Abstract
Clostridioides difficile infections are associated with gut microbiome dysbiosis and are the leading cause of hospital-acquired diarrhoea. The infectious process is strongly influenced by the microbiota and successful infection relies on the absence of specific microbiota-produced metabolites. Deoxycholate and short-chain fatty acids are microbiota-produced metabolites that limit the growth of C. difficile and protect the host against this infection. In a previous study, we showed that deoxycholate causes C. difficile to form strongly adherent biofilms after 48 h. Here, our objectives were to identify and characterize key molecules and events required for biofilm formation in the presence of deoxycholate. We applied time-course transcriptomics and genetics to identify sigma factors, metabolic processes and type IV pili that drive biofilm formation. These analyses revealed that extracellular pyruvate induces biofilm formation in the presence of deoxycholate. In the absence of deoxycholate, pyruvate supplementation was sufficient to induce biofilm formation in a process that was dependent on pyruvate uptake by the membrane protein CstA. In the context of the human gut, microbiota-generated pyruvate is a metabolite that limits pathogen colonization. Taken together our results suggest that pyruvate-induced biofilm formation might act as a key process driving C. difficile persistence in the gut.
Similar content being viewed by others
Introduction
Clostridioides difficile, formerly known as Clostridium difficile, causes infections associated with gut microbiome dysbiosis and is the leading cause of nosocomial diarrhoea and colitis following antibiotic therapy [1]. While infections are typically associated with dysbiosis, recent epidemiological studies indicate that 5–15% of the population asymptomatically carry C. difficile despite having a healthy microbiota [1]. There is also increasing evidence that C. difficile causes community-acquired infections and is a zoonotic pathogen. Pets and farm animals asymptomatically carry this pathogen and, as a result, it is detected in retail meat. Based on these findings, C. difficile can now be viewed as a quintessential “One Health” pathogen [2].
The C. difficile infectious cycle depends on the ability of this anaerobic Gram-positive rod to sporulate. Once ingested from the surrounding environment or food, spores germinate in the ileum and then vegetative cells colonize the caecum and the colon [1, 2]. Successful colonization relies on the disruption of the host’s microbiota [3,4,5,6]. The microbiota protects against C. difficile infection by producing metabolites including deoxycholic acid (DOCA) and short-chain fatty acids (SCFA) [7,8,9,10,11]. During dysbiosis specific members of the microbiota are missing, which results in altered SFCA and DOCA production, thus allowing C. difficile to grow unimpeded [3]. Following successful treatment of C. difficile infections (via antibiotic therapy or fecal transplant), the production of protective metabolites by the microbiota is slowly restored. Yet, relapses occur in more than 30% of patients that received antibiotics to treat their first C. difficile infection and this rate increases to 50% after an initial relapse [1]. The exact cause of relapse has not been fully elucidated and represents a major challenge for managing C. difficile infections. In 40% of relapse cases, patients are infected with the same strain that caused the initial infection, suggesting C. difficile persists in the gastrointestinal tract [1].
Persistence was initially associated with sporulation during antibiotic treatment, followed by germination after treatment. Evidence for this hypothesis was based on the ability of spores to enter epithelial cells and the inability of a non-sporulating spo0A-inactivated strain to persist and cause relapse in a murine infection model [12, 13]. Inactivation of spo0A has pleiotropic effects influencing flagellar motility, metabolism, and biofilm formation [14, 15]. This suggests that factors other than sporulation may also contribute to C. difficile persistence. Recent studies have demonstrated that multi-species biofilms formed by the gut microbiota can harbour and drives persistence and recurrence [16,17,18,19]. In support of these observations, recent data from our laboratory showed that co-culture of C. difficile with Clostridium scindens, a bacterium that converts primary bile salts to secondary bile salts, promotes dual-species biofilm formation in the presence of cholate [20].
Typically, biofilms are defined as community of bacterial cells encased in self-produced polymeric matrix [21]. This matrix is usually composed of any combination of proteins, extracellular DNA, or exopolysaccharides and matrix composition will vary from species to species. Biofilms provide a microenvironment that decrease the susceptibility of bacteria towards different environmental stressors, including antimicrobial compounds, to promote bacterial persistence. In C. difficile, biofilm formation is mediated by several surface structures (e.g. pili and the S-layer), environmental triggers (e.g. DOCA and sub-MIC of antibiotics), quorum sensing (e.g. luxS), in addition to other determinants (e.g. c-di-GMP) [22,23,24,25,26,27,28].
The aim of this study is to identify key factors that contribute to biofilm formation by C. difficile in response to DOCA. We used time-course transcriptomics and genetic techniques to identify sigma factors, metabolic processes and adhesins that drive biofilm formation. We then demonstrate that extracellular pyruvate is the key metabolite that triggers biofilm formation and identify a C. difficile pyruvate importer involved in this process.
Methods
Bacterial strains and culture conditions
Bacterial strains and plasmids used in this study are listed in Table S1. E. coli strains were grown in LB broth with chloramphenicol (15 μg/ml). C. difficile strains were grown anaerobically (5% H2, 5% CO2, 90% N2) in BHISG (BHI supplement with 0.5% (w/v) yeast extract, 0.01 mg/mL cysteine and 100 mM glucose). In addition, 10 ng/ml of anhydrotetracycline (Atc) was used to induce the Ptet promoter of pRPF185 vector derivatives in C. difficile.
The final composition of C. difficile semi-defined biofilm medium (BM) is as follow: Oxoid casein hydrolysate (10 mg/mL), L-Tryptophane (0.5 mg/mL), L-Cysteine (0.01 mg/mL), L-Leucine (0.0033 mg/mL), L-Isoleucine 0.0033 mg/mL), L-Valine (0.0033 mg/mL), Na2HPO4 (5 mg/mL), NaHCO3 (5 mg/mL), KH2PO4 (0.9 mg/mL) NaCl (0.9 mg/mL), (NH4)2SO4 (0.04 mg/mL), CaCl2·2H2O 0,026 MgCl2·6H2O (0,02 mg/mL), MnCl2·4H2O (0,01 mg/mL), CoCl2·6 H2O (0.001 mg/mL) FeSO4·7 H2O (0.004 mg/mL) D-biotine (0.001 mg/mL), calcium-D-panthothenate (0.001 mg/mL) and pyridoxine (0.0001 mg/mL). The desired sugars and/or DOCA were added, as necessary.
Biofilm assays
Overnight cultures of C. difficile were diluted 1/100 into the desired medium (BHIS or BM) containing the desired supplements (100 mM glucose, 240 μM DOCA and/or 50 mM or 100 mM sodium pyruvate) and 1 ml of the dilution was aliquoted in each well of a 24-well polystyrene tissue culture-treated plates (Costar, USA). Plates were incubated at 37 °C in an anaerobic environment for 48 h. Biofilm biomass was measured using established methods [20]. Briefly, spent media was removed by inverting the plate and wells were washed twice by pipetting phosphate-buffered saline (PBS) at 45° angle. Biofilms were air dried and stained with crystal violet (CV; 0.2% w/v) for 2 min. CV was removed by inversion; wells were washed twice with PBS then air-dried. Dye bound to the biofilm biomass was solubilized by adding 1 ml of a 75% ethanol solution and the absorbance, corresponding to the biofilm biomass, was measured at a λ600nm with a plate reader (Promega GloMax Explorer, Promega, France). Sterile medium was used as a negative control and a blank for the assays.
RNA isolation and quantitative reverse-transcriptase PCR
A 24-well plate was used to produce one replicate for one condition. At 9, 14 and 24 h, the total bacterial population was collected, and cells were harvested by centrifugation (10 min, 4000 × g, 4 °C). The pellet was frozen (−80 °C) until used. For the 48 h biofilm sample, the supernatant was removed by inverting the plate, the biofilm was washed twice and resuspended in 20 mL of PBS. The recovered biofilm cells were centrifuged and the pellet was frozen until RNA was extracted. Total RNA was extracted from cell pellets as previously described [29]. cDNA synthesis and qRT-PCR were carried as described before [29] using primers listed in Table S2.
Whole transcriptome sequencing and analysis
Transcriptomic analysis for each condition was performed using three independent RNA preparations using methods described before [20]. Briefly, the RNA samples were first treated using the Epicenter Bacterial Ribo-Zero kit. This depleted rRNA fraction was used to construct cDNA libraries using TruSeq Stranded Total RNA sample prep kit (Illumina, France). Libraries were then sequenced by HiSeq2500 sequencer (Illumina). Cleaned sequences were aligned to the reannotated C. difficile strain 630 [30] for the mapping of the sequences using Bowtie 2 (Version 2.1.0). DEseq2 (version 1.8.3) was used to perform normalization and differential analysis using the 9 h time point values as a reference for reporting the expression data of the 14, 24 and 48 h. Genes were considered differentially expressed if the fold changes were ≥ Log2 1.5 and their adjusted p-value was ≤0.05.
Gene deletion in C. difficile
Gene deletions were carried as described in Peltier et al. [31]. Briefly, regions upstream and downstream of the gene of interest were PCR-amplified using primer pairs listed in Table S2. PCR fragments and linearized pDIA6754 [31] were then mixed and assembled using IVA cloning [32] or Gibson assembly (NEB, France) and transformed by heat shock into E. coli NEB 10β. Constructions were verified by sequencing and the selected plasmid were introduced into E. coli HB101 (RP4). Plasmids were transferred by conjugation into the desired C. difficile strains and deletion mutants were obtained using counter-selection described elsewhere [31].
Treatment with pyruvate dehydrogenase
The pyruvate depletion assay was adapted from Goodwine et al. [33]. Briefly, biofilms were prepared in BM with 100 mM glucose and 240 µM DOCA as described above. After 24 h, 250 µL containing 20 mU of pyruvate dehydrogenase and cofactors (2 mM CoA, 2 mM ß-NAD+, 20 µM thiamine pyrophosphate and 50 µM MgSO4) was added to the biofilm. For control purposes, pyruvate dehydrogenase was heat inactivated at 100 °C for 10 min and mixed with its cofactors. A cofactor-only control and a non-treated control were also included. The values are reported as a percent of the biofilm formed using the following formula: (Treated biofilm (Enzyme, heat-inactivated enzyme or buffer only)/untreated biofilm) × 100.
Quantification of pyruvate in culture supernatant
C. difficile strains were grown in 1 mL of BM with 100 mM pyruvate or BM with 100 mM glucose and 50 mM pyruvate aliquoted in individual wells of a 24-well plate. After 24 h and 48 h, 1 mL of each sample was recovered and the supernatant was recovered by centrifugation (1 min, 14,000 × g). The clarified supernatant was transferred to a new tube and stored −20 °C until used. The sterile medium was used as a control to quantify the amount of pyruvate at time 0 h. Pyruvate was quantified using the EnzyChrom Pyruvate Assay kit (BioAssay Systems Hayward, CA). The values are reported as a percent of the pyruvate remaining in the supernatant calculated with the following formula: (Concentration of pyruvate in culture supernatant/Concentration of pyruvate in sterile medium) × 100.
Statistical analysis
Biofilm assays, effect of treatment and effect of genetic inactivation or deletion were analysed using a Kruskal–Wallis test followed by an uncorrected Dunn’s test. Effect of pyruvate supplementation was compared and analysed using a two-way ANOVA followed by a Fisher LSD test.
Results
Overview of the time course transcriptomic analysis of C. difficile grown in the presence of DOCA
In our previous study, we observed that when grown in the presence of sub-inhibitory concentrations of DOCA, C. difficile enters the stationary phase between 14 and 20 h and by 48 h has formed a strong biofilm [20]. To identify key events leading to biofilm formation in the presence of DOCA, we performed a time-course transcriptomic analysis on C. difficile grown in BHI supplemented with yeast extract, cysteine, glucose (BHISG) and 240 µM DOCA. For this analysis, time points were selected based on the growth curve. Planktonic bacterial cells were harvested in the logarithmic phase (9 h), transition phase/early stationary phase (14 h), and mid-stationary phase (24 h); biofilm cells were harvested at 48 h. We then compared the expression profile of the cells at 14, 24 and 48 h using the 9 h time point as our reference point. Relative to the 9 h time point, a total of 262, 659 and 659 genes were downregulated at 14, 24 and 48 h, respectively, whereas a total of 218, 794 and 997 genes were upregulated at 14, 24 and 48 h, respectively (Supplementary data). We started our analysis by focusing on cell surface structures known to contribute to biofilm formation. The genes encoding type IVa pili (T4aP) machinery [pilA2 (CD630_32940), pilW (CD630_23050) and the pilA1 cluster (CD630_35030-CD630_35130)] were upregulated in the DOCA-induced biofilm cells (Supplementary data). In addition, the F1 (late), F2 (glycolysation) and F3 (early) gene clusters of the flagellum biogenesis, were downregulated at 48 h whereas the collagen-binding protein gene (CD630_28310), and bcsA (CD630_25450), which encodes a protein with a cellulose synthase domain, were upregulated at 48 h (Supplementary data)
Cell surface proteins and structures play a limited role in DOCA-induced biofilm formation
To see if the T4aP and other surface proteins were critical for biofilm formation in the presence of DOCA, the pilA2 cluster (CD630_32910-CD630_32970), the major pilin pilW the pilA1 cluster, and bcsA were deleted. We also tested the biofilm formation for strains lacking the CD630_28310 adhesin, pilA1, the flagellin fliC and the alternative sigma factor for the flagellar operon sigD. Deletion of the pilA1 cluster resulted in a significant decrease in DOCA-induced biofilm formation but deletion of pilA1, pilW or the pilA2 cluster had no effect (Fig. 1A and S1A). In addition, inactivation of bscA, CD630_28310, fliC or sigD had no effect on DOCA-induced biofilm formation or its kinetics (Fig. 1B and S1A).
Biofilm formation was assessed in strains lacking genes (A) encoding pili (48 h), (B) or involved in flagella motility (24 and 48 h). Asterisks indicate statistical significance determined with a Kruskal–Wallis test followed by an uncorrected Dunn’s test (**p ≤ 0.01, vs 630Δerm). Dots represent biological replicates from experiments performed on different days, the bars represent the mean and the error bars represent the SEM.
T4aP gene expression is also under the control of c-di-GMP and overproduction of c-di-GMP results in measurable auto-aggregation and biofilm formation [33]. Several c-di-GMP associated genes were upregulated in biofilms (48 h) suggesting this signalling molecule might have a role (Table S3 and Supplementary data). We then decided to test the effect of c-di-GMP overproduction on auto-aggregation in the presence of DOCA. To do so, a C. difficile strain (CDIP634) with an inducible diguanylate cyclase (CD630_1420) that overproduces c-di-GMP was grown in the presence or absence of DOCA. When we compared CDIP634 to a parental control strain, we observed high levels of auto-aggregation in BHISG exclusively when c-di-GMP was overproduced (Fig. S1B). However, auto-aggregation was not observed in BHISG supplemented with DOCA, suggesting that cells do not aggregate in response to c-di-GMP overproduction in the presence of DOCA (Fig. S1B).
Altogether, we show that the T4aP machinery encoded by the pilA1 cluster is required for biofilm formation in the presence of DOCA. Other surface proteins associated with biofilm formation in the absence of DOCA [22, 34] are dispensable, suggesting a distinct biofilm-formation mechanism.
Biofilm formation in the presence of DOCA is associated with profound metabolic rearrangement
We then analysed our time-course transcriptomic data using the Biocyc database for changes in metabolic pathways (Fig. S2). As observed in our previous study (Dubois et al. [20]), genes encoding enzymes associated with glycolysis were downregulated from 14 h onward (Fig. S2 and Supplementary data). Furthermore, enzymes involved in reductive and oxidative Stickland reactions were upregulated at 14 and 24 h, but their expression was unchanged at 48 h. Genes associated with fermentation pathways such as butanoate were upregulated at 24 h but downregulated at 48 h (Fig. S2 and Supplementary data). Interestingly, C. difficile appears to down-regulate genes associated with ethanolamine degradation (Fig. S2 and Supplementary data), a valuable nutrient source that can be converted to acetyl-CoA [35].
We also observed significant up- and downregulation in the transcriptional profiles of PTS-dependent transporters with different predicted affinity (Fig S2). Furthermore, the genes encoding proteins that import and process N-acetylneuraminic acid (Neu5Ac) were upregulated at 48 h (Fig. S2 and Supplementary data). This could indicate that C. difficile might use different carbon source once glucose is depleted, probably near the 14 h time point.
We then compared our transcriptome to a published omics analysis of C. difficile cells in different growth phases [36]. Based on these comparisons, we found evidence that the cells harvested at 24 h have a transcriptome profile of stationary phase cell, (Fig. 2). Specifically, transcription of genes associated with protein degradation, butanoate fermentation, acetate fermentation, glycine metabolism, and oxidative Stickland reaction of branched chained amino acids were upregulated at 24 h (Fig. 2 and Supplementary data) as observed for cells in the stationary phase [36]. The expression of these genes decreased at 48 h (Fig. 2). Other genes that were upregulated during the stationary phase were those associated with cysteine biosynthesis, pantothenate biosynthesis, riboflavin biosynthesis, ferrous iron transport, flavodoxin and chaperones (Fig. 2) but these genes remain induced at 48 h in our analysis (Supplementary data). On the other hand, genes associated with glycolysis were downregulated at 24 h as observed for the stationary phase analysis and their expression remains downregulated in biofilm cells at 48 h (Fig. 2). Interestingly, genes involved in protein synthesis were downregulated at 24 h but their expression increased at 48 h (Fig. 2 and Supplementary data).
Overall, long-term exposure to DOCA results in metabolic profile remodelling by C. difficile and our observations support previous data demonstrating that DOCA induces metabolic changes in C. difficile [37]. Consequently, these changes in metabolic profiles will influence lifestyle switching during the stationary phase. In support of this hypothesis, the 24 h transcriptomic signature of C. difficile grown in BHISG in the presence of DOCA resembles cells in a stationary phase but at 48 h, once C. difficile is in a biofilm state, metabolic activity and transcriptional activity increase.
The transition phase regulator SigH is required for DOCA-induced biofilm formation
In C. difficile, SigH and SinR modulate Spo0A expression and activity in addition to mediating the transition from exponential to stationary phase [38, 39]. Furthermore, the absence of spo0A decreases biofilm formation regardless of the conditions ([15, 20]; Fig. 3A) Therefore, based on the role of spo0A in DOCA-induced biofilm formation in combination with our time-course transcriptomic analysis indicating the effect of major transcriptional changes associated with the stationary phase, we tested the effect of SigH and SinR on biofilm formation. Inactivation of sigH results in decreased biofilm-formation whereas inactivation of sinR did not have an effect (Fig. 3A and Fig. S3A).
Biofilm formation was assessed 48 h after inoculation in strains lacking genes involved in (A) the transition to stationary phase, (B) the sigH regulon, (C) cysteine metabolism, (D) PTS transport and (E) other metabolic pathways. Asterisks indicate statistical significance determined with a Kruskal–Wallis test followed by an uncorrected Dunn’s test or a two-way ANOVA followed by a Fisher’s least significant difference test (**p ≤ 0.001 ***p ≤ 0.001, ****p ≤ 0.0001vs 630Δerm). Dots represent biological replicates from experiments performed on different days, the bars represent the mean and the error bars represent the SEM.
Inactivation of sigH decreased biofilm formation to a lower level than observed in the spo0A-inactivated strain. Given that our time-course transcriptome did not show changes in spo0A expression, our data suggest that genes other than spo0A that are under SigH control contribute to biofilm formation. To identify SigH controlled genes that contribute to biofilm formation, we compared the list of genes with a SigH binding site in their promoter [38] with our transcriptome and identified specific genes that were upregulated at 24 h (Table S4). We selected 4 genes (CD630_08650, CD630_12640, cwp29 and CD630_34580) that were not involved in sporulation, transcription or metabolism and we tested their expression in the sigH::erm strain and the parental strain grown in BHISG with DOCA at 24 h. The expression of CD630_08650 and cwp29 was greatly reduced in the absence of SigH whereas the expression of CD630_12640 and CD630_34580 was only moderately reduced (Fig. S3B). This confirmed that the expression of CD630_08650 and cwp29 is controlled by SigH in our biofilm-inducing condition.
Based on these results, we deleted CD630_08650 and cwp29 and tested the ability of the resulting strains to form biofilm. Deletion of either gene did not have an effect on DOCA-induced biofilm formation (Fig. 3B). Unfortunately, efforts to generate a strain lacking both CD630_08650 and cwp29 were unsuccessful. When we tested the viably of the sigH::erm strain, this strain had a one log reduction compared to the parental strain in the number of viable cells in the total population (Fig. S3C). The viability of the spo0A::erm strain is not affected in our conditions [20]. The decrease in viability of the sigH::erm strain provides a possible explanation for the reduction in DOCA-induced biofilm formation. Based on our previous study, we know that biofilm formation in the presence of DOCA is not dependent on sporulation [20]. In addition, SigH and Spo0A contribute to metabolic adaptation of C. difficile [14, 38]. Taken together, the sub-optimal metabolic profile of these strains might result in a decrease in DOCA-induced biofilm formation highlighting the impact of metabolism in C. difficile persistence.
PTS mediated transport and cysteine metabolism are required for DOCA-induced biofilm formation
Given that metabolism appears to be crucial for DOCA-induced biofilm formation, we next tested the effect of inactivating different metabolism-associated genes. We first tested the role of cysteine metabolism because several genes associated with this metabolic pathway were differentially regulated in our transcriptomic analysis (Fig. 2 and Supplementary data). Inactivation of the OAS-thiol-lyase encoding gene cysK, which participates in cysteine biosynthesis, did not alter DOCA-induced biofilm formation (Fig. S4A). We then tested the inactivation of the sigma factor encoding gene, sigL and deletion of the SigL-controlled cysteine desulfidase encoding gene, cdsB, important for amino acid degradation and cysteine catabolism resulting in the production of pyruvate and sulfide, respectively [40, 41]. Inactivation of sigL greatly reduced DOCA-induced biofilm formation by C. difficile, whereas deletion of cdsB had an intermediate phenotype that was highly variable (Fig. 3C). Complementation of the sigL::erm and ΔcdsB strain with sigL or cdsB expressed from their native promoter on a plasmid restored biofilm formation (Fig. S4B).
We then targeted the phosphotransferase system (PTS) and carbon catabolite repression (CCR) regulatory network by deleting ptsI that encodes enzyme I of the PTS and deleting hprK that encodes the Hpr kinase which is involved in carbon metabolism regulation and transport. DOCA-induced biofilm formation was abolished in the ΔptsI strain whereas only a slight reduction in biofilm formation was observed in the ΔhprK strain (Fig. 3D). Complementation of the ΔptsI strain with inducible ptsI on a plasmid restored biofilm formation (Fig. S4B). These observations support our previously published results [20] showing that the presence of glucose and carbon metabolism are essential for DOCA-induced biofilm formation. However, there might be some redundancy within the carbon metabolism regulation network because deletion of hprK had limited and variable effects. These findings are in line with our transcriptomic analysis indicating that a profound metabolic reorganisation occurs prior to DOCA-induced biofilm formation. Given that DOCA-induced biofilm formation and glucose transport are both PTS dependent, our data suggest that C. difficile preferentially uses glucose first and then adapts its metabolism to use different nutrient sources, including pathways controlled by SigL, SigH and Spo0A, to induce biofilm formation in the presence of DOCA.
To elucidate the downstream metabolic pathways driving biofilm-formation, we targeted genes identified in our transcriptomic analysis associated with the conversion of fumarate to pyruvate (fumAB-CD630_10500), Neu5Ac transport and metabolism (nanEAT), proline metabolism (prdB), low intracellular NADH/NAD + activated regulator (rex) and the ferric uptake regulator (fur). We then tested the ability of these gene deletion and inactivation strains to form biofilms. Every strain tested formed biofilms similar to the parental strain (Fig. 3E) with the exception of the fur::erm strain that had a slight growth delay in BHISG with DOCA while biofilm levels reached that of the parental strain by 72 h (data not shown). Taken together, our results suggest that BHISG provides multiple and redundant nutrient sources and only specific nutrients such as glucose and cysteine are essential for DOCA-induced biofilm formation in C. difficile.
Branched-chain amino acids and mucus-derived sugars potentiate the effect of DOCA
The use of a complex medium (BHISG) in the biofilm formation assay made it difficult to identify specific metabolic processes and metabolites involved in DOCA-induced biofilm formation. Therefore, we sought to optimize a minimal medium that could support biofilm formation in the presence of DOCA. We first tested the C. difficile minimal medium (CDMM) described by Cartman and Minton [42] with some modifications. Specifically, glucose and cysteine were increased to 100 mM and 0.1%, respectively, to match the concentrations present in our complex medium. We also tested different sources of amino acids and/or peptides. To increase the biofilm biomass, we used our transcriptomic data to identify key metabolic pathways that were upregulated during DOCA-induced biofilm formation and observed that predicted branched-chain amino acid (BCAA) transporters (CD630_12590, CD630_12600, CD630_27020) were upregulated at 24 h. Therefore, we added BCAA to our semi-defined medium and observed an increase in biofilm biomass when the medium was supplemented with BCAA, cysteine and a carbohydrate source, such as glucose (Fig. 4A, B and Fig. S5A). We noted that only casein hydrolysate from Oxoid supported DOCA-induced biofilm formation while casamino acids from Difco or a mixture of individual essential amino acids did not (Fig. S5A). The resulting medium that supported DOCA-induced biofilm formation was named C. difficile semi-defined biofilm medium (BM).
Biofilm formation was assessed 48 h after incubation when the medium was supplemented with 1% (w/v) BCAA supplementation (A), without cysteine supplementation (B),100 mM mucus-derived sugars (C) or glucose (Glu) and N-acetyl-glucosamine (GluNAc; D). Neu5Ac: N-acetylneuraminic acid; GalNAc; N-acetylgalactosamine. Experiments presented in (B) and (C) were performed at the same time. Biofilm formation by ΔptsI and ΔnanEAT in medium supplemented with 100 mMNeu5Ac (E). Asterisks indicate statistical significance determined with a Kruskal–Wallis test followed by an uncorrected Dunn’s test (*p ≤ 0.05, **p ≤ 0.001 vs no sugars, 100 mM glucose or 630Δerm). Dots represent biological replicates from experiments performed on different days, the bars represent the mean and the error bars represent the SEM.
In addition to BCAA transporters being upregulated during biofilm formation, the Neu5Ac transporter was also upregulated in our transcriptomic analysis (Supplementary data). Thus, we tested if mucus-derived sugars potentiate DOCA-induced biofilm formation. Mucus is typically broken down into different hexose sugars including glucose, GlcNAc, fucose, Neu5Ac, galactose and N-acetylgalactosamine (GalNAc) and GlcNac and Neu5Ac acquisition is important for C. difficile growth in the intestinal tract [6, 43]. We tested the effect of these sugars on biofilm formation. When BM was supplemented with DOCA and 100 mM of each sugar; the addition of glucose, GlcNAc or Neu5Ac induced biofilm formation, whereas the addition of fucose, galactose and GalNAc had no effect (Fig. 4C). We then sought to see if combining different sugars has additive effects on biofilm formation. When glucose and GlcNAc were mixed at concentrations below those required for biofilm formation (100 mM), the mixture supported biofilm formation at levels equivalent to those with a single sugar (Fig. 4D). We then tested the biofilm-formation ability of a strain lacking the nanEAT operon in the presence of 100 mM Neu5Ac. This operon encodes a non-PTS transporter (nanT), an acetylneuraminate lyase (nanA) and the N-acetylmannosamine-6-phosphate 2-epimerase (nanE), involved in the metabolism of Neu5Ac (Fig. S2). In the absence of nanEAT but not ptsI, C. difficile failed to form a biofilm in BM supplemented with DOCA and 100 mM Neu5Ac (Fig. 4E). Taken together, our results reinforce the idea that metabolized sugars (glucose, GlcNAc and Neu5Ac) potentiate the effect of DOCA (see [20]).
To further characterize the biofilm formed in BM supplemented with glucose (BMG) and DOCA, we analysed the composition of the biofilm matrix by enzymatic dispersion of preformed biofilms and by gel electrophoresis of the isolated matrix. As observed previously with biofilm formed in BHISG supplemented with DOCA [20], NaIPO4 treatment, which denatures polysaccharides, failed to disperse preformed biofilms, while DNase-treatment dispersed preformed biofilms (Fig. S5B). Unlike what was observed in BHISG with DOCA, proteinase K treatment dispersed preformed biofilms from cells grown in BMG with DOCA. When we analysed biofilm matrix of cells grown in BMG with DOCA, we observed extracellular DNA (eDNA) and patterns of proteins, glycoproteins and DNase/proteinase resistant material (e.g. polysaccharides or glycosylated amyloid-like fibres) that was similar to that of the biofilm matrix of C. difficile grown in BHISG with DOCA (Fig. S5C, D and E). However, the high molecular weight smear observed in BHISG with DOCA disappeared from the matrix of biofilms from cells grown in BMG with DOCA (Fig. S5E and S5F). This disappearance suggests that either C. difficile produces high molecular weight glycol molecules in BHISG with DOCA or that the smear is from component(s) of the medium in our samples. Taken together, our data indicate that the biofilm formed in BMG with DOCA is similar to the one formed in BHISG with DOCA but proteins play a larger role in maintaining biofilm matrix stability when C. difficile is grown in BMG with DOCA.
Extracellular pyruvate is required for biofilm formation in BMG with DOCA
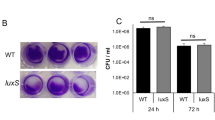
Given that we detect DOCA-induced biofilm formation at 48 h but not 24 h, we hypothesized that C. difficile detects high-cell density via quorum sensing. In support of this hypothesis, we observed up-regulation at 24 h of the genes encoding the autoinducer agr and its associated transporter protein, while luxS was downregulated (Supplementary data). Deletion of the autoinducer agr operon or inactivation of luxS did not alter DOCA-induced biofilm formation (Fig. S4A).
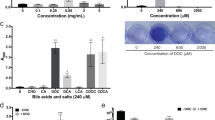
Since deletion of typical quorum sensing molecules did not appear to alter DOCA-induced biofilm formation, we hypothesized that a metabolite could drive this lifestyle switch. We analysed the volatile and non-volatile acid content of spent culture supernatants. After 24 h of growth in BHISG with DOCA, pyruvic acid levels were high and these levels were decreased at 48 h. In the absence of DOCA, pyruvic acid levels were lower, but the level did not decrease over time (Fig. S6A).
Based on this analysis, we hypothesized that a high level of extracellular pyruvate was important for initiating DOCA-induced biofilm formation. To build on this observation, we tested the effect of pyruvate depletion on biofilm formation by adding pyruvate dehydrogenase to C. difficile cultures. When extracellular pyruvate was enzymatically depleted by pyruvate dehydrogenase addition at 24 h in BMG with DOCA, biofilm formation was inhibited. This effect was not observed when heat-inactivated pyruvate dehydrogenase or buffer were added to BMG with DOCA at 24 h (Fig. 5A). Based on these data, we conclude that pyruvate might act as a critical molecule triggering C. difficile’s switch from a planktonic to a biofilm lifestyle in the presence of DOCA.
A Effect of enzymatic depletion of extracellular pyruvate on 48 h biofilm formation in C. difficile. Pyruvate dehydrogenase was added to C. difficile grown in BMG after 24 h of growth. Control culture were treated with heat-inactivated enzyme or buffer are shown. Effect of pyruvate on 48 h biofilm formation in the absence of DOCA by C. difficile grown in BM (B) or BMG (C). Where indicated, pyruvate was added at inoculation (0 h) or after 8 hours of growth (8 h). D Biofilm formation for C. difficile 630Δerm in BM in the presence of 240 µM DOCA and glucose (glu) and/or pyruvate (pyr). For (A), and (D), asterisks indicate statistical significance determined with a Kruskal–Wallis test followed by an uncorrected Dunn’s test (*p ≤ 0.05, vs heat-inactivated enzyme; *p ≤ 0.05, ***p ≤ 0.001 vs 100 mM glucose). For (B), and (C), asterisks indicate statistical significance determined with a two-way ANOVA followed by Fisher’s least significant difference test (****p ≤ 0.001 vs 0 mM pyruvate). Dots represent biological replicates from experiments performed on different days, the bars indicate the mean and the error bars represent the SEM.
Pyruvate induces biofilm formation in the absence of DOCA
To test if pyruvate alone could induce biofilm formation, we supplemented BM or BMG with increasing concentrations of pyruvate (25, 50 and 100 mM). When added at inoculation, pyruvate-induced biofilm formation in BM and BMG at 100 and 50 mM respectively (Fig. 5B and C). When it was added after 8 h of growth (log to stationary phase transition), pyruvate only induced biofilm formation in BMG at 100 mM (Fig. 5C). We also added pyruvate after 24 h of growth but this failed to induce biofilm formation (Data not shown). This indicates the metabolic state of the bacteria partially determines whether pyruvate will induce biofilm formation. We then tested if pyruvate could cooperate with DOCA to support biofilm formation and observed that 25 mM, instead of 100 mM pyruvate was sufficient to support biofilm formation in BM with DOCA (Fig. 5D). However, glucose and pyruvate did not have a cooperative effect when added at 50 and 12.5 mM, respectively (Fig. 5D). When we consider our biofilm formation assay, the gas chromatography data and our pyruvate depletion assay, our results indicate that biofilm-formation depends on the amount of available pyruvate and suggests that this metabolite is a key factor driving DOCA-induced biofilm formation.
Induction of biofilm formation requires pyruvate sensing by the CD630_26010-26020 TCS and pyruvate importer CstA
In a previous study, we identified a LytRS two-component regulatory system (TCS) homologue (CD630_26020-CD630_26010) that regulated toxin gene expression in response to pyruvate [40]. In other Gram-positive bacteria, genes encoding TCSs sensing pyruvate are associated with a pyruvate importer and a gene encoding a potential importer (CD630_26000) that is located immediately downstream of CD630_26010. CD630_26000 encodes a CstA homologue that was initially annotated as a peptide transporter, but homologues were recently shown to be pyruvate importers in Escherichia coli [44]. To determine if CstA or TCS CD630_26020-26010 are involved in biofilm formation and respond to pyruvate availability, we deleted cstA and tested whether the ΔcstA strain and the previously described CD630_26020::erm strain [40] form biofilms in BM with 100 mM pyruvate and BMG with 50 mM pyruvate. Pyruvate was added at inoculation and the ΔptsI strain, unable to uptake PTS-dependent sugars, was used as a positive or negative control when pyruvate was added alone or with glucose, respectively, to the growth medium.
In BM supplemented with 100 mM pyruvate as the sole carbon source, the CD630_26020::erm and ΔcstA strains did not form biofilm whereas, as expected, the ΔptsI and parental strain formed biofilms (Fig. 6A). In BMG supplemented with 50 mM pyruvate, the CD630_26020::erm and ΔcstA strains formed biofilms whereas the ΔptsI strain did not form a biofilm (Fig. 6B). The inability of the ΔptsI strain to form a biofilm in the presence of glucose confirms that glucose metabolism is required for biofilm formation. This might be due to the production of pyruvate by glycolysis which would increase the level of extracellular pyruvate above the threshold required to induce biofilm-formation (Fig. 5A and C). The absence of biofilm formation for the ΔcstA strain or the CD630_26020::erm strain when pyruvate is the sole carbon source suggests that the CstA importer and the CD630_26020-CD630_26010 TCS are primarily involved in the uptake and sensing of extracellular pyruvate, respectively. Biofilm formation by the ΔptsI strain indicates that the PTS system is not involved in pyruvate uptake.
Biofilm formation was assessed in strains lacking the CD630_26020-CD630_26010 TCS, cstA, ptsI, ccpA or ccpA and cstA in BM with pyruvate (A) and BMG with pyruvate (B). Pyruvate used during growth by the parental, ΔcstA, ΔccpA or ΔcstAΔccpA strains grown in BMG (D), BM with 100 mM pyruvate (D) or CDMOG with 50 mM pyruvate (E) (% remaining = 100 × pyruvate in media after growth (24 h or 48 h)/starting pyruvate concentration). For (A), and (B), asterisks indicate statistical significance determined with a Kruskal–Wallis test followed by an uncorrected Dunn’s test (**p ≤ 0.001 ***p ≤ 0.001, vs 630Δerm). For (C), (D) and (E), asterisks indicate statistical significance determined with a two-way ANOVA test followed by Fisher’s least significant difference test (**p ≤ 0.001, ***p ≤ 0.001 vs 630Δerm). Dots represent biological replicates from experiments performed on different days, the bars represent the mean and the error bars represent the SEM.
The ability of the ΔcstA strain or the CD630_26020::erm strain (Fig. 6B) to form biofilms in the presence of glucose and pyruvate suggests that at least in the presence of glucose, CstA or CD630_26020-CD630_26010 are not solely required for the uptake of pyruvate and that other importer(s) and/or regulatory mechanisms might be involved. In other bacteria, multiple proteins have been identified as pyruvate importers and the expression of these importers are often controlled by the CCR regulatory network [45, 46]. In C difficile, CcpA is the major regulator of CCR, which controls the use of alternate carbon sources. In support of its role in controlling pyruvate metabolism, CcpA binds to the region upstream of cstA [47]. To test if the catabolic repression system was involved in pyruvate transport, we deleted cstA in our ΔccpA strain. The resulting double deletion strain (ΔccpAΔcstA) was tested for its ability to form biofilms in BMG supplemented with 50 mM pyruvate. The ΔccpAΔcstA strain did not form a biofilm whereas the ΔccpA or ΔcstA strains formed biofilms, although the amount of biofilm formed by the ΔccpA strain was more variable (Fig. 6B). These data support that extracellular pyruvate could also be imported by a second importer controlled by CcpA in the presence of glucose.
To test if extracellular pyruvate was used by the parental and deletion strains, pyruvate levels in the culture supernatant was measured from cells grown in BM with 100 mM pyruvate, BMG or BMG with 50 mM pyruvate for 24 and 48 h. In BMG, our assay did not detect significant changes in extracellular pyruvate concentration in the parental strains but the ΔcstA strain accumulated six times more pyruvate in its supernatant (Fig. 6C). This accumulation suggests that the ΔcstA strain can excrete pyruvate in a CstA-independent manner. In BM with 100 mM pyruvate, the ΔcstA strain used ~50% of the pyruvate present in the growth medium but did not deplete pyruvate to the same extent as the parental strain (10% of starting concentration; Fig. 6D). In BMG with 50 mM pyruvate, the ΔcstA and ΔccpAΔcstA strains did not reduce the amount pyruvate to the levels of the parental or the ΔccpA strains after 48 h (Fig. 6E). However, the ΔcstA and the ΔccpAΔcstA strain were able to reduce the amount of pyruvate present in the growth medium. Interestingly, the ΔccpA strain was able to use 90% of the pyruvate by 24 h. The rapid depletion of pyruvate in the ΔccpA strain compared to the parental strain and the reduction in the amount of extracellular pyruvate used by the ΔccpAΔcstA strain suggest that CstA, and not another importer, is highly active in the absence of CcpA. This is consistent with previous data showing that CcpA binds to the region upstream of cstA and supports the hypothesis that cstA expression is repressed by CcpA in the presence of glucose [47].
The decrease in extracellular pyruvate in the absence of CstA suggests that pyruvate is also imported independently of CstA (Fig. 6E). In other bacteria such as Bacillus subtilis, there are additional pyruvate importers encoded by pftAB that belong to the LrgAB holin anti-holin family. When these predicted holin anti-holin systems act as pyruvate importers, their coding genes are invariably located next to a TCS. While an lrgAB-family gene (~35% amino acid identity to PftB and no PftA homologs) exists in C. difficile strain 630, the genes are not associated with a TCS and were downregulated at 24 h in BHISG supplemented with DOCA (Supplementary data), suggesting that this lgrB gene homolog is unlikely to encode the additional pyruvate importer in C. difficile.
In addition to the lrgAB homolog, we noted the presence of a second cstA-homolog encoded by CD630_23730. This gene was highly expressed in C. difficile at 24 h in BHISG supplemented with DOCA (Supplementary data); however, unlike cstA it is not located next to a TCS. It is possible that CD630_23730 is the importer responsible for the partial pyruvate uptake by the ΔcstA and ΔccpAΔcstA strains when grown in BMG with 50 mM pyruvate (Fig. 6D). This would be consistent with previous data showing that CD630_23730 is repressed by CcpA in TY medium, but not in TY medium supplemented with glucose [47]. The expression of additional pyruvate importers in the presence of preferred carbon sources, such as glucose are in agreement with our findings and those in other bacteria, such as E. coli [48].
In the absence of glucose, our data indicate that pyruvate uptake leading to biofilm formation is dependent on CstA. However, in the presence of glucose, biofilm formation is dependent on glucose metabolism followed by a critical metabolic shift requiring pyruvate uptake by CstA and other importers. In support of this, we observed that the ΔccpAΔcstA strain used the same amount of extracellular pyruvate as the parental strain after 24 h, but pyruvate uptake in the ΔccpAΔcstA strain was five times less (onefold decrease in extracellular pyruvate) than the parental strain (fivefold decrease in extracellular pyruvate) between 24 and 48 h (Fig. 6E). This suggests the 24–48h period is when the critical shift in metabolism occurs. In support, the ΔccpA and ΔcstA that formed biofilms also had ~10-fold and ~2-3-fold decreases, respectively, in extracellular pyruvate levels between 24 and 48 h. Taken together, active pyruvate uptake in the stationary phase, a process that is partially CstA-dependent, drives a metabolic shift in C. difficile and leads to biofilm formation. These data confirm the importance of the role of pyruvate uptake in biofilm formation. This lifestyle switch can be driven by efficient pyruvate uptake and/or carbon metabolism.
Discussion
We previously identified DOCA as an inducer of biofilm formation by C. difficile. DOCA-induced biofilm took more than 24 h to form, suggesting the specifics steps and pathways controlling the shift from planktonic to biofilm could be elucidated. In this study, our objectives were to identify and characterize key molecules and events required for DOCA-induced biofilm formation. Using a combination of time-course transcriptomics and deletion strains (Table S5), we identified several metabolic processes as a key drivers of DOCA-induced biofilm formation. We also show that DOCA-induced biofilm formation does not require quorum sensing and surface proteins previously associated with biofilm formation in the absence of DOCA (Table S5). However, the T4aP machinery encoded by the pilA1 cluster is required for biofilm formation. Using these findings, we decided to analyze culture supernatants for key metabolites involved in DOCA-induced biofilm formation. We identify extracellular pyruvate as a potential metabolite and then demonstrated its importance and integration for biofilm formation.
In some bacteria, pyruvate is known to be excreted during overflow metabolism [45, 46, 49, 50]. In our study, C. difficile is grown in BHISG and this medium provides an environment rich in proteins with an excess of glucose, which is suited for overflow metabolism [51]. Based on our transcriptomic analysis, DOCA has a profound impact on metabolism-associated genes, specifically glycolysis (Fig. S2). The presence of DOCA induces a metabolic stress that probably leads to overflow metabolism between inoculation and 14 h, the excretion of pyruvate and, once glucose is exhausted, biofilm formation (see the proposed model in Fig. 7). The mechanism by which pyruvate is excreted remains to be elucidated in bacteria and this appears to be independent of pyruvate importers [52]. Furthermore, the absence of the metabolic regulatory factors SigL, CcpA, and CodY resulted in decreased DOCA-induced biofilm formation as shown in this work, and Dubois et al. [20]. Specifically, these regulators are important to control flux between different metabolic pathways and help C. difficile transition to different sources of energy [47, 53, 54]. Interestingly, the sigL inactivated strain was previously shown to require glucose for optimal growth and does not excrete pyruvate [40, 54]. Given that the sigL inactivated strain did not form biofilm, this is consistent with our hypothesis that pyruvate excretion drives DOCA-induced biofilm formation. Consumption of a preferred carbon source (e.g. glucose) is also important since a strain lacking ptsI was unable to form DOCA-induced biofilms in the presence of glucose. In addition to glucose, other sugars used by C. difficile that require their own uptake systems, such as Neu5Ac and the NanEAT system, might also potentiate the metabolic shift required for biofilm formation. Taken together, our data indicate that in presence of DOCA, C. difficile must control its metabolic activity to ensure overflow metabolism is activated to promote stationary phase survival and biofilm formation (Fig. 7).
A Important cellular processes that contribute to metabolic adaptation and biofilm formation are highlighted for each time interval. Green outer ring indicates the concentration of extracellular pyruvate; darker shades of green are indicative of increased pyruvate concentrations (i.e. light green is the lowest concentration and dark green is the highest concentration). The undefined arc indicates when C. difficile forms a biofilm. Arrows indicate cellular processes that occur, and the thickness of the arrows indicates how essential they are for biofilm formation (i.e. dashed arrows: least essential, thick, full arrows: absolutely required). B Line graph showing hypothetical model of the changes in the quantity of extracellular nutrients, biomass, and biofilm over time. Briefly, during the exponential phase (light blue shading), the PTS will import glucose leading to the excretion of pyruvate. Glucose is preferentially used until the bacteria enter stationary phase (light green shading) after ~14 h of growth. After entry into the stationary phase, there is a metabolic shift driven by CcpA, SigH, SigL and CodY, and cells start using alternative sources of energy. As C. difficile progresses through stationary phase, it will sense extracellular pyruvate via CD630_26010-26020 TCS and will start actively importing extracellular pyruvate via CstA and other importers such as CD630_23730. The use of extracellular pyruvate maintains C. difficile viability during the stationary phase and the type IVa pili (T4aP) machinery will start assembly pili to enhance adherence. A sub-population of the cells will undergo lysis contributing eDNA for the biofilm matrix and, as time passes, the biofilm biomass will increase reaching its peak at ~48 h (biofilm phase indicated in yellow).
The importance of overflow metabolism and the excretion of pyruvate likely explain why casein hydrolysate supported biofilm formation whereas casamino acids and specific essential amino acids did not. Casein hydrolysate is a richer source of small peptides and amino acids than casamino acids or purified amino acids. This explains the additive effect of BCAA supplementation on biofilm formation as adding BCAAs may replace the BCAA synthesized as end-products of pyruvate metabolism. BCAAs are likely used in the oxidative Stickland reaction to produce energy during late stationary phase to promote biofilm formation. Overall, it appears that our semi-defined medium was optimized to support overflow metabolism and biofilm formation by C. difficile.
The key role of extracellular pyruvate for biofilm formation is not limited to C. difficile. Recently, it was demonstrated that Staphylococcus aureus requires the presence of extracellular pyruvate to form and maintain a biofilm [33]. In Streptococcus mutans, pyruvate improves stationary phase survival and protects against microbiota-generated oxidative stresses in a density- and CcpA-dependent fashion [55,56,57]. Furthermore, pyruvate fermentation is important for Pseudomonas aeruginosa microcolony formation and long-term survival in anaerobic environments [58, 59]. In this case, lactate is oxidized to pyruvate by the cells in the oxygen-rich top layer of the biofilm and the secreted pyruvate is then converted to acetate by the cells in the anoxic lower layers [58, 59]. This metabolic cooperation within the biofilm community is crucial, as it provides a means for the cells to produce enough ATP to survive, but not grow, in the nutrient-poor environment of the deep layer of the biofilm [60]. Unlike P. aeruginosa, B. subtilis uses lactate to promote optimal biofilm formation [61]. However, C. difficile is a strict anaerobe and does not need to switch from an aerobic-based metabolism to an anaerobic-based metabolism. We must consider that the cells in the deep layers of the biofilm have limited resources. Our transcriptomic analysis supports that biofilm cells reprioritise their metabolism, as we observed major shifts in the expression of several genes associated with metabolic pathways at 48 h. In our transcriptional analysis, genes associated with fermentation were upregulated (Fig. 2) at 24 h but not 48 h. We also found evidence that butyric acid and lactic acid production occurs between 24 h and 48 h (Fig. S6B). Therefore, C. difficile probably starts using extracellular pyruvate after 24 h of incubation, which helps long-term survival of the cells during the stationary phase. In support of this, Haemophilus influenzae uses pyruvate as a pivotal point in metabolic adaptation for biofilm cells [62]. This reprioritization is critical for metabolic adaptation and long-term survival of biofilm cells.
A recent study provides some interesting insight into biofilm formation that is dependent on eDNA [63]. In this study, sub-inhibitory concentrations of antibiotics that target the cell envelope enhance biofilm formation by increasing the amount of DNA release from lysing cells without affecting the overall viability of the population. From their data, the authors build a mathematical model that accounts for cell lysis and death, viability, growth and aggregation or binding provided by eDNA. Using this model, we can understand the importance of metabolism in the induction of biofilm in the presence of DOCA or pyruvate given that the DOCA-induced biofilm formed by C. difficile is eDNA-dependent [20]. In our proposed model, extracellular pyruvate improves long-term viability of C. difficile during the stationary phase which compensates for autolysis (Fig. 7). As DNA is released by lysis, viable cells start aggregating, which may involve the T4aP, as time passes, the biomass increases and C. difficile adapts its metabolism to this sedentary lifestyle. Overall, our growth conditions allow C. difficile to pass the “biofilm threshold” and any disturbance to the metabolism (SigL, CcpA, CodY, PtsI), lysis (Cwp19), binding or aggregation (eDNA, T4aP) would prevent C. difficile from crossing this threshold (Fig. 7).
Extracellular pyruvate is also produced by the gut microbiota [64,65,66,67] and provides protection against colonization by Salmonella [64]. In this case, pyruvate concentrations were higher in the luminal content of specific pathogen-free (SPF) mice than those of germ-free mice or SPF mice treated with oral vancomycin or neomycin. Furthermore, colonization by the gut commensal bacterium Lactobacillus helveticus, which excretes high concentrations of pyruvate in vitro, increased pyruvate concentrations in SPF mice. Therefore, it is possible that C. difficile encounters extracellular pyruvate during colonization of the intestinal tract containing a normal microbiota or when the microbiota is restored after antibiotic therapy.
Depending on the composition of the microbiota, C. difficile could encounter favourable conditions that would include sub-inhibitory concentrations of DOCA, and availability of specific amino acids, mucus-derived sugars, and pyruvate [3,4,5,6]. Under these favourable conditions, sub-inhibitory concentration of DOCA would trigger a metabolic adaptation in C. difficile to use the available metabolites produced by the microbiota. Interestingly, the molecules that trigger biofilm formation also repress sporulation (DOCA; [20]) and toxin production (DOCA and pyruvate; [20, 40]). Overall, conditions that are favourable for long-term colonization and biofilm formation promote C. difficile persistence rather than sporulation or virulence. Any disturbance to this balance, such as the disappearance of inhibitory molecules (e.g. DOCA), increase nutrient availability and change in SCFA profiles, could trigger blooms of C. difficile as observed recently [68]. Furthermore, other signals, such as change in amino acids availability or increase SCFA production, could prevent C. difficile from entering a biofilm or persistence state and promote sporulation. These hypotheses are supported by recent in silico modelling of the metabolism in the context of sporulation and virulence where each context has distinct metabolic intake and efflux [69].
In summary, we have identified key determinants of DOCA-induced biofilm by C. difficile. These determinants are unique to DOCA-induced biofilm formation suggesting a distinct mechanism. These include regulator of lifestyle and metabolism and T4aP but the most interesting finding is the importance of extracellular pyruvate and its integration in promoting biofilm formation. Early in our growth conditions (before 14 h), pyruvate is probably excreted as a result of overflow metabolism and, as C. difficile progresses from exponential to stationary phase, extracellular pyruvate is imported using CstA and other pyruvate importers (Fig. 7). This prevents rapid cell death and allows C. difficile to generate eDNA through autolysis and pass the “biofilm threshold”. Interestingly, extracellular pyruvate is produced in the gut by commensal bacteria and this could act as a source of pyruvate for C. difficile. In conclusion, extracellular pyruvate in the presence of other microbial metabolite could act as a key molecule driving C. difficile persistence in the intestinal tract or in response to DOCA.
Data availability
RNA-Seq data generated in this study are available in the NCBI-GEO with accession no GSE165116.
References
Crobach MJT, Vernon JJ, Loo VG, Kong LY, Péchiné S, Wilcox MH, et al. Understanding Clostridium difficile colonization. Clin Microbiol Rev. 2018;31:e00021–17.
Lim SC, Knight DR, Riley TV. Clostridium difficile and one health. Clin Microbiol Infect. 2020;26:857–63.
Abbas A, Zackular JP. Microbe-microbe interactions during Clostridioides difficile infection. Curr Opin Microbiol. 2020;53:19–25.
Ghimire S, Roy C, Wongkuna S, Antony L, Maji A, Keena MC, et al. Identification of Clostridioides difficile inhibiting gut commensals using culturomics, phenotyping, and combinatorial community assembly. mSystems. 2020;5:e00620–19.
Girinathan BP, DiBenedetto N, Worley JW, Peltier J, Arrieta-Ortiz ML, Immanuel R, et al. In vivo commensal control of Clostridioides difficile virulence. bioRxiv 2020.01.04.894915 [Preprint] 2021. Available from: https://www.biorxiv.org/content/10.1101/2020.01.04.894915v2.
Pereira FC, Wasmund K, Cobankovic I, Jehmlich N, Herbold CW, Lee KS, et al. Rational design of a microbial consortium of mucosal sugar utilizers reduces Clostridiodes difficile colonization. Nat Commun. 2020;11:5104.
Buffie CG, Bucci V, Stein RR, McKenney PT, Ling L, Gobourne A, et al. Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature. 2015;517:205.
Studer N, Desharnais L, Beutler M, Brugiroux S, Terrazos MA, Menin L, et al. Functional intestinal bile acid 7α-dehydroxylation by Clostridium scindens associated with protection from Clostridium difficile infection in a gnotobiotic mouse model. Front Cell Infect Microbiol. 2016;6:191.
McDonald JAK, Mullish BH, Pechlivanis A, Liu Z, Brignardello J, Kao D, et al. Inhibiting growth of Clostridioides difficile by restoring valerate, produced by the intestinal microbiota. Gastroenterology. 2018;155:1495.
Solbach P, Chhatwal P, Woltemate S, Tacconelli E, Buhl M, Gerhard M, et al. baiCD gene cluster abundance is negatively correlated with Clostridium difficile infection. PLoS ONE. 2020;13:e0196977.
Seekatz AM, Theriot CM, Rao K, Chang Y-M, Freeman AE, Kao JY, et al. Restoration of short chain fatty acid and bile acid metabolism following fecal microbiota transplantation in patients with recurrent Clostridium difficile infection. Anaerobe. 2020;53:64.
Deakin LJ, Clare S, Fagan RP, Dawson LF, Pickard DJ, West MR, et al. The Clostridium difficile spo0A gene is a persistence and transmission factor. Infect Immun. 2012;80:2704.
Castro-Córdova P, Mora-Uribe P, Reyes-Ramírez R, Cofré-Araneda G, Orozco-Aguilar J, Brito-Silva C, et al. Entry of spores into intestinal epithelial cells contributes to recurrence of Clostridioides difficile infection. Nat Commun. 2021;12:1140.
Pettit LJ, Browne HP, Yu L, Smits WK, Fagan RP, Barquist L, et al. Functional genomics reveals that Clostridium difficile Spo0A coordinates sporulation, virulence and metabolism. BMC Genomics. 2014;15:160.
Dawson LF, Valiente E, Faulds-Pain A, Donahue EH, Wren BW. Characterisation of Clostridium difficile biofilm formation, a role for Spo0A. PLoS ONE. 2012;7:e50527.
Donelli G, Vuotto C, Cardines R, Mastrantonio P. Biofilm-growing intestinal anaerobic bacteria. FEMS Immunol Med Microbiol. 2012;65:318–25.
Crowther GS, Chilton CH, Todhunter SL, Nicholson S, Freeman J, Baines SD, et al. Comparison of planktonic and biofilm-associated communities of Clostridium difficile and indigenous gut microbiota in a triple-stage chemostat gut model. J Antimicrob Chemother. 2014;69:2137–47.
Semenyuk EG, Poroyko VA, Johnston PF, Jones SE, Knight KL, Gerding DN, et al. Analysis of bacterial communities during Clostridium difficile infection in the mouse. Infect Immun. 2015;83:4383–91.
Normington C, Moura IB, Bryant JA, Ewin DJ, Clark EV, Kettle MJ, et al. Biofilms harbour Clostridioides difficile, serving as a reservoir for recurrent infection. npj Biofilms Microbiomes. 2021;7:1–10.
Dubois T, Tremblay YDN, Hamiot A, Martin-Verstraete I, Deschamps J, Monot M, et al. A microbiota-generated bile salt induces biofilm formation in Clostridium difficile. npj Biofilms Microbiomes. 2019;5:14.
Hall-Stoodley L, Costerton JW, Stoodley P. Bacterial biofilms: from the natural environment to infectious diseases. Nat Rev Microbiol. 2004;2:95–108.
Ðapa T, Leuzzi R, Ng YK, Baban ST, Adamo R, Kuehne SA, et al. Multiple factors modulate biofilm formation by the anaerobic pathogen Clostridium difficile. J Bacteriol. 2013;195:545–55.
Soutourina OA, Monot M, Boudry P, Saujet L, Pichon C, Sismeiro O, et al. Genome-wide identification of regulatory RNAs in the human pathogen Clostridium difficile. PLoS Genet. 2013;9:e1003493.
Boudry P, Gracia C, Monot M, Caillet J, Saujet L, Hajnsdorf E, et al. Pleiotropic role of the RNA chaperone protein Hfq in the human pathogen Clostridium difficile. J Bacteriol. 2014;196:3234–48.
Pantaléon V, Soavelomandroso AP, Bouttier S, Briandet R, Roxas B, Chu M, et al. The Clostridium difficile protease Cwp84 modulates both biofilm formation and cell-surface properties. PLoS ONE. 2015;10:e0124971.
Walter BM, Cartman ST, Minton NP, Butala M, Rupnik M. The SOS response master regulator LexA is associated with sporulation, motility and biofilm formation in Clostridium difficile. PLoS ONE. 2015;10:e0144763.
Maldarelli GA, Piepenbrink KH, Scott AJ, Freiberg JA, Song Y, Achermann Y, et al. Type IV pili promote early biofilm formation by Clostridium difficile. Pathog Dis. 2016;74:ftw061.
Vuotto C, Moura I, Barbanti F, Donelli G, Spigaglia P. Subinhibitory concentrations of metronidazole increase biofilm formation in Clostridium difficile strains. Pathog Dis. 2016;74:ftv114.
Saujet L, Pereira FC, Serrano M, Soutourina O, Monot M, Shelyakin PV, et al. Genome-wide analysis of cell type-specific gene transcription during spore formation in Clostridium difficile. PLoS Genet. 2013;9:e1003756.
Monot M, Boursaux-Eude C, Thibonnier M, Vallenet D, Moszer I, Medigue C, et al. Reannotation of the genome sequence of Clostridium difficile strain 630. J Med Microbiol. 2011;60:1193–9.
Peltier J, Hamiot A, Garneau JR, Boudry P, Maikova A, Hajnsdorf E, et al. Type I toxin-antitoxin systems contribute to the maintenance of mobile genetic elements in Clostridioides difficile. Commun Biol. 2020;3:718.
García-Nafría J, Watson JF, Greger IH. IVA cloning: A single-tube universal cloning system exploiting bacterial In Vivo Assembly. Sci Rep. 2016;6:27459.
Goodwine J, Gil J, Doiron A, Valdes J, Solis M, Higa A, et al. Pyruvate-depleting conditions induce biofilm dispersion and enhance the efficacy of antibiotics in killing biofilms in vitro and in vivo. Sci Rep. 2019;9:3763.
Poquet I, Saujet L, Canette A, Monot M, Mihajlovic J, Ghigo J-M, et al. Clostridium difficile biofilm: remodeling metabolism and cell surface to build a sparse and heterogeneously aggregated architecture. Front Microbiol. 2018;9:2084.
Nawrocki KL, Wetzel D, Jones JB, Woods EC, McBride SM. Ethanolamine is avaluable nutrient source that impacts Clostridium difficile pathogenesis. Environ Microbiol. 2018;20:1419–35.
Hofmann JD, Otto A, Berges M, Biedendieck R, Michel A-M, Becher D, et al. Metabolic Reprogramming of Clostridioides difficile during the stationary phase with the induction of toxin production. Front Microbiol. 2018;9:1970.
Sievers S, Metzendorf NG, Dittmann S, Troitzsch D, Gast V, Tröger SM, et al. Differential view on the bile acid stress response of Clostridioides difficile. Front. Microbiol. 2019;10:258.
Saujet L, Monot M, Dupuy B, Soutourina O, Martin-Verstraete I. The key sigma factor of transition phase, SigH, controls sporulation, metabolism, and virulence factor expression in Clostridium difficile. J Bacteriol. 2011;193:3186–96.
Girinathan BP, Ou J, Dupuy B, Govind R. Pleiotropic roles of Clostridium difficile sin locus. PLoS Pathog. 2018;14:e1006940.
Dubois T, Dancer-Thibonnier M, Monot M, Hamiot A, Bouillaut L, Soutourina O, et al. Control of Clostridium difficile physiopathology in response to cysteine availability. Infect Immun. 2016;84:2389–3405.
Gu H, Yang Y, Wang M, Chen S, Wang H, Li S, et al. Novel cysteine desulfidase CdsB involved in releasing cysteine repression of toxin synthesis in Clostridium difficile. Front Cell Infect Microbiol. 2018;7:531.
Cartman ST, Minton NP. A mariner-based transposon system for in vivo random mutagenesis of Clostridium difficile. Appl Environ Microbiol. 2010;76:1103–9.
Ng KM, Ferreyra JA, Higginbottom SK, Lynch JB, Kashyap PC, Gopinath S, et al. Microbiota-liberated host sugars facilitate post-antibiotic expansion of enteric pathogens. Nature. 2013;502:96–99.
Hwang S, Choe D, Yoo M, Cho S, Kim SC, Cho S, et al. Peptide transporter CstA imports pyruvate in Escherichia coli K-12. J Bacteriol. 2018;200:e00771–17.
Charbonnier T, Le Coq D, McGovern S, Calabre M, Delumeau O, Aymerich S, et al. Molecular and physiological logics of the pyruvate-induced response of a novel transporter in Bacillus subtilis. mBio. 2017;8:e00976–17.
van den Esker MH, Kovács ÁT, Kuipers OP. YsbA and LytST are essential for pyruvate utilization in Bacillus subtilis. Environ Microbiol. 2017;19:83–94.
Antunes A, Camiade E, Monot M, Courtois E, Barbut F, Sernova NV, et al. Global transcriptional control by glucose and carbon regulator CcpA in Clostridium difficile. Nucleic Acids Res. 2012;40:10701–18.
Ogasawara H, Ishizuka T, Yamaji K, Kato Y, Shimada T, Ishihama A. Regulatory role of pyruvate-sensing BtsSR in biofilm formation by Escherichia coli K-12. FEMS Microbiol Lett. 2019;366:fnz251.
Tomlinson GA, Hochstein LI. Studies on acid production during carbohydrate metabolism by extremely halophilic bacteria. Can J Microbiol. 1972;18:1973–6.
Ruby EG, Nealson KH. Pyruvate production and excretion by the luminous marine bacteria. Appl Envir Microbiol. 1977;34:164–9.
Sonenshein AL. Control of key metabolic intersections in Bacillus subtilis. Nat Rev Microbiol. 2007;5:917–27.
Gasperotti A, Göing S, Fajardo-Ruiz E, Forné I, Jung K. Function and regulation of the pyruvate transporter CstA in Escherichia coli. Int J Mol Sci. 2020;21:9068.
Dineen SS, McBride SM, Sonenshein AL. Integration of metabolism and virulence by Clostridium difficile CodY. J Bacteriol. 2010;192:5350–62.
Soutourina O, Dubois T, Monot M, Shelyakin PV, Saujet L, Boudry P, et al. Genome-wide transcription start site mapping and promoter assignments to a sigma factor in the human enteropathogen Clostridioides difficile. Front Microbiol. 2020;11:1939.
Ahn S-J, Deep K, Turner ME, Ishkov I, Waters A, Hagen SJ, et al. Characterization of LrgAB as a stationary phase-specific pyruvate uptake system in Streptococcus mutans. BMC Microbiol. 2019;19:223.
Ishkov IP, Ahn S-J, Rice KC, Hagen SJ. Environmental triggers of lrgA expression in Streptococcus mutans. Front Microbiol. 2020;11:18.
Redanz S, Treerat P, Mu R, Redanz U, Zou Z, Koley D, et al. Pyruvate secretion by oral streptococci modulates hydrogen peroxide dependent antagonism. ISME J. 2020;14:1074–88.
Eschbach M, Schreiber K, Trunk K, Buer J, Jahn D, Schobert M. Long-term anaerobic survival of the opportunistic pathogen Pseudomonas aeruginosa via pyruvate fermentation. J Bacteriol. 2004;186:4596–5604.
Petrova OE, Schurr JR, Schurr MJ, Sauer K. Microcolony formation by the opportunistic pathogen Pseudomonas aeruginosa requires pyruvate and pyruvate fermentation. Mol Microbiol. 2012;86:819–35.
Stewart PS, White B, Boegli L, Hamerly T, Williamson KS, Franklin MJ, et al. Conceptual model of biofilm antibiotic tolerance that integrates phenomena of diffusion, metabolism, gene expression, and physiology. J Bacteriol. 2019;201:e00307–19.
Chai Y, Kolter R, Losick R. A widely conserved gene cluster required for lactate utilization in Bacillus subtilis and its involvement in biofilm formation. J Bacteriol. 2009;191:2423–30.
Harrison A, Hardison RL, Wallace RM, Fitch J, Heimlich DR, Bryan MO, et al. Reprioritization of biofilm metabolism is associated with nutrient adaptation and long-term survival of Haemophilus influenzae. npj Biofilms Microbiomes. 2019;5:33.
Yu W, Hallinen KM, Wood KB. Interplay between antibiotic efficacy and drug-induced lysis underlies enhanced biofilm formation at subinhibitory drug concentrations. Antimicrob Agents Chemother. 2018;62:e01603–17.
Morita N, Umemoto E, Fujita S, Hayashi A, Kikuta J, Kimura I, et al. GPR31-dependent dendrite protrusion of intestinal CX3CR1(+) cells by bacterial metabolites. Nature. 2019;566:110–4.
Jacobs DM, Deltimple N, Velzen E, van, Dorsten FA, van, Bingham M, Vaughan EE, et al. 1H NMR metabolite profiling of feces as a tool to assess the impact of nutrition on the human microbiome. NMR Biomedicine. 2008;21:615–26.
Theriot CM, Koenigsknecht MJ, Carlson PE, Hatton GE, Nelson AM, Li B, et al. Antibiotic-induced shifts in the mouse gut microbiome and metabolome increase susceptibility to Clostridium difficile infection. Nat Commun. 2014;5:3114.
Shankar V, Homer D, Rigsbee L, Khamis HJ, Michail S, Raymer M, et al. The networks of human gut microbe–metabolite associations are different between health and irritable bowel syndrome. ISME J. 2015;9:1899–903.
VanInsberghe D, Elsherbini JA, Varian B, Poutahidis T, Erdman S, Polz MF. Diarrhoeal events can trigger long-term Clostridium difficile colonization with recurrent blooms. Nat Microbiol. 2020;5:642–50.
Jenior ML, Leslie JL, Powers DA, Garrett EM, Walker KA, Dickenson ME, et al. Conserved virulence-linked metabolic reprogramming in Clostridioides difficile identified through genome-scale metabolic network analysis. bioRxiv:2020.11.09.373480. [Preprint]. 2020. Available from: https://www.biorxiv.org/content/10.1101/2020.11.09.373480v2.full.
Acknowledgements
We would like to thank Johann Peltier, Imane El Meouche and Laurent Bouillaut for generously providing the CDIP634, fliC::erm, rex::erm and prdB::erm strains. This work was funded by the Institut Pasteur and the “Integrative Biology of Emerging Infectious Diseases” (LabEX IBEID) funded in the framework of the French Government’s “Programme Investissements d’Avenir”. YDNT postdoctoral fellowship was funded by the LabEX IBEID.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Tremblay, Y.D.N., Durand, B.A.R., Hamiot, A. et al. Metabolic adaption to extracellular pyruvate triggers biofilm formation in Clostridioides difficile. ISME J 15, 3623–3635 (2021). https://doi.org/10.1038/s41396-021-01042-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41396-021-01042-5
This article is cited by
-
The cell wall lipoprotein CD1687 acts as a DNA binding protein during deoxycholate-induced biofilm formation in Clostridioides difficile
npj Biofilms and Microbiomes (2023)