomic data are available at the NCBI (BioProject number PRJNA517416, BioSample number SAMN10830013). This Whole Genome Sequencing (WGS) Project has been deposited at DDBJ/ENA/GenBank under the accession number SELE00000000. The raw genome resequencing data obtained have been deposited at the NCBI in the Short Read Archive database under the accession numbers SRR8568788-SRR8568862. Raw VCF files (SNP and indel variants) are available via Data Dryad (https://doi.org/10.5061/dryad.zs7h44j6x). The alignment and ML tree of mat1-2-1 were deposited in TreeBase (http://purl.org/phylo/treebase/phylows/study/TB2:S27214).
References
Tigano A, Colella JP, MacManes MD. Comparative and population genomics approaches reveal the basis of adaptation to deserts in a small rodent. Mol Ecol. 2020;29:1300–14.
Gladieux P, Ropars J, Badouin H, Branca A, Aguileta G, de Vienne DM, et al. Fungal evolutionary genomics provides insight into the mechanisms of adaptive divergence in eukaryotes. Mol Ecol. 2014;23:753–73.
Martin F, Aerts A, Ahrén D, Brun A, Danchin EG, Duchaussoy F, et al. The genome of Laccaria bicolor provides insights into mycorrhizal symbiosis. Nature. 2008;452:88–92.
Weiß M, Waller F, Zuccaro A, Selosse MA. Sebacinales-one thousand and one interactions with land plants. New Phytol. 2016;211:20–40.
Knapp DG, Németh JB, Barry K, Hainaut M, Henrissat B, Johnson J, et al. Comparative genomics provides insights into the lifestyle and reveals functional heterogeneity of dark septate endophytic fungi. Sci Rep. 2018;8:6321.
Martino E, Morin E, Grelet GA, Kuo A, Kohler A, Daghino S, et al. Comparative genomics and transcriptomics depict ericoid mycorrhizal fungi as versatile saprotrophs and plant mutualists. New Phytol. 2018;217:1213–29.
Arnold AE. Understanding the diversity of foliar endophytic fungi: progress, challenges, and frontiers. Fungal Biol Rev. 2007;21:51–66.
Carroll G. Fungal endophytes in stems and leaves: from latent pathogen to mutualistic symbiont. Ecology. 1988;69:2–9.
Miller JD, Sumarah MW, Adams GW. Effect of a rugulosin-producing endophyte in Picea glauca on Choristoneura fumiferana. J Chem Ecol. 2008;34:362–8.
White JF Jr, Torres MS. Is plant endophyte-mediated defensive mutualism the result of oxidative stress protection? Physiol Plant. 2010;138:440–6.
May G, Nelson P. Defensive mutualisms: do microbial interactions within hosts drive the evolution of defensive traits? Funct Ecol. 2014;28:356–63.
Carroll G. The foraging ascomycete, in: Abstracts of the 16th International Botanical Congress. St Louis, Missouri, USA, 1999.
Müller MM, Valjakka R, Suokko A, Hantula J. Diversity of endophytic fungi of single Norway spruce needles and their role as pioneer decomposers. Mol Ecol. 2001;10:1801–10.
Thomas DC, Vandegrift R, Ludden A, Carroll GC, Roy BA. Spatial ecology of the fungal genus Xylaria in a tropical cloud forest. Biotropica. 2016;48:381–93.
Naranjo-Ortiz MA, Gabaldón T. Fungal evolution: major ecological adaptations and evolutionary transitions. Biol Rev Camb Philos Soc. 2019;94:1443–76.
Oono R, Lutzoni F, Arnold AE, Kaye L, U’Ren JM, May G, et al. Genetic variation in horizontally transmitted fungal endophytes of pine needles reveals population structure in cryptic species. Am J Bot. 2014;101:1362–74.
Shao S, Jin Z. In Species Diversity and Extinction (ed. Tepper, GH) Ch. 15. Nova Science Publishers. 2010.
Yuan ZL, Rao LB, Chen YC, Zhang CL, Wu YG. From pattern to process: species and functional diversity in fungal endophytes of Abies beshanzuensis. Fungal Biol. 2011;115:197–213.
Yuan ZL, Verkley GJM. Pezicula neosporulosa sp. nov. (Helotiales, Ascomycota), an endophytic fungus associated with Abies spp. in China and Europe. Mycoscience. 2014;56:205–13.
Sieber T. Endophytic fungi in forest trees: are they mutualists? Fungal Biol Rev. 2007;21:75–89.
Levis NA, Martin RA, O’Donnell KA, Pfennig DW. Intraspecific adaptive radiation: competition, ecological opportunity, and phenotypic diversification within species. Evolution. 2017;71:2496–509.
Grigoriev IV, Nikitin R, Haridas S, Kuo A, Ohm R, Otillar R, et al. MycoCosm portal: gearing up for 1000 fungal genomes. Nucleic Acids Res. 2014;42:D699–D704.
Lombard V, Golaconda Ramulu H, Drula E, Coutinho PM, Henrissat B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014;42:D490–D495.
Zhang H, Yohe T, Huang L, Entwistle S, Wu P, Yang Z, et al. dbCAN2: a meta server for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 2018;46:W95–W101.
Blin K, Wolf T, Chevrette MG, Lu XW, Schwalen CJ, Kautsar SA, et al. antiSMASH 4.0-improvements in chemistry prediction and gene cluster boundary identification. Nucleic Acids Res. 2017;45:W36–W41.
Emms DM, Kelly S. OrthoFinder: solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol. 2015;16:157.
Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–7.
Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 2000;17:540–52.
Enright AJ, Dongen SV, Ouzounis CA. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res. 2002;30:1575–84.
Han MV, Thomas GW, Lugo-Martinez J, Hahn MW. Estimating gene gain and loss rates in the presence of error in genome assembly and annotation using CAFE 3. Mol Biol Evol. 2013;30:1987–97.
Walkowiak S, Rowland O, Rodrigue N, Subramaniam R. Whole genome sequencing and comparative genomics of closely related Fusarium Head Blight fungi: Fusarium graminearum, F. meridionale and F. asiaticum. BMC Genomics. 2016;17:1014.
Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–95.
Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–9.
Van der Auwera GA, Carneiro MO, Hartl C, Poplin R, Del Angel G, Levy-Moonshine A, et al. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinformatics. 2013;43:11.10.1–11.10.33.
Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–95.
Fu YX, Li WH. Statistical tests of neutrality of mutations. Genetics. 1993;133:693–709.
Hutter S, Vilella AJ, Rozas J. Genome-wide DNA polymorphism analyses using VariScan. BMC Bioinform. 2006;7:409.
Richards JK, Stukenbrock EH, Carpenter J, Liu Z, Cowger C, Faris JD, et al. Local adaptation drives the diversification of effectors in the fungal wheat pathogen Parastagonospora nodorum in the United States. PLoS Genet. 2019;15:e1008223.
Huson DH, Bryant D. Application of phylogenetic networks in evolutionary studies. Mol Biol Evol. 2006;23:254–67.
Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 2015;31:3210–2.
Looney B, Miyauchi S, Morin E, Drula E, Courty PE, Kohler A, et al. Evolutionary priming and transition to the ectomycorrhizal habit in an iconic lineage of mushroom-forming fungi: is preadaptation a requirement? bioRxiv. 2021. https://doi.org/10.1101/2021.02.23.432530.
Wey T, Schlegel M, Stroheker S, Gross A. MAT-gene structure and mating behavior of Hymenoscyphus fraxineus and Hymenoscyphus albidus. Fungal Genet Biol. 2016;87:54–63.
Zijlstra JD, Van’t Hof P, Baar J, Verkley GJM, Summerbell RC, Paradi I, et al. Diversity of symbiotic root endophytes of the Helotiales in ericaceous plants and the grass, Deschampsia flexuosa. Stud Mycol. 2005;53:147–62.
Almario J, Jeena G, Wunder J, Langen G, Zuccaro A, Zuccaro A, et al. Root-associated fungal microbiota of nonmycorrhizal Arabis alpina and its contribution to plant phosphorus nutrition. Proc Natl Acad Sci USA. 2017;114:E9403–E9412.
Gazis R, Kuo A, Riley R, LaButti K, Lipzen A, Lin J, et al. The genome of Xylona heveae provides a window into fungal endophytism. Fungal Biol. 2016;120:26–42.
Perotto S, Daghino S, Martino E. Ericoid mycorrhizal fungi and their genomes: another side to the mycorrhizal symbiosis? New Phytol. 2018;220:1141–7.
Wrzosek M, Ruszkiewicz-Michalska M, Sikora K, Damszel M, Sierota Z. The plasticity of fungal interactions. Mycol Prog. 2017;16:101–8.
Parrent JL, James TY, Vasaitis R, Taylor AF. Friend or foe? Evolutionary history of glycoside hydrolase family 32 genes encoding for sucrolytic activity in fungi and its implications for plant-fungal symbioses. BMC Evol Biol. 2009;9:148.
Zhang F, Anasontzis GE, Labourel A, Champion C, Haon M, Kemppainen M, et al. The ectomycorrhizal basidiomycete Laccaria bicolor releases a secreted β-1,4 endoglucanase that plays a key role in symbiosis development. New Phytol. 2018;220:1309–21.
Mesny F, Miyauchi S, Thiergart T, Pickel B, Atanasova L, Karlsson M, et al. Genetic determinants of endophytism in the Arabidopsis root mycobiome. Nat Commun. 2021;12:7227.
Schulz B, Sucker J, Aust HJ. Biologically active secondary metabolites of endophytic Pezicula species. Mycol Res. 1995;99:1007–15.
Tanney JB, McMullin DR, Miller JD. Toxigenic Foliar Endophytes from the Acadian Forest. In: Pirttilä A, Frank A (eds) Endophytes of Forest Trees. Forestry Sciences, vol 86. Springer, Cham. 2018;343–81.
Yue Q, Li Y, Chen L, Zhang X, Liu X, An Z, et al. Genomics-driven discovery of a novel self-resistance mechanism in the echinocandin-producing fungus Pezicula radicicola. Environ Microbiol. 2018;20:3154–67.
Rogers RL, Grizzard SL, Titus-McQuillan JE, Bockrath K, Patel S, Wares JP, et al. Gene family amplification facilitates adaptation in freshwater unionid bivalve Megalonaias nervosa. Mol Ecol. 2021;30:1155–73.
Mäkinen M, Kuuskeri J, Laine P, Smolander OP, Kovalchuk A, Zeng Z, et al. Genome description of Phlebia radiata 79 with comparative genomics analysis on lignocellulose decomposition machinery of phlebioid fungi. BMC Genomics. 2019;20:430.
Yang Y, Liu X, Cai J, Chen Y, Li B, Guo Z, et al. Genomic characteristics and comparative genomics analysis of the endophytic fungus Sarocladium brachiariae. BMC Genomics. 2019;20:782.
Franco MEE, Wisecaver JH, Arnold AE, Ju YM, Slot JC, Ahrendt S, et al. Secondary metabolism drives ecological breadth in the Xylariaceae. bioRxiv. 2021. https://doi.org/10.1101/2021.06.01.446356.
Matsuda Y, Yamakawa M, Inaba T, Obase K, Ito S. Intraspecific variation in mycelial growth of Cenococcum geophilum isolates in response to salinity gradients. Mycoscience. 2017;58:369–77.
Taylor JW, Branco S, Gao C, Hann-Soden C, Montoya L, Sylvain I, et al. Sources of fungal genetic variation and associating it with phenotypic diversity. Microbiol Spectr. 2017;5:1–21.
Chen ECH, Morin E, Beaudet D, Noel J, Yildirir G, Ndikumana S, et al. High intraspecific genome diversity in the model arbuscular mycorrhizal symbiont Rhizophagus irregularis. New Phytol. 2018;220:1161–71.
McCutcheon TL, Carroll GC, Schwab S. Genotypic diversity in populations of a fungal endophyte from Douglas Fir. Mycologia. 1993;85:180–6.
Perotto S, Girlanda M, Martino E. Ericoid mycorrhizal fungi: some new perspectives on old acquaintances. Plant Soil. 2002;244:41–53.
Müller MM, Valjakka R, Hantula J. Genetic diversity of Lophodermium piceae in South Finland. For Pathol. 2007;37:329–37.
Morgenstern K, Polster J-U, Krabel D. Genetic variation between and within two populations of Rhabdocline pseudotsugae in Germany. Can J Res. 2016;46:716–24.
Atwell S, Corwin JA, Soltis NE, Subedy A, Denby KJ, Kliebenstein DJ. Whole genome resequencing of Botrytis cinerea isolates identifies high levels of standing diversity. Front Microbiol. 2015;6:996.
Gasca-Pineda J, Velez P, Hosoya T. Phylogeography of post-Pleistocene population expansion in Dasyscyphella longistipitata (Leotiomycetes, Helotiales), an endemic fungal symbiont of Fagus crenata in Japan. MycoKeys. 2020;65:1–24.
Groenewald M, Linde CC, Groenewald JZ, Crous PW. Indirect evidence for sexual reproduction in Cercospora beticola populations from sugar beet. Plant Pathol. 2008;57:25–32.
Nordborg M, Charlesworth B, Charlesworth D. Increased levels of polymorphism surrounding selectively maintained sites in highly selfing species. Proc R Soc B. 1996;263:1033–9.
Koenig D, Hagmann J, Li R, Bemm F, Slotte T, Neuffer B, et al. Long-term balancing selection drives evolution of immunity genes in Capsella. Elife. 2019;8:e43606.
Carbone I, Jakobek JL, Ramirez-Prado JH, Horn BW. Recombination, balancing selection and adaptive evolution in the aflatoxin gene cluster of Aspergillus parasiticus. Mol Ecol. 2007;16:4401–17.
Drott MT, Debenport T, Higgins SA, Buckley DH, Milgroom MG. Fitness cost of aflatoxin production in Aspergillus flavus when competing with soil microbes could maintain balancing selection. mBio. 2019;10:e02782–18.
Chen F, Goodwin PH, Khan A, Hsiang T. Population structure and mating-type genes of Colletotrichum graminicola from Agrostis palustris. Can J Microbiol. 2002;48:427–36.
Acknowledgements
Research in Yuan’s lab is financially supported by the Fundamental Research Funds for the Central Non-profit Research of the Chinese Academy of Forestry (CAFYBB2019ZA001-3) and the National Natural Science Foundation of China (No. 31722014 and 31772032). Research in Martin’s lab is supported by the Laboratory of Excellence ARBRE (ANR-11-LABX-0002-01) and the Beijing Advanced Innovation Center for Tree Breeding by Molecular Design. We extend our sincere gratitude to Dr. Primrose Boynton (Wheaton College, USA) and Dr. Fengyan Bai (State Key Laboratory of Mycology, Chinese Academy of Sciences) for useful discussions. We thank Dr Yougui Wu (Fengyangshan-Baishanzu National Nature Reserve, China) for providing basic information about the wild relict fir trees.
Author information
Authors and Affiliations
Contributions
ZLY and QW conceived and designed this study. XYW conducted molecular and microbiological work. LXX performed HPLC analysis. ZLY, ISD, GHS completed bioinformatics, evolutionary, and phylogenetic analyses and interpreted the results with guidance from QW, JYW, and ZJL. ISD and XLS helped ZLY and QW with figure preparation. ZLY wrote the manuscript, and BPS, FMM, ZHZ, ISD, and CPK contributed improvements to the text. ZLY and ISD prepared the revised version of the manuscript. LP contributed text to the supplementary file. FMM, QW, BPS, CPK, and CF aided in the discussion of results. All authors approved the final version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Yuan, Z., Wu, Q., Xu, L. et al. Genomic landscape of a relict fir-associated fungus reveals rapid convergent adaptation towards endophytism. ISME J 16, 1294–1305 (2022). https://doi.org/10.1038/s41396-021-01176-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41396-021-01176-6